ISSN: 0973-7510
E-ISSN: 2581-690X
Legionellosis is a neglected disease due to the absence of well-defined clinical symptoms and difficulties in isolating the causal organism. Legionella spp. is known to colonize the lumen of respiratory therapy equipment(RTE) and evade conventional detection by entering the viable but non-culturable state. Monitoring these surfaces for Legionella pneumophila in addition to routine monitoring of water could aid in decreasing incidences of hospital-acquired infections by this pathogen. In this study swabs of different respiratory therapy equipment were tested for the presence of Legionella by conventional culture-based methods versus molecular detection of culture-independent template by polymerase chain reaction (PCR). Genetic diversity of the genes amplified were studied using bioinformatic tools. The dotA genes were genetically diverse indicating no clonality. This communication highlights that the persistence of virulence genes like dotA on abiotic surfaces can result in the mobilization of these genes to other species and give rise to virulent forms especially in a healthcare setting.
Legionellosis, Nosocomial pneumonia, Respiratory therapy equipment, Tertiary care hospitals, Horizontal gene transfer, dot A
Legionella is a Gram-negative aerobic bacterium which is a part of the natural aquatic environment1. The bacteria are ubiquitous in nature and are known for their ability to jump from the natural aquatic habitats into artificial water systems which can result in life threatening respiratory health risks2 . An estimated 3 to 8% community acquired pneumonia are caused by Legionella spp and the greater risk of death due to this infection is common in immuno- compromised and elderly patients3. The World Health Organization deems that the problem of Legionella is underestimated in developing countries due to the lack of awareness or failure in diagnosis. Legionnaire’s disease (LD) is primarily spread via inhalation of contaminated aerosols. The sources of sporadic hospital-acquired infections caused by this pathogen are hospital showers and respiratory therapy equipment (RTE) 4,5. Patients in hospitals are at greater risk of contracting respiratory infections when forced to inhale aerosols from respiratory devices that may contain Legionellae6,7 . The water distribution system serves as a major source for Legionella but the mode of transmission continues to remain unclear8. Though Legionella comes under the purview of water quality, no guidelines are issued for the monitoring of Legionella on surfaces of respiratory therapy equipment where they can continue to reside even in the absence of water. This study was initiated to determine if Legionella can persist on respiratory therapy equipment in the absence of water.
Sample collection
A total of 35 swab samples from respiratory therapy devices were collected aseptically using polyester sterile swabs. The sites for sampling were identified as follows: the compressor air outlet of nebulizers, the port of attaching mouthpiece of spirometer, the insertion tube surface of bronchoscope, and air inlet and outlet vents of ICU ventilators. Collected swabs were immediately immersed in sterile water and transported to the laboratory for analysis within 2 hours.
Detection of Legionella by culture method
The isolation of Legionella was performed as per US Centers for Disease Control and Prevention guidelines 2005. Buffered charcoal yeast extract (BCYE) agar containing 0.1% alpha- ketoglutarate and supplemented with glycine, vancomycin, polymyxin and cycloheximide (GVPC) was used for isolation of Legionella pneumophila and other related species. A volume of 0.1 ml of each sample suspension was plated on GVPC agar and incubated at 35°C with 2.5% carbon dioxide. The isolates that grew on GVPC were further enriched in BCYE broth which was used for DNA extraction by Cetyl trimethylammonium bromide (CTAB) method9. The quality and quantity of the extracted DNA was quantified using a UV nanodrop spectrophotometer(IMPLEN) and then used as templates for molecular detection.
Molecular detection by polymerase chain reaction (PCR)
The isolated cultures were confirmed as members of genus Legionella by polymerase chain reaction (PCR) targeting genus specific 16S rRNA coding region using the JFP-JRP primer pair10 and the species L. pneumophila by the presence of gene coding for the virulence factor dotA11. Alternatively, the swab suspension was directly used as template for
culture-independent detection by PCR. All samples that were dotA positive by direct PCR detection were tested for amplification with eubacterial 16S rDNA universal primers12 .
Genetic relatedness of the dotA gene
The amplified PCR products of dotA were purified using QIAquick PCR purification kit and outsourced for Sanger sequencing (Eurofins Genomics India). A phylogenetic analysis of the dotA sequences was performed to compare the genetic relatedness of this gene among the reads obtained in this study to representative sequences from around the world available in NCBI nucleotide database. Sequences were aligned using Clustal Omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the phylogenetic tree was constructed using the Neighbor Joining algorithm available in MEGA X (version 10.0) with bootstrap tested for 100 replicates13.
From a total of 35 swab samples, 23 isolates were obtained on GVPC agar. Of the 23 isolates that grew on GVPC only two isolates from different nebulizer outlets were positive for the JFP-JRP region. None of the 23 isolates were positive for dotA. The two Legionella specific 16S rRNA sequences obtained in this study were deposited in GenBank and assigned the following accession numbers: MN784440 and MN795709. Sequence analysis of the JFP_JRP amplicons showed >98% identity with Legionella pneumophila ATCC 33152 thus confirming the presence of L. pneumophila on dry surfaces of RTE.
Twelve of 35 swab samples were positive for the dotA gene when the swab suspensions(culture-independent) were directly used as templates for PCR. All amplicons were confirmed as the dotA gene by aligning the reads obtained by Sanger sequencing with nucleotide reads in GenBank database using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
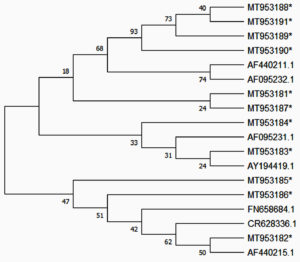
The sequence reads were deposited in GenBank and assigned the accession numbers MT953181 to MT953191. The details of sequence used in construction of phylogenic tree is provided (Suplementary data1). A phylogenetic tree for dotA region was constructed using the Neighbor-Joining method in MEGA X (Figure 1). The evolutionary distances were computed using the Maximum Composite Likelihood method. It could be inferred that there was no clonality among the L. pneumophila on the RTE surfaces even from the same healthcare facility.
Fig. 1. DotA Phylogenetic tree: dotA gene sequences obtained in this study (marked with *) were aligned with sequences from various geographical locations retrieved from GenBank. The individual reads did not cluster together indicating that a diverse population of remnant Legionella pneumophila adhere onto respiratory equipment
Eubacterial 16S rDNA was amplified in all samples which were positive for dotA. However, eubacterial 16S rDNA amplicons from the swab samples yielded mixed reads.
Legionella has emerged as major pathogen and may consequence to death. Knowledge of its affliction in India is limited to a few clinical studies14-17 and fewer environmental studies15,18. There is a need to prioritize studies on this unexplored pathogen as the rates of ventilator associated pneumonia is fairly larger in India19,20. Though conventional culture technique is considered the gold standard for detection, use of molecular techniques like PCR have proven to increase the sensitivity for clinical diagnosis of Legionella21-24 . Respiratory equipment like nebulizers and humidifiers have been implicated as sources of Legionella resulting in hospital-acquired Legionellosis. Surveillance of hospital water has been a primary prevention method of hospital-acquired legionellosis. L. pneumophila cell populations can potentially survive as free organisms for long periods by maintaining metabolic activity but temporarily losing cultivability25. Considering the fact that, L. pneumophila and other respiratory pathogens are able to enter into the viable but non culturable (VBNC) state easily, conventional culture techniques may no longer be the ideal method for detection. A lot of speculation over false positives being reported due to PCR have limited the applications of this technique.
Swabs from surfaces have yielded better recovery of Legionella than water samples14. A comparative account for the detection of Legionella by conventional and direct PCR method is our study depicted in table 1. Higher sensitivity for detection was observed in the direct PCR detection of genes from samples compared to culture technique which is in concordance to many previous reports26.
Table (1):
Isolation of Legionella by conventional method and detection by molecular methods.
| Sample type | Total no of Samples | Detected in culture | Direct PCR of sample | ||||
|---|---|---|---|---|---|---|---|
| JFP | Dot A | Both | JFP | Dot A | Both | ||
| Nebulizers | 15 | 2 | 0 | 0 | 1 | 5 | 1 |
| Spirometers | 4 | 0 | 0 | 0 | 0 | 2 | 0 |
| Bronchoscopes | 3 | 0 | 0 | 0 | 1 | 2 | 1 |
| ICU ventilators (inlet & outlet) | 13 | 0 | 0 | 0 | 0 | 3 | 0 |
The JFP-JRP primer pair has been repeatedly reported to be specific for the genus Legionella and the same is proven based on sequence alignment (Supplementary Figure S1). Many studies have reported the concomitant occurrence of both JFP-JRP region and dotA in clinical samples27 . In our study, we observed many samples that were positive for dotA but not JFP-JRP region, hence the eubacterial 16S rDNA was amplified and attempted for sequencing. Mixed reads were obtained which indicated that these surfaces contain a mixture of bacteria. The non-specific growth observed in 23 of 35 samples on GVPC reiterates that these surfaces harbor many viable bacteria. The detection of dotA gene alone and absence of L. pneumophila 16S rRNA in some samples brings to light the need to elucidate the importance of this virulence gene in other potentially infectious organisms. The phylogenetic tree analysis of the dotA sequences revealed that this gene was not clonal and have been derived from many different strains. There was no geographical clonality among these sequences since though 4 of 11 (MT953188 to MT953191) clustered together they were not 100% identical. The other sequences were similar to dotA sequences reported from many other geographical locations of the world including Europe and America. The higher rate of detection of dotA compared to JFP-JRP region can be attributed to the fact that dotA detection was by a semi-nested PCR method.
Horizontal gene transfer has come to be of prime importance in the rise of new age pathogens. The Dot/Icm is the major virulence mechanism present in the pathogens like, Helicobacter pylori, Bordetella pertussis, Brucella sp, L. pneumophila and Coxiella burnetii utilize type IV secretion system for pathogenesis28. Of these, L. pneumophila and Coxiella burnetii are evolutionarily closely related organisms and both are human intracellular pathogens transmitted to humans through aerosols. Both organisms reside in alveolar macrophages inside an intact phagosome during infection which is mediated by the Dot/Icm.
Though in this study Legionella species have been detected only in two samples by the conventional culture technique, it is necessary to highlight that the presence of virulence genes like dotA on the therapy equipment surfaces may indicate the presence of VBNC forms of the pathogen L.pneumophila. Additionally, the persistence of the virulence gene on these surfaces can result in the mobilization of these genes to other species and hence give rise to virulent forms. The dotA regions from the different samples were not identical indicating that these genes were from a variety of strains of L. pneumophila that are present on the various RTE within a healthcare setup.
RTEs are often used in individuals with compromised immunity and hence better cold sterilization techniques and routine monitoring using culture-independent techniques have to be made mandatory in hospitals.
The present study reveals the occurrence of Legionella pneumophila and associated species in respiratory equipment. To the best of our knowledge, this is the first report on the occurrence Legionella spp. in respiratory therapy equipment in tertiary care hospitals from this geographical location. This evidences that these RTE may serve as potential reservoirs of Legionella species and the usage of such respiratory equipment could transmit pathogenic Legionella spp in to the respiratory tract of patients.
Our observations support the need for concerted efforts to improve the proper maintenance of respiratory equipment as well as awareness among hospital personnel on the potential for aerosolizing pulmonary pathogens. Apart from routine monitoring of water systems for Legionella, we recommend monitoring swabs of respiratory therapy instruments for Legionella and other respiratory pathogens by culture-independent methods as an additional preventive measure against Legionellosis.
Additional file: Additional Table S1. Additional Figs. S1a and S1b.
ACKNOWLEDGMENTS
The authors are grateful to Nitte (Deemed to be University) for supporting this work through the faculty research grant.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RV and JRMR did the experimentation and acquired funding. BKK and JRMR performed methodology. BKK verified the study. RV drafted the manuscript. BKK, IK and JRMR reviewed the manuscript. IK and JRMR did the final editing of the manuscript. All authors approved the manuscript for publication.
FUNDING
Nitte (Deemed to be University) faculty research grant NURG/STF/07/7-2015.
ETHICS STATEMENT
Approval was obtained from the Institutional Ethics Committee (Ref INST.EC/EC/021/2015-16 dated 26.03.2015).
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Sanden GN, Fields BS, Barbaree JM, Feeley JC. Viability of Legionella pneumophila in choline-free water at elevated temperatures. Curr Microbiol. 1989; 18:61-65.
Crossref - Bartram J, Chartier Y, Lee JV, Pond K, Surman-Lee S. Legionella and the prevention of legionellosis. World Health Organization. 2007. https://www.who.int/water_sanitation_health/emerging/legionella.pdf
- Viasus D, Di Yacovo S, Garcia-Vidal C, et al. Community-acquired Legionella pneumophila pneumonia: a single-center experience with 214 hospitalized sporadic cases over 15 years. Medicine. 2013;92(1): 51-60.
Crossref - Bradley BT, Bryan A. Emerging respiratory infections: The infectious disease pathology of SARS, MERS, pandemic influenza, and Legionella. Semin Diag Pathol.2019 : 36(3) : 152-159.
Crossref - Joly JR, Dery P, Gauvreau L, Cote L, Trepanier C. Legionnaires’ disease caused by Legionella dumoffii in distilled water. Can Med Assoc J. 1986; 135(11):1274-1277.
- Woo AH, Goetz A, Yu VL. Transmission of Legionella by respiratory equipment and aerosol generating devices. Chest. 1992; 102(5):1586-90.
Crossref - Kashif M, Patel R, Bajantri B, Diaz-Fuentes G. Legionella pneumonia associated with severe acute respiratory distress syndrome and diffuse alveolar hemorrhage-A rare association. Respir Med Case Rep. 2017; 21:7-11.
Crossref - Kollef MH. Prevention of hospital-associated pneumonia and ventilator-associated pneumonia, Crit Care Med. 2004; 32(6):1396-1405.
Crossref - Wilson K. Preparation of Genomic DNA from Bacteria. Current protocols in molecular biology .2001; 241-245.
Crossref - Jonas D, Rosenbaum A, Weyrich S,Bhakdi S. Enzyme linked immunoassay for detection of PCR amplified DNA of legionellae in bronchoalveolar fluid. J Clin Microbiol .1995; 33(5): 1247-1252.
Crossref - Wójcik-Fatla A, Stojek NM, Dutkiewicz J. Efficacy of the detection of Legionella in hot and cold-water samples by culture and PCR. I. Standardization of methods. Ann Agric Environ Med. 2012; 19(2): 289-293.
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. Analysis of actinomycete communities by specific amplification of genes encoding 16SrRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol .1997; 63(8): 3233–3241.
Crossref - Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sciences. Protein Sci. 2018; 27(1):135-145.
Crossref - Agrawal L, Dhunjibhoy KR, Nair KG.Isolation of Legionella pneumophilafrom patients of respiratory tract disease and environmental samples. Ind J Med Res. 1991; 93:364-365.
- Anbumani S, Gururajkumar A, Chaudhury A. Isolation of Legionella pneumophila from clinical and environmental sources in a tertiary care hospital. Ind J Med Res. 2010; 131(6):761-764.
- Javed S, Chaudhry R, Passi K, et al.Sero diagnosis of Legionella infection in community acquired pneumonia. Ind J Med Res. 2010;131(1): 92-96.
- Chaudhry R, Dhawan B,Dey AB. The incidence of Legionella pneumophila: A prospective study in a tertiary care hospital in India. Tropical Doctor. 2000;30(4):197-200.
Crossref - Jinna S, Gaikwad UN. Environmental surveillance of Legionella pneumophila in distal water supplies of a hospital for early identification & prevention of hospital-acquired legionellosis. Ind J Med Res.2018; 147(6), 611-614.
Crossref - Khurana S, Mathur P, Kumar S, Soni KD, Aggrawal R, Batra P et al. Incidence of ventilator-associated pneumonia and impact of multidrug-resistant infections on patient’s outcome: Experience at an Apex Trauma Centre in North India. Ind J Med Microbiol. 2017;35(4): 504-51
Crossref - Chaudhry R, Sreenath K, Agrawal SK, Valavane A. Legionella and Legionnaires’ disease: time to explore in India. Ind J Med Microbiol. 2018;36(3):324-333.
Crossref - Reischl U, Linde HJ, Lehn N, Landt O, Barratt K,Wellinghausen N. Direct detection and differentiation of Legionella spp. and Legionella pneumophila in clinical specimens by dual-color real-time PCR and melting curve analysis. J Clin Microbiol. 2002;40(10): 3814-3817.
Crossref - Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, Procop GW.Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol. 2003;41(7): 3327-3330.
Crossref - Angrup A, Chaudhry R, Sharma S, et al. Application of real-time quantitative polymerase chain reaction assay to detect Legionella pneumophilain patients of community-acquired pneumonia in a tertiary care hospital. Ind J Med Microbiol. 2016;34(4):539-543.
Crossref - Para RA, Fomda BA, Jan RA, Shah S, Koul PA. Microbial etiology in hospitalized North Indian adults with community-acquired pneumonia. Lung India. 2018;35(2):108-115.
Crossref - Ohno A, Kato N, Yamada K, Yamaguchi K. Factor’s influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water. Appl Environ Microbiol. 2003;69(5):2540-2547.
Crossref - Whiley H, Taylor M. Legionella detection by culture and qPCR: comparing apples and oranges. Crit Rev Microbiol. 2016;42(1):65-74.
Crossref - Cloud JL, Carroll KC, Pixton P, Erali M, Hillyard DR. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J Clin Microbiol. 2000;38(5):1709-1712.
Crossref - Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000; 8(8):354-360.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.