ISSN: 0973-7510
E-ISSN: 2581-690X
Tannase enzyme (EC 3.1.1.20) is an enzyme used in many biotechnological applications as in chemical, beverage, pharmaceutical and food industries. was isolated and purified from marine Aspergillus nomius GWA5 by 75% acetone fractional precipitation, followed by gel filtration in Sephadex G-100 and ion exchange chromatography on DEAE-Sephadex A-50 yielding 4.48-fold purification. Estimation of tannase molecular weight was carried out using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) showing a molecular weight of 30 kDa. The highest activity (291 U/mg protein) were at pH 6.0 and 50 °C, respectively. Tannase stability was observed in acidic range (4-6) and was stable to heat treatment. In absence of its substrate it retained about 84.5% of its activity at 80 °C for 15 min. Effect of some metal ions and chelator on tannase activity was investigated. Mg2+ activated as activator of the pure enzyme while EDTA, Cd2+, Pb2+ and Hg2+ inhibited its activity and retained about 40.78, 51.55, 30.24 and 24.55% of its activity, respectively. Promising activity of Tannase was shown in removing tannin stains of tea from clothes.
Tannase; Aspergillus nomius GWA 5; Purification; Characterization; Application
Tannin is water-soluble polyphenolic compound found in plants as secondary metabolites 1. Many of phenolic hydroxyl groups present in tannins permit them to form complexes with proteins and to a lesser amount with other macromolecules like pectin and cellulose 2. Tannins also have toxic effects on some organisms, but they can resist tannins by degrading them into useful derivatives and oligomeric tannins such as gallic acid or pyrogallol using tannase enzyme 3.
Tannase enzyme is tannin acyl hydrolase (EC 3.1.1.20) which can only hydrolyze the galloyl residues of galloyl esters of tannins and can be used in different industrial applications 4. It finds a wide range of applications such as in the production of gallic acid, an intermediate for trimethoprim, pharmaceutical industry, food industry through chemical synthesis of pyrogallol or ester galates which are used as food preservatives, clarification of beer and beverages, manufacture of instant tea, manufacture of coffee flavored drinks, reduction of anti-nutritional effects of tannins in animal feed and high grade leather tanning 5. Also, antioxidant in fats, oils and beverages (6) and in decontamination of tannery effluents 7-10.
Tea preparation is one of the most promising application of this enzyme as it help in solubilization of polyphenolic complexes which are insoluble in cold water by hydrolyses the ester linkage between galloyl groups and various compounds present in unconverted tea leaves 11.
The enzyme can be produced by many microorganisms such as fungi, bacteria and yeast. Fungi are predominant tannase producers and are extensively studied 12-16. Aspergillus sp. is the most abundant species of fungi which can grow in a wide types of environments with very little nutrients available and used for the tannase production17. Tannase usage is severally restricted due to high production costs, despite of its several important applications. Therefore, there is a continuous search for new sources of tannase 18, 19 as well as improved methods for production, recovery, and application of the enzyme.
The present investigation was aimed to purification, characterization and investigation of valuable application of tannase produced by the locally isolated Aspergillus nomius GWA5 as a new source for tannase production.
Organism and growth condition
Aspergillus nomius GWA 5 was isolated from marine sediment of Western Harbour, Alexandria, Egypt. The isolated fungus was identified as Aspergillus according to morphological and microscopically examination, Further identification to A.nomius GWA 5 was done using 18s rRNA sequence analysis (accession No. KU359224). The medium used for cultivation and production of enzyme was composed of (g/l) :(NH4)2SO4, 1; K2HPO4, 1; KH2PO4, 0.5; MgSO4.7H2O, 0.5; CaCl2.H2O, 0.02; FeSO4.7H2O, 0.001; NaCl, 20. The pH of the medium was adjusted to 6.0 using 0.1M HCl or 0.1 N NaOH before sterilizing in an autoclave at 121oC for 20 min. After sterilization, one ml of a spore suspension (2×106 spore/ml) prepared from 5-day old slants of the test organism was used to inoculate 50 ml of sterilized medium dispensed in 250 ml Erlenmeyer flasks. Thereafter, the flasks were incubated for 72 hr at 30 oC under static condition 20.
Protein estimation
The protein content preparation was measured according to the method of Lowry et al. 21 using bovine serum albumin as the standard.
Enzyme assay
Tannase activity was determined using the method of rhodanine 22. The reaction mixture containing 0.25 ml enzyme preparation and 0.25 ml methyl gallate as a substrate (0.01M methyl gallate prepared in 0.05 M citrate buffer, pH 5.0). The mixture was incubated for 5 min at 30°C. Add 0.3 ml of methanolicrhodanine solution (0.667% w/v) for stopping the reaction and for formation of complex between gallate and rhodanine. The tubes were kept at 30°C for 5 min, followed by addition of 0.2 ml KOH solution (0.5M) and the tubes were again kept at 30°C for 5 min. All the tubes were diluted and kept again at 30oC for 10 min. The pink color developed was read at 520 nm using a spectrophotometer. One unit of enzyme activity (U) was defined as the amount of enzyme required to produce 1 µmol of gallic acid per minute under assay conditions.
Spectrophotometric estimation of gallic acid
Estimation of gallic acid was carried out by the method described by Sharma et al. 22.
Partial purification
Tannase enzyme produced from A.nomius GWA5 cultures was purified by three steps purification involving precipitation (either by ammonium sulphate, ethanol or acetone), gel filtration and ion exchange.
Acetone or ethanol
Both acetone and ethanol were cooled at 4°C one day before the precipitation. A certain volume of acetone or ethanol was added slowly to a definite volume of crude enzyme with continuous stirring until the required concentration of acetone was reached. The obtained fractions at 25, 35, 50, 65, 75, 85 and 95% were dried over anhydrous calcium chloride under reduced pressure at room temperature, followed by precipitation and dialysis against distilled water in a refrigerator for 24h. The enzyme activity and protein content of each fraction were estimated 23.
Ammonium sulphate fractionation
Ammonium sulfate was added to definite volume of the culture filtrate at different concentrations to obtain various fractions at 25, 35, 50, 65, 75, 85 and 95% saturation. Each precipitate was dissolved in a certain amount of distilled water and dialyzed against distilled water in a refrigerator overnight after dialysis 16.
Purification of tannase
Gel-filtration chromatography on Sephadex G-100
Sephadex G-100 (Sigma) was packed in a glass column (2.5 cm × 45.0 cm) and equilibrated with 400 ml of 0.02 M phosphate buffer at pH 8.0. 30 ml of 0.02 M phosphate buffer was used to dissolve the precipitate resulting from acetone fractionation 75% saturation and dialyzed for 24 h at 4 °C in 0.02 M phosphate buffer after dialysis. Elution of the enzyme protein was performed at a flow rate of about 30ml/h by using a peristaltic pump. The eluent was collected in 3ml fractions. The protein content and tannase activity of each fraction were estimated.
Ion-exchange chromatography DEAE Sephadex A-50
A slurry of diethylaminoethyl (DEAE) Sephadex A-50 was packed in a column (2.5 cm×45.0 cm). The sephadex bed (30 cm long) was equilibrated with 0.05M acetate buffer (pH 5.2). The fractions of highest specific activity obtained from gel filtration on Sephadex G-100 column pooled and applied to DEAE- Sephadex A-50 column. Elution was performed with 0.05M acetate buffer, followed by 0.05M NaCl in 0.05M acetate buffer at pH 5.2, at a flow rate of 60 ml h-1. Fractions (about five ml) were collected, tannase activity and protein content for each fraction were estimated. The fractions had higher specific activity were pooled.
Characterization of purified tannase enzyme
The effect of different temperatures, pH, pH and thermal stability of the enzyme fractions obtained after column chromatography was studied 24. Also, effect of some metal ions and chelators on the activity of purified tannase was studied.
Determination of molecular weight of pure tannase
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS- PAGE) was performed to determine the molecular weight of pure tannase according to the method decribed by Laemmli 25.
Effect of pH and temperature on the tannase activity
The effect of pH and temperature on the purified A.nomius GWA5 tannase enzyme was determined. Tannase activity was studied in the pH range of 3.0 to 9.0 using the following buffers: (0.05 M citrate buffer for pH 3-6, 0.05 M phosphate buffer for pH 7–8 and 0.05 M Tris-HCl for pH 9). The optimum temperature for tannase activity was determined by incubating enzyme and substrate at various reaction temperatures ranging from 20 to 70 oC. Tannase activity was performed as described before under the optimal conditions.
pH stability and thermal stability of tannase
The purified A.nomius GWA5 tannase was incubated at various pH values at 50oC for 30 min in absence of its substrate and then the residual activity was determined at the optimum pH (6). The thermal stability of the purified enzyme preparation was studied at the optimum pH. Identical enzyme solutions in acetate buffer were preheated separately (in absence of its substrate) at different temperatures (40- 80 oC) for (15, 30, 60 min). Tannase residual activity was assayed each time under the optimum conditions.
Effect of some metal ions and chelator on tannase activity
The purified enzyme solution was preincubated for 2 h at room temperature with the tested substance, MgSO4.7H2O, ZnSO4, NaCl, PbCl2, HgCl2, CdCl2, BaCl2, and EDTA (10 mM). The residual enzyme activity was measured by adding the substrate and carrying out the enzyme assay under the optimum conditions.
Application of tannase in removing tannin stains of tea
Two small square pieces (5×5 cm) of clean cotton cloth were treated with tea extract. The stained pieces were then subjected to the following treatment: First piece was washed with tap water while the second piece was immersed in 5ml of 0.05M citrate buffer pH 5.0 and treated with crude tannase and left under the observation26.
Partial purification of tannase
Tannases are enzymes used in different industrial applications as in food and pharmaceutical. In the last decade, there have been efforts looking for new sources for tannase enzyme production 27. Many researches on tannase synthesis showed clearly that almost all species of Aspergillus are the predominant source of tannase for industrial production and application 28. Most of the tannase producing fungi are of terrestrial origin and till date no reports are available on utilization of marine fungi for tannase production except the work done by Beena et al. 29 who isolated Aspergillus awamori, from marine environment as potential source for tannase. The present investigation aimed to purified and characterized the tannase produced from locally isolated marine A. nomius GWA5 20 for used in many biotechnological applications.
Table (1):
Fractional precipitation of tannase from A. nomius GWA5 cultures using different agents.
| Agent concentration (%) | Acetone | Ethanol | Ammonium sulphate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC | RP | TA | RA | PC | RP | TA | RA | PC | RP | TA | RA | |
| Culture filtrate | 844 | 100 | 41.8 | 100 | 844 | 100 | 41.8 | 100 | 844 | 100 | 41.8 | 100 |
| 25 | 9.22 | 1.092 | 22.94 | 0.599 | 8.11 | 0.961 | 12.62 | 0.29 | 8.98 | 1.064 | 20.09 | 0.511 |
| 35 | 11.65 | 1.38 | 40.86 | 1.347 | 10.23 | 1.212 | 42.91 | 1.243 | 12.55 | 1.487 | 43.77 | 1.555 |
| 50 | 18.88 | 2.237 | 44.65 | 2.386 | 15.65 | 1.854 | 49.85 | 2.208 | 18.34 | 2.173 | 45.28 | 2.35 |
| 65 | 23.76 | 2.815 | 52.23 | 3.512 | 23.99 | 2.842 | 51.73 | 3.512 | 22.11 | 2.62 | 52.15 | 3.264 |
| 75 | 26.8 | 3.175 | 66.49 | 5.043 | 22.78 | 2.699 | 42.85 | 2.763 | 20.11 | 2.383 | 48.54 | 2.763 |
| 85 | 14.9 | 1.765 | 62.89 | 2.652 | 16.44 | 1.948 | 34.3 | 1.596 | 18.95 | 2.245 | 35.22 | 1.889 |
| 95 | 11.45 | 1.357 | 47.86 | 1.551 | ||||||||
| Total | 116.66 | 13.822 | 17.09 | 97.2 | 11.517 | 11.611 | 101.04 | 11.972 | 20.09 | 12.331 | ||
PC: protein content (mg), RP: relative protein (%), TA: tannase activity (U/mg protein), RA: relative activity (%).
Purification of A. nomius GWA5 tannase was efficient and effective. The partially purification was carried out by fractional precipitation with (NH4)2SO4, acetone and ethanol, each used separately (Table 1). A total of 18 fractions were obtained and the highest recovered protein (13.82 %) was present in the fractions precipitated with acetone followed by (NH4)2SO4 sulphate (11.972 %) and ethanol (11.517 %). Using of acetone (17.09 %) followed by (NH4)2SO4 (12.331 %) and ethanol (11.611 %) recorded the highest tannase activity. Some of the obtained fractions showed a specific activity higher than that of the crude enzyme, indicating the presence of purification. Among all the obtained fractions, the 75% acetone fraction showed the highest recovered protein (3.175%). Also, 75% acetone fraction showed the highest tannase activity as it gave about 1.59-fold specific activity higher than that obtained from culture filtrate. Therefore, the fraction 75% acetone was selected for further purification and was considered as the partially purified enzyme. Many investigators have used acetone for precipitation of tannase12,30. On the other hand, (NH4)2SO4 with different concentrations was used by several researchers for precipitating tannase from the fermentation broth 30-33, 16.
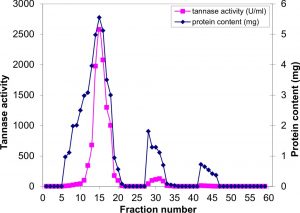
Fig. 1. Gel filtration in Sephadex-G100 of the semi purified tannase obtained from A.nomius GWA5 culture
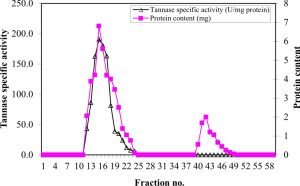
The concentrated partially purified tannase was exposed to dialysis and gel filtration on a SephadexG-100 column. The protein and elution profiles for tannase from the Sephadex G-100 column are shown in Figure 1, which indicated that there are three peaks of protein. The first peak contains the highest protein content (16.65 mg) which represented about 82.2% of the total recovered protein from the column, and the highest specific activity (154.9 U/mg protein) showing about 2.3-fold purification. The most active fractions (6 to 20) obtained from the Sephadex G-75 column were collected and then purified by using DEAE-Sephadex A-50 column chromatography (Figure 2). The specific activity of the pooled fractions reached (187.315 U/mg protein), showing about 4.48-fold purification of the crude enzyme produced by A. nomius GWA5, 2.8-fold purification of the 75% acetone fraction (semi-purified) and 1.39-fold purification of gel filtration fractions.
Fig. 2. Ion-exchange chromatography on DEAE-Sephadex A-50 of the major active tannase component obtained from gel filtration
Roushdy et al. 34 purified tannase from Aspergillus flavus var. columnaris using Sephadex G-200 column chromatography technique and achieved a purification with 5.8-fold. Beena et al. 28 observed that, purification of tannase from A. awamori when applied on Sephadex G-200 column chromatography raised the purification folds to 6.73 times, while Mahendran et al. 35 noted that, purification of tannase from Paecilomyces variotii by Sephadex G- 200 column was 30.5- fold purification. Also, Paranthaman et al. 36 purified tannase from A. niger by using DEAE-Sephadex A-50. On the line with our results, Bagga et al. 37 used two chromatographic techniques DEAE-Cellulose column followed by Sephadex G-100 column for purification of tannase from A. aculaetus and a specific activity of purified enzyme was about 79.7 U/mg protein.
A summary of the purification of tannase from the culture medium of A. nomius GWA5 is presented in Table 2.
Table (2):
A summary of tannase from A. nomius GWA5 cultures.
Purification step |
Total protein(mg) |
Total activity (U) |
Specific tannase activity (U/mg protein) |
Purification (Fold) |
Yield (%) |
|---|---|---|---|---|---|
Culture filtrate |
844 |
35333 |
41.8 |
1 |
100 |
Acetone fraction (75%) |
268 |
17820 |
66.49 |
1.59 |
50.04 |
Sephadex G-100 |
66.45 |
8933.37 |
134.44 |
3.21 |
25.28 |
DEAE-Sephadex A-50 |
20.82 |
3899.9 |
187.315 |
4.48 |
11.04 |
Characterization of purified tannase
Determination of molecular weight of pure tannase
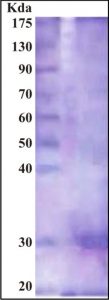
The purity of the enzyme was analyzed by SDS-PAGE electrophoresis. SDS-PAGE revealed a single homogenous band with an approximate molecular mass of 30 kDa as illustrated in Figure 3. Many investigators have used SDS-PAGE for detection of tannase enzyme molecular mass 28, 38, 39.
Fig. 3. SDS-PAGE of purified tannase from A. nomius GWA5. Lane A: molecular weight markers; Lane B: purified tannase
Effect of pH and temperature of the reaction mixture
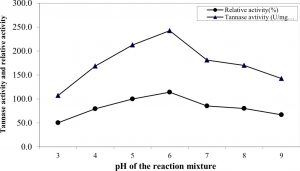
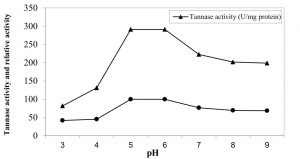
The influence of pH value (ranged from 3 to 9) of the reaction on the purified enzyme activity (Figure 4) indicated that the optimum pH value for highest activity was 6.0 at which tannase activity reached its maximal value (242.5 U/mg protein). The lowest activity (107 U/mg) was obtained at the alkaline range and the enzyme lost about 49.7% of its activity at pH 9. The obtained results were in good agreement with those reported by Sabu et al. 24 and Renovato et al. 40. Many authors reported that the optimal pH values of tannase were ranged from 5.0 to 6.0 41, 42, 33, 37.
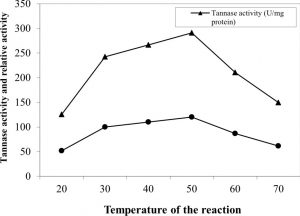
Enzyme showed an increase in its activity parallel to the increase in reaction temperature reaching a maximum value at 50oC (Figure 5). Further rise of temperature resulted in gradual loss in the activity. At high temperatures (60 and 70oC), the enzyme activity was decreased by about 72.3% and 51.4%, respectively, of that at 50oC. Also, the enzyme showed a stability in a broad temperature range of 40-70°C. These results are in comparable to the purified tannase preparation from Paecilomyces variotii 31. However, this optimum temperature is much higher than that reported for purified tannase enzymes produced by A. tamari 43, Penicillium sp. EZ-ZH190 30, A. flavus var. columnaris 34 and Kluyveromyces marxianus 16.
Fig. 5. Effect of the reaction temperature on the activity of pure tannase produced by A. nomius GWA5
pH stability and thermal stability
pH stability of purified A. nomius GWA 5 tannase was studied using pH range of 3 – 9 (Figure 6). Also, the enzyme activity decrease with increasing in pH. However enzyme activity exhibited about 42% and 68.4% at pH 3 and 9, respectively of activity with respect to the activity of optimal pH. Our results are in agreement with Andrea et al. 44 and Costa et al. 43. Also, Batra and Saxena 41 reported that tannase from A. fumigatus and A. flavus was stable at pH 4.0 and doesn’t show tannase activity at alkaline pH of 8.0. In contrast, A. versicolor tannase showed less stability at pH 3.0 with a maximum (100%) activity at pH 6.0.
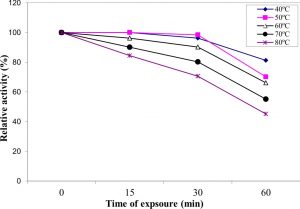
Thermal stability of the pure enzyme in absence of its substrate depended upon temperature of heating and time of exposure (Figure 7). The enzyme retained about 96.2% and 66.2% of its activity when treated at 60 °C for 15 and 60 min, respectively. At higher temperature, the enzyme lost about 15.5% of its activity by heating at 80 °C for 15 min and about 54.8 % of its original activity by treatment for 60 min at the same temperature. The results of the present investigation showed also that the purified enzyme was fairly stable. The obtained results are in good with Bagga et al. 37 who reported that Aspergillus aculaetus tannase was stable up to 50 °C and lost only 5% activity at 60°C which was the optimum temperature of the enzyme. The thermal stability of A. nomius GWA5 tannase is comparable to the tannase isolated from Aspergillus ochraceus 45 which was stable in a wide range of temperatures from 40oC to 70oC.
Effect of some metal ions and chelator on tannase activity
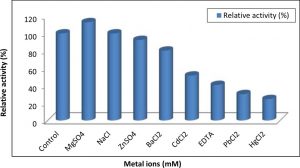
The purified A. nomius GWA5 tannase was subjected to different metal ions to study their activation and inhibition effects on its activity. The results illustrated in Figure 8 indicated that only Mg2+ increased activity of tannase enzyme. Also, the results showed that Zn2+ and Ba2+ partially inhibited tannase activity. On the other hand, the enzyme was strongly inhibited by Cd2+, EDTA, Pb2+ and Hg2+ retained about 51.55%, 40.78%, 30.24% and 24.55% of its activity, respectively. The effect of some ions on the activity of the purified tannase preparation was studied. The results reveled that only Mg+2 activated the activity which in good agreement with many authors 46-48, 4. Also, metal ions Zn2+, Cd2+, Hg2+, Pb2+, Ba2+ and chelator EDTA were partially inhibited tannase activity. Many investigators reported that chelator EDTA disodium salt at a concentration 1 mM decreased tannase activity 45, 48, 50, 51. On the constract, Beniwal et al. 47 found that EDTA at a concentration of 1 mM did not affect tannase activity after 5 min of incubation period; however a slight loss was observed when the incubation period was extended to 30 min for thermophilic tannase from Enterobacter cloacae. The decrease detected in the presence of EDTA could be due to its influence on the interfacial area between the substrate and enzyme 52.
Maximum inhibition of the tannase enzyme occurred in the presence of Hg2+ 45,47 because Hg2+ may interact with SH and S-S groups of proteins causing conformational changes in proteins. Also ions of mercury react with protein thiol groups as well as histidine and tryptophan residues. Moreover, the disulfide bond could also be hydrolytically degraded by the action of mercury. Also the decreased activity in the presence of divalent cations could be due to the non-specific binding or aggregation of the enzyme 45,50.
Application of tannase in removing tannin stains of tea
Application of A.nomius GWA5 tannase in removing tannin stains of tea was investigated. It is obvious from Figure 9 that tea stains removal was better in cloth piece soaked in solution containing purified tannase when compared with control which was washed with tap water only .This is due to the activity tannase on tannins. The results were also indicated that the purified A. nomius GWA5 tannase can remove the tea stain and indicates that, tannase from A. nomius GWA5 can be used in formulation of tannin stain remover or detergents. These results in line with Sharma and Saxena 26 who worked on tannases from A. niger and P. variable.
The present study may append some new data on the production of tannase from Aspergilli and confirmed the capability of the marine A. nomius GWA5 to produce an active tannase for degradation of tannin. Moreover this enzyme can be used in different industrial and pharmaceutical uses. Good stability of pure tannase in wide range of pH and temperature was obtained. These properties collectively point that the tannase enzyme obtained from A.nomius GWA5 is a good candidate for removal of tannin stain of tea. Large scale production of tannase by using various recombinant engineering techniques will be tracked in the future study.
- Girdhari SN, Peshwe SA. Isolation and screening of tannase producing fungi. Int J Curr. Microbiol. Appl. Sci, 2015; 4(7): 765-774.
- Bele AA, Jadhav VM, Kadam V. Potential of tannins: A Review. Asian. J. Plant. Sci, 2010; 9(4): 209-214.
- Chávez-González M, Rodríguez-Durán LV, Balagurusamy N, PardoBarragán A, Rodríguez R, Contreras JC, Aguilar CN. Biotechnological advances and challenges of tannase: an overview. Food. Bioprocess Technol, 2012; 5: 445–459.
- Lata S, Rani KP. Production of tannase enzyme by Penicillium duclauxii, screened and isolated from soil. Res. J. Pharm. Biol. Chem. Sci, 2016; 7(1): 1471-1485.
- Lal D, Gardner J J. Production, characterization and purification of tannase from Aspergillus niger. European. J. Exp. Biol, 2012; 2 (5): 1430-143.
- Rodríguez H, de las Rivas B, Gómez-Cordovés C, Muñoz R. Characterization of tannase activity in cell-free extracts of Lactobacillus plantarumCECT 748T. Int. J. Food. Microbiol, 2008; 121: 92-98.
- Belmares R, Contreras-Esquivel JC, RodriguezHerrera R, Coronel AR, Aguilar CN .Microbial production of tannase: an enzyme with potential use in food industry. Lebensm-Wiss U-Techno, 2004; 37: 857-864.
- BAuthor%D&cauthor=true&cauthor _uid=17530245″AguilarHYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Aguilar%20CN%5 BAuthor%5D&cauthor= true&cauthor_uid=17530245” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Aguilar%20CN%5BAuthor %5D&cauthor=true&cauthor_uid=17 530245”CN, 3%ADguez%20R%5BAutho r%5D&cauthor=true& cauthor_uid=17530245″Rodríguez R, 3%A9rrez-S%C3%A1nchez%20G%5B Author%5D&cauthor =true&cauthor_uid =17530245″Gutiérrez-Sánchez G, 5BAuthor%5D&cauthor= true&cauthor_uid=17530245″Augur C, 5BAuthor%5D&cauthor= true&cauthor_uid=17530245″Favela-TorresHYPERLINK “https://www.ncbi.nlm.nih. gov/pubmed/?term= Favela-Torres %20E%5 BAuthor%5D &cauthor= true&cauthor_uid =17530245” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Favela-Torres%20E%5BAuthor%5D&c author=true&cauthor_uid=17530245” E, 5BAuthor%5D&cauthor=true &cauthor_uid=17530245″Prado-BarraganHYPERLINK “https://www.ncbi.nlm. nih.gov/pubmed/?term=Prado-Barragan%20LA%5 BAuthor%5D& cauthor=true& cauthor_ uid=17530245” HYPERLINK “https://www.ncbi. nlm.nih.gov/pubmed/?term= Prado- Barragan%20LA%5B Author%5D &cauthor=true&cauthor_uid=17530245” LA, Ramírez-Coronel A, 5BAuthor%5D& cauthor=true &cauthor_ uid=17530245 “Contreras-Esquivel HYPERLINK “https://www.ncbi. nlm.nih.gov/pubmed/?term=Contreras-Esquivel%20JC%5B Author%5D& cauthor=true &cauthor_ uid=17530245” HYPERLINK “https://www. ncbi.nlm. nih.gov/pubmed/?term =Contreras-Esquivel%20JC%5B Author% 5D&cauthor =true&cauthor _uid= 17530245”JC (2007) Microbial tannases: Advances and perspectives. Appl. Microbiol. Biotechnol , 2007; 76: 47–59.
- Srivastava A, Kar R. Characterization and application of tannase produced by Aspergillus niger ITCC 6514.07 on Pomegranate rind. Braz. J. Microbiol, 2009; 40(4):782-9.
- Malgireddy, N.R., Nimma, L.N.R. Optimal conditions for production of Tannase from newly isolated Aspergillus terrus under solidstate fermentation. European. J. Biotechnol. Biosci, 2015; 3 (2): 56-64.
- Mehta, M., Muddapur, U.M., ShanmugaPriya, V.G. Fungal production of tannase : a review. Int. J. Sci. Eng. Technol, 2013; 2: 752-755.
- Costa A M, Ribeiro WX, Kato A, Monteiro A R G, Peralta RM. Production of tannase by Aspergillus tamarii in submerged cultures. Braz. Arch. Biol. Technol, 2008; 51(2):399-404.
- Manjit, A., Yadav, N.K., Aggarwal, K., Kumar, K., Kumar, A. Tannase production by Aspergillus fumigatusMA under solid-state fermentation. World J. Microbiol. Biotechnol, 2008; 24: 3023–3030.
- Enemuor SC, Odibo FJC. Culture conditions for the production of a tannase of Aspergillus tamariiIMI388810. Afr. J. Biotechnol, 2009; 8: 2554–2557.
- Aboubkar HA, El-Sahn MA, El-Banna AA. Some factors affecting tannase production by Aspergillus niger Van Tieghem. Braz. J. Microbiol, 2013; 44: 559-567.
- BAuthor%D&cauthor= true&cauthor_uid=29055707″Mahmoud AE, BAuthor%D&cauthor= true&cauthor_uid= 29055707″FathyHYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Fathy%20SA%5BAuthor %5D&cauthor=true&cauthor_uid=29055707” SA, BAuthor%D&cauthor=t rue&cauthor_uid=29055707″Rashad MM, Ezz, M K, BAuthor%D&cauthor=true&cauthor_uid= 29055707″MohammedHYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Mohammed%20AT%5BAuthor%5 D&cauthor=true&cauthor_uid=29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Mohammed%20AT%5BAuthor%5D& cauthor=true&cauthor_ uid=29055707”AHYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Mohammed%20AT%5BAuthor% 5D&cauthor=true&cauthor_uid=29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/?term=Mohammed%2 0AT%5BAuthor% 5D& cauthor =true& cauthor _ uid= 29055707”T. Purification and characterization of a novel tannase produced by Kluyveromyces marxianus using olive pomace as solid support, and its promising role in gallic acid production. Int.HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707” J.HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707”Biol.HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707” HYPERLINK “https://www.ncbi.nlm.nih.gov/pubmed/29055707”Macromol, 2018; 2342-2350
- Philip D C, Lavanya B, Latha S. Purification of tannase from Aspergillus niger under solid state fermentation. World. J. Pharm. Pharma Sci, 2015; 4(2): 993-1001.
- Kasieczka-Burnecka M, Kuc K, Kalinowska H, Knap M, Turkiewicz M. Purification and characterization of two cold-adapted extracellular tannin acyl hydrolases from an Antarctic strain Verticillium sp. Appl. Microbiol. Biotechnol, 2007; 77(1): 77–89.
- Kumar R, Sharma J , Singh R. Production of tannase from Aspergillus ruber under solid-state fermentation using jamun (Syzygium cumini) leaves. Microbiol. Res, 2007; 162: 384-390.
- El-Says AMI. Degradation of tannin by a marine microorganism. Master of Science in Microbiology, 2016. Thesis submitted to the Faculty of Science, Alexandria University, Egypt.
- Lowry O H, Rosenberg WJ, Farr, AL, Randell, R J. Quantitation of protein using Folin Ciocalteu reagent. J. Biol. Chem, 1951; 193: 265–75.
- Sharma S, Bhat TK, Dawra RK. A spectrophotometric method for assay of tannase using rhodanine. Analy Biochem, 2000; 279: 85-89.
- Farag AM, Ibrahim H AH, Abd-Elnaby HM, El-Shenawy M. Purification, characterization and antimicrobial activity of chitinase from marine-derived Aspergillus terreus. Egyp. J. Aquatic. Res, 2016; 42(2):185-192.
- Sabu A, Kiran GS, Pandey A. Purification and Characterization of Tannin Acyl Hydrolase from Aspergillus niger ATCC 16620. Food. Technol. Biotechnol, 2005; 43 (2):133–138.
- Laemmli UK. Cleavage of structural protein during the assembly of heat of bacteriophage T4. 1970. Nature (London) 227, 680–685.
- Sharma S, Saxena R . Evaluation of the versatility of the tannase produced from Aspergillus niger and penecilium variable with respect to gallic acid production, gallate ester synthesis, animal feed improvement, tannery effluent degradation and tannin stain removal. Res. Biotechnol, 2012; 3(5): 09-20.
- Belur PD, Mugeraya G, Nainegali B. Release of cell-associated tannase of Serratia ficariaDTC by sonication, surfactants and solvents. Asian. J. Biotechnol, 2011; 3: 91-97.
- Beena PS, Soorej MB, Elyas KK, Sarita GB, Chandrasekaran M. Acidophilic tannase from marine Aspergillus awamori BTMFW032. J. Microbiol. Biotechnol, 2010; 20(10): 1403–1414.
- Beena PS, Basheer SM, Bhat SG, Bahkali AH, Chandrasekaran M. Propyl gallate synthesis using acidophilic tannase and simultaneous production of tannase and gallic acid by marine Aspergillus awamori BTMFW032. Appl .Biochem. Biotechnol, 2011; 164: 612–628.
- Zakipour-Molkabadi E, Hamidi-Esfahani Z, Sahari MA, Azizi MH. A new native source of tannase producer, Penicillium sp. EZ-ZH190: Characterization of the enzyme. Iran. J. Biotechnol, 2013; 11(4): 244-50.
- Battestin V, Macedo GA. Effects of temperature, pH and additives on the activity of tannase produced by Paecilomyce svariotii. Electronic. J. Biotechnol, 2007; 10(2): 191-199.
- Sivashanmugam K, Jayaraman G. Production and partial purification of extracellular tannase by Klebsiella pneumonia MTCC 7162 isolated from tannery effluent. Afr. J. Biotechnol, 2011; 10(8): 1364-1374.
- Gayen S, Ghosh U. Purification and characterization of tannin acyl hydrolase produced by mixed solid state fermentation of wheat bran and marigold flower by Penicillium notatum NCIM 923. Bio. Med. Res. Int, 2013; 1-6.
- Roushdy MM, Desouky SE, Esmael ME, El-Louboudy SS, Elshikh HH. Optimization and characterization of tannin acyl hydrolase produced by Aspergillus flavus var. columnar is using solid state fermentation technique. New York Sci J, 2014; 7(3):88-98.
- Mahendran B, Raman N, Kim D J. Purification and characterization of tannase from Paecilomyce svariotii: hydrolysis of tannic acid using immobilized tannase. Appl. Microbiol. Biotechnol, 2006; 70: 444–450.
- Paranthaman R, Vidyalakshmi R, Murugesh S, Singaravadivel K. Optimization of various culture media for tannase production in submerged fermentation by Aspergillus flavus. Adv. Biol. Res, 2009; 3 (1-2): 34-39.
- Bagga J, Pramanik SK, Pandey V. Production and purification of tannase from Aspergillus aculeatus using plant derived raw tannin. Int. J. Sci. Eng. Technol, 2015; 4(2): 50-55.
- Anitha A, Arunkumar D. Extraction, partial purification and characterization of tannase enzyme from Mucor sp. J. Nehru. Arts. Sci. Coll, 2013; 1: 25-29.
- Valera LS, Jorge JJ, Guimarães LHS. Characterization of a multi-tolerant tannin acyl hydrolase II from Aspergillus carbonarius produced under solid-state fermentation. Electronic. J. Biotechnol, 2015; 18: 464–470.
- Renovato J, Gutiérrez-Sánchez G, Rodríguez-Durán LV, Bergman C, Rodríguez, R, Aguilar CN. Differential properties of Aspergillus niger tannase produced under solid-state and submerged fermentations. Appl. Biochem. Biotechnol, 2011;165: 382-395.
- Batra A, Saxena R K. Potential tannase producers from the genera Aspergillus and Penicillium. Process Biochem, 2005; 40:1553-1557.
- Mahapatra K, Nanda RK, Bag SS, Banerjee R , Pandey A, Szakacs G. Purification, characterization and some studies on secondary structure of tannase from from Aspergillus awamori nakazawa. Process Biochem, 2005; 40(10): 3251–3254.
- Costa A M, Kadowaki MK, Minozzo MC, Cristina Souza, GM, Boer, CG, Bracht A, Peralta R M. Production, purification and characterization of tannase from Aspergillus tamarii. Afri. J. Biotechnol, 2012; 11(2): 391-398.
- Andrea MC, Marina KK, Monique CM, Cristina GM, Cinthia GB, Adelar B, Rosane M P. Production, purification and characterization of tannase from Aspergillus tamari. Afr. J. Biotechnol, 2012; 11(2): 391-398.
- Gonçalves HB, Riul AJ, Quiapim AC, Jorge JA, Guimarães L H S. Characterization of a thermostable extracellular tannase produced under submerged fermentation by Aspergillus ochraceus. Electronic. J. Biotechnol, 2012; 15(5): 1-12.
- Mukherjee G, Banerjee R. Effects of temperature, pH and additives on the activity of tannase produced by a co-culture of Rhizopus oryzae and Aspergillus foetidus. World. J. Microbiol. Biotechnol, 2006; 22(3): 207-212.
- Beniwal V, Kumar A, Goel G, Chhokar V (2013) A novel low molecular weight acido- thermophilic tannase from Enterobacter cloacae MTCC 9125. Biocat Agric. Biotechnol, 2013; 2: 132–137.
- Yao J, Chen Q, Zhong G, Cao W, Yu A, Liu Y. Immobilization and characterization of tannase from a metagenomic library and its use for removal of tannins from green tea infusion. J. Microbiol. Biotechnol, 2014; 24(1): 80–86.
- Marco, M.G. Rodríguez, L.V., Ramos, E.L., Renovato, J., Cruz-Hernández, M. A., Rodriguez, R., Contreras, J., Aguilar, C.N. A novel tannase from Xerophyllic fungus Aspergillus niger GH1. J. Microbiol. Biotechnol, 20089; 19: 987–996.
- El-Fouly MZ, El-Awamry Z, Shahin A AM, El-Bialy HA, Naeem E, El-Saeed GE. Biosynthesis and characterization of Aspergillus niger AUMC 4301 tannase. J. American. Sci, 2010; 6(12): 709-721.
- Nadaf N H, Ghosh J S. Production, purification and characterization of tannase from Rhodococcus NCIM 2891. Curr. Res. J.Biol. Sci, 2013; 3(3): 246-253.
- Jinwal UK, Roy U, Chowdhury A R, Bhaduri AP, Roy PK. Purification and characterization of an alkaline Lipase from a newly isolated Pseudomonas mendocina PK-12CS and chemo-selective hydrolysis of fatty acid ester. Bioorganic. Medicinal. Chem, 2003; 11(6):1041-1046.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.