ISSN: 0973-7510
E-ISSN: 2581-690X
Saliva samples could be used as a non-invasive method to diagnose COVID-19. We aimed to assess the results of the reverse transcriptase-polymerase chain reaction (RT-PCR) of saliva specimens in the detection of COVID-19. We collected saliva and nasopharyngeal (NP) samples from consecutive COVID-19 suspects in Al-Fallujah Teaching Hospital, Anbar, Iraq from November 29, 2021 to February 15, 2022. The results of the two specimens were compared using RT-PCR. For the positive saliva tests, repetition of the test was undertaken at weekly intervals for four weeks from the time of the presentation. There were 55% men and 60% people ≤ 35 years. The majority of cases presented within 2-5 days (92%) and were of mild severity (89%). A hundred pairs of samples were taken. COVID-19 was diagnosed by NP swab RT-PCR in 56% and 31% of the saliva samples. The saliva samples had 100% sensitivity (95% confidence interval [CI] 60.4% e96.6%), 63.8% specificity (95% CI 96.1% e99.9%), and mild coefficient agreement (kappa coefficient = 0.522). The positive test for the saliva samples remained as such in all examined cases in the first and second weeks after the first test, 31/31 and 30/30, respectively. While half of them were positive in the third week (15/30). All cases became negative in the fourth week (0/15). We recommend not using the saliva swab as an alternative to the NP swab in the detection of the SARS-CoV-2 by RT-PCR. However, saliva sample can be used for the follow-up of the COVID-19 subjects, in children, elderly, and handicapped patients.
COVID-19, Saliva RT-PCR, Nasopharyngeal Swab, Oropharyngeal Swab, SARS-CoV-2, Saliva Self-sampling
The causative agent of the coronavirus disease-19 (COVID-19) is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. On March 11, 2020, the World Health Organization (WHO) considered this infection a pandemic with new cases of 508,041,253 and deaths of 6,224,220 all over the world as registered on 26-4-2022 (https://covid19.who.int/).
The main step in controlling the COVID-19 pandemic is the detection of SARS-CoV-2 in suspicious cases or in an individual with whom they have contact with infected subjects. Besides, obtaining an ideal clinical specimen to find the virus is the main effect in controlling the pandemic.1
The gold standard technique for the diagnosis of COVID-19 is the examination of the nasopharyngeal (NP) and oropharyngeal swabs by reverse transcriptase polymerase chain reaction (RT-PCR).2 Nevertheless, this technique has many drawbacks, it needs some experience in taking the sample, poor sensitivity, causes discomfort to the suspected COVID-19 individuals, and it increases the risk of exposure to the healthcare workers.3
Although swabs from the nasopharynx and oropharynx are mostly used for detection of the SARS-CoV-2, other body fluids can be used for this purpose, like blood, sputum, urine, nasal secretion, etc.4 Saliva is one of the biological fluids used for the detection of the virus by RT-PCR test. The validity of diagnostic accuracy has been investigated by previous studies.5 The saliva sampling carries the following advantages; self-collection way, non-invasive, and the saliva contains various salivary metabolomics which aid in differentiating the severity of COVID-19 from asymptomatic to severe cases.6 Moreover, the diagnosis of the viruses in the saliva specimens depends on the detection of the viral RNA, DNA, microRNA, and antigens. SARS-CoV-2 has an RNA genome that is quite similar to that of the severe acute respiratory syndrome coronavirus (SARS-CoV), which is the causal agent of SARS.7,8 SARS-CoV-2 uses the host cell angiotensin-converting enzyme 2 (ACE-2) as the major host receptor for cellular entrance, just like SARS-CoV.9 Previous research has found that salivary glands have higher levels of ACE-2 expression than the lungs,10 and that the epithelial cells lining the salivary gland duct were early targets of SARS-CoV infection in rhesus macaques.11 SARS-CoV was found in saliva samples as well.12 As a result, salivary glands could be a potential target for SARS-CoV-2 infection, and saliva could be a sample for SARS-CoV-2 detection.
The viral load of SARS-CoV-2 has recently been shown to be high just before the start of the disease. Utilizing a saliva sample as a specimen for disease screening seems to be an interesting concept.13 Another interesting issue in virology, some viruses remain in the saliva for up to 29 days, indicating that the saliva can be used as a useful diagnostic tool for early diagnosis, follow-up, and treatment of viral diseases.14,15
Several studies from various countries examine the role of saliva as a valuable specimen in the detection of SARS-CoV-2 and compare its results with NP or oropharyngeal swabs in the same individual.3,16,17 A recent study from Iran reported that the saliva swabs cannot substitute NP samples, but they can increase the detection rate and can be used together with NP samples.18 To our best knowledge, there is no study from Iraq concerning this issue. The major goal of the investigation was to contrast the findings of the RT-PCR tests on the identical COVID-19 suspects’ saliva and NP swabs. The secondary goal was to test the saliva, at weekly intervals for four weeks from the time of the presentation, in those with positive saliva RT-PCR test.
A cross-sectional study was performed on 100 COVID-19 suspects who visited an acute respiratory infection clinic at Al-Fallujah Teaching Hospital in Al-Anbar Governorate, Iraq, between November 29, 2021 and February 15, 2022. Those who had suspicious symptoms of COVID-19 like fever, cough, dyspnea, etc. for 2-8 days, of both sexes, and over the age of 15 years met the inclusion criteria. Subjects ≤ 15 years of age, those who had previous COVID-19, oral or salivary gland diseases or surgeries, or previous radiotherapy to the head and neck, patients who were taking drugs that increase or decrease the saliva secretion, and those who didn’t wish to participate in the study were excluded.
Data were gathered from every participant regarding the patient demographics, duration of the disease, symptoms at the time of presentation, and residence.
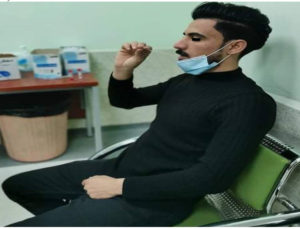
Under full aseptic technique and precautions (personal protective equipment), NP and salivary swabs were obtained using separate Nylon Swabs and sterile tubes containing VTM (Virus Transport Medium-TM) for each specimen. Individuals were asked to produce a volume of 1-2 ml of saliva before the swabs were collected (Figure 1).
Figure 1. After stroking the submandibular gland by the subject for one minute, he collects the saliva using a nylon swab
The subjects were divided into two groups according to the severity of the disease (mild and moderate to severe).
For the positive RT-PCR of the saliva at the time of the presentation, we took four saliva specimens from all individuals at weekly intervals. The Ethical Approval Committee of the University Of Anbar examined and approved the study protocol (reference number 20 on 8-5-2022). Informed consent was taken from every participant.
Specimen Preparation
The Central Health laboratory in the Al Anbar Health Directorate labeled the pairs of specimens; 1) NP and saliva swabs in two tubes, which were processed by technicians. We use a lysis buffer to treat samples from the collection container. 2) To inactivate SARS-CoV-2. A total of 200 ml of viral RNA was isolated from the samples. 3) Nucleic Acid Extraction Kit (“Magnetic Beads, Muenster, Germany) and a completely automated nucleic acid extraction system (AeHealth) in 10 minutes according to the manufacturer’s instruction.
Reverse Transcriptase-Polymerase Chain Reaction Work
The SARS-CoV-2 Nucleic Acid Detection Kit (Multiplex PCR Florescent Probe Method) detects SARS-CoV-2 in respiratory specimens such as oropharyngeal swabs, sputum, bronchoalveolar lavage fluid, NP, and saliva swabs in vitro. Primer sets and FAM labeled probes are designed for specific detection of the ORF1ab gene of SARS-CoV-2, VIC labeled probe for the N gene of SARA-CoV-2. The human RNase P gene extracted concurrently with the test sample provides an internal targeting human RNase P gene labeled with CY5. The CFX96 Real-Time Detection System was used to perform RT-PCR (Bio-Rad, Optics, Singapore). The used PCR primers are shown in the Table 1.
Table (1):
The used polymerase chain reaction (PCR) primers.
Name |
Description |
Oligonucleotide Sequence (5´ >3´ ) |
|---|---|---|
2019-nCoV_N1-F |
2019- nCoV_N1 Forward Primer |
GAC CCC AAA ATC AGC GAA AT |
2019-nCoV_N1-R |
2019- nCoV_N1 Reverse Primer |
TCT GGT TAC TGC CAG TTG AAT CTG |
2019- nCoV_N1 -P |
2019- nCoV_N1 Probe |
FAM-ACC CCG CAT TAC GGT TGG TGG ACC- |
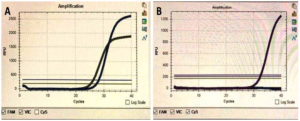
When the cycle threshold (Ct) values of both FAM and VIC, which target genes ORF1ab and N gene, respectively were ≤ 38 and the amplification curve was S-shaped, the result was regarded as positive, and when the Ct values of both targets were > 38, the result was considered negative (Figure 2). The samples with discordant Ct values were retested; that is, samples with one target gene with a Ct value of > 38 and another with a Ct value of ≤ 38. The specimens with recurring discordancy were declared as negative during the retesting. The diagnostic process took around 2 hours to complete. The technician who did the RT-PCR tests for the samples was unaware of the patient’s detail.
Statistical Analysis
The data were entered and analyzed using SPSS version 26 for the window. For continuous variables, the mean and standard deviation (SD) were used. While the categorical variables were presented in simple tables as frequencies and percentages. The Chi-Square test was used in the comparison of the categorical variables. The result of the RT-PCR of the NP was considered the reference standard to determine the sensitivity, specificity, and predictive value of salivary samples, and the accuracy measures of salivary samples were expressed as percentages with a 95% confidence interval. The Kappa coefficient was used to measure the agreement between the results of the RT-PCR of NP and saliva samples. A P-value of less than 0.05 is considered a statistically significant difference.
The following equations were used to determine the sensitivity, specificity, positive predictive value, and negative predictive value:
Sensitivity = true positive/(true positive+ false negative)
Specificity = true negative/(true negative + false positive)
Positive predictive value = true positive/(true positive+ false positive)
Negative predictive value = true negative/(true negative + false negative)
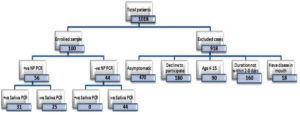
Of 1018, 100 sample pairs of NP and saliva swabs were collected (Figure 3). Fifty-five (55%) individuals were men. The age range of the participants was from 16 to 85 years (mean 35.95 ± 14.457). Sixty subjects were from the age group of ≤ 35 years. The duration of the disease ranged from 2 to 8 days, with a mean duration of 3.77 ± 1.462 days. Around three-quarters were from urban areas. The majority of the cases (92%) presented within 2-5 days and were of mild severity (89%) (Table 2).
Table (2):
Demographic and clinical characteristics of the 100 COVID-19 suspects.
Variable |
Frequency |
Percent |
|---|---|---|
Age groups per years |
||
≤ 35 years |
60 |
60% |
> 35 years |
40 |
40% |
Gender |
||
Male |
55 |
55% |
Female |
45 |
45% |
Residence |
||
Urban |
74 |
74% |
Rural |
26 |
26% |
Duration groups per days |
||
2-5 |
92 |
92% |
6-8 |
8 |
8% |
Severity |
||
Mild |
89 |
89% |
Moderate to severe |
11 |
11% |
We choose this division to put the COVID-19 suspects into two age groups, the first one (≤ 35 yeas) referred to children and young youth and the other group (> 35 years) referred to relatively older youth, middle age group, and elderly. I hope this answer is satisfactory for you. This will help us to compare the incidence of COVID-19 disease among different age group which relatively differs according to the social activity and exposure to the infection.
Figure 3. Flowchart of the COVID-19 suspects who were tested paired with saliva and NP samples for detection of SARS-CoV-2 by RT-PCR
Table (3):
A 2 X 2 table of the results of RT-PCR of the NP and saliva swabs.
| Saliva swab | Nasopharyngeal swab | P-value | ||
|---|---|---|---|---|
| Positive (n/%) | Negative (n/%) | Total (n/%) | ||
| Positive | 31 (100%) | 0 (0%) | 31 (100%) | 0.000 |
| Negative | 25 (36.2%) | 44 (63.8%) | 69 (100%) | |
| Total | 56 (56%) | 44 (44%) | 100 (100%) | |
Table 3 revealed that 56% of the NP swabs were positive by RT-PCR. While 31% of saliva swabs were positive. The sensitivity, specificity, positive predictive value, and negative predictive value were as below:
Sensitivity = true positive/(true positive+ false negative) = 31/(31+0) =100%
Specificity = true negative/(true negative + false positive) = 44/(44+25) = 63.8%
Positive predictive value = true positive/(true positive+ true positive) = 31/(31+25) = 55.4%
Negative predictive value = true negative/(true negative + false negative) = 44/(44+0) =1=100%
There was a mild agreement between the two specimens (kappa coefficient 0.522, 95% CI 0.723e0.979; P-value = 0.000).
Cough was the most common symptom (94%) and conjunctivitis was the least common (1%). There were no statistically significant differences between the results of the saliva RT-PCR and the symptoms (P-value > 0.05) (Table 4).
Table (4):
Relationship between the results of the saliva RT-PCR and the clinical symptoms of the 100 COVID-19 suspects.
| Variable | Saliva RT-PCR results | P-value | ||
|---|---|---|---|---|
| Positive (n, %) | Positive (n, %) |
Total (n, %) |
||
| Cough | 0.09 | |||
| Yes | 31 (33%) | 63 (67%) | 94 (100%) | |
| No | 0 (0%) | 0 (100%) | 0 (100%) | |
| Headache | 0.540 | |||
| Yes | 25 (29.8%) | 59 (70.2%) | 84 (100%) | |
| No | 6 (37.5%) | 10 (62.5%) | 16 (100%) | |
| Fever | 0.744 | |||
| Yes | 26 (31.7%) | 56 (68.3%) | 82 (100%) | |
| No | 5 (27.8%) | 13 (72.2%) | 18 (100%) | |
| Malaise | 0.538 | |||
| Yes | 23 (29.5%) | 55 (70.5%) | 78 (100%) | |
| No | 8 (36.4%) | 14 (63.6%) | 22 (100%) | |
| Arthralgia | 0.680 | |||
| Yes | 13 (28.9%) | 32 (71.1%) | 45 (100%) | |
| No | 18 (32.7%) | 37 (67.3%) | 55 (100%) | |
| Sneezing | 0.373 | |||
| Yes | 16 (35.6%) | 29 (64.4%) | 45 (100%) | |
| No | 15 (27.3%) | 40 (72.7%) | 55 (100%) | |
| Rhinorrhea | 0.482 | |||
| Yes | 9 (26.5%) | 25 (73.5%) | 34 (100%) | |
| No | 22 (33.3%) | 44 (66.7%) | 66 (100%) | |
| Olfactory dysfunction | 0.533 | |||
| Yes | 5 (38.5%) | 8 (61.5%) | 13 (100%) | |
| No | 26 (29.9%) | 61 (70.1%) | 87 (100%) | |
| Dyspnea | 0.777 | |||
| Yes | 3 (100%) | 8 (72.7%) | 11 (100%) | |
| No | 28 (31.5%) | 61 (68.5%) | 89 (100%) | |
| Gustatory dysfunction | 0.683 | |||
| Yes | 4 (36.4%) | 7 (63.6%) | 11 (100%) | |
| No | 27 (30.3%) | 62 (69.7%) | 89 (100%) | |
| Diarrhea | 0.338 | |||
| Yes | 0 (0%) | 2 (100%) | 2 (100%) | |
| No | 31 (31.6%) | 67 (68.4%) | 98 (100%) | |
| Conjunctivitis | 0.501 | |||
| Yes | 0 (0%) | 1 (100%) | 1 (100%) | |
| No | 31 (31.3%) | 68 (68.7%) | 99 (100%) | |
Out of 31 positive RT-PCR tests of the saliva, there were 18 (30%) from the age group ≤ 35 years, 21 (38.2%) male, 28 (30.4%) with a duration of 2-5 days, and 28 (31.5%) with mild COVID-19 severity. There were no statistically significant differences between the saliva RT-PCR test results and the above-mentioned variables (P-value > 0.05) (Table 5). The positive RT-PCR test results remained positive for the first and second weeks (31 and 30 respectively). In the third week, 50% (15/30) were positive. While no specimen was positive in the fourth week (Table 6).
Table (5):
The relationship between the saliva RT-PCR results and certain variables.
| Variables | Saliva swab | P-value | ||
|---|---|---|---|---|
| Positive (n/%) | Negative (n/%) | Total (n/%) | ||
| Age groups per years | 0.791 | |||
| ≤ 35 | 18 (30%) | 42 (70%) | 60 (100%) | |
| > 35 | 13 (32.5%) | 27 (67.5%) | 40 (100%) | |
| Total | 31 (31%) | 69 (69%) | 100 (100%) | |
| Gender | 0.086 | |||
| Male | 21 (38.2%) | 34 (61.8%) | 55 (100%) | |
| Female | 10 (22.2%) | 35 (77.8%) | 45 (100%) | |
| Total | 31 (31%) | 69 (69%) | 100 (100%) | |
| Duration per days | 0.679 | |||
| 2-5 | 28 (30.4%) | 64 (69.6%) | 92 (100%) | |
| 6-8 | 3 (37.5%) | 5 (62.5%) | 8 (100%) | |
| Total | 31 (31%) | 69 (69%) | 100 (100%) | |
| Severity | 0.777 | |||
| Mild | 28 (31.5%) | 61 (69.6%) | 89 (100%) | |
| Moderate to severe | 3 (27.3%) | 8 (72.7%) | 11 (100%) | |
| Total | 31 (31%) | 69 (69%) | 100 (100%) | |
Table (6):
The results of the PCR examination of the saliva according to the time of the presentation.
Timing |
Positive PCR N (%) |
Negative PCR N (%) |
Loss of follow-up N (%) |
Total |
|---|---|---|---|---|
At presentation |
31 (100) |
0 (0) |
0 (0) |
31 (100) |
One week |
31 (100) |
0 (0) |
0 (0) |
31 (100) |
Two weeks |
30 (96.8) |
0 (0) |
1 (3.2) |
31 (100) |
Three weeks |
15 (50) |
13 (43.3) |
2 (6.7) |
30 (100) |
Four weeks |
0 (0) |
10 (66.7) |
5 (33.3) |
15 (100) |
Detection of the causative agent in an epidemic or pandemic infection, such as the COVID-19 pandemic, has a crucial role in controlling the infection and preventing unwanted complications or even mortality. At the beginning of the COVID-19 pandemic, researchers from all over the world tried to detect the SARS-CoV-2 in various body fluids or tissues.19 They succeeded in the isolation of the virus in the nasopharynx, oropharynx, sputum, bronchoalveolar fluids, saliva, urine, etc. The RT-PCR test on NP swabs is the gold standard for detecting SARS-CoV-2. However, these swabs have some drawbacks, including the risk of infection to healthcare personnel, a difficult sampling technique, inconvenience to patients, and insufficient sampling, which may result in a lower swab quantity and a low sensitivity rate of the test.20 Therefore, searching for non-invasive samples like saliva is of utmost importance for the diagnosis of this novel coronavirus. The role of testing a saliva sample by RT-PCR has been assessed by several researchers, and they found that it can be used as an alternative to the conventional NP swab.21 The current study found that saliva cannot be used as an alternative sample to the NP swab in the detection of SARS-CoV-2. However, the saliva sample can be used in the follow-up of COVID-19 patients and monitoring of treatment.
Saliva expresses a homogenous and wide evaluation of the genomic characteristics of the SARS-CoV-2, therefore, it is useful for determining the COVID-19 status.22,23 Besides, saliva specimens for the detection of SARS-CoV-2 by RT-PCR serve five benefits. First, these specimens are easily collected from the subject without an invasive method. Therefore, it can greatly reduce the nosocomial infection of the COVID-19 among healthcare workers, visitors, and patients. Second, the collection of saliva specimens can be performed in an outpatient clinic or at home. Third, they can be used as a screening test owing to the non-invasive nature of the sample collection. Fourth, these samples are not needed for healthcare provisional, and hence there is no need for a waiting time in collecting the samples. Therefore, it is particularly useful in overcrowded clinics.24 Fifth, when NP samples are unavailable, saliva samples can be used in certain individuals, such as children or the disabled.5,25
There are four ways of collecting saliva samples, including drooling, coughing out, self-collecting, and directly from the salivary duct. Previous studies reported that self-collecting of samples was a reasonable method.3 In this study, patients self-generated a saliva sample without coughing, resulting in reduced aerosol formation and a lower risk of infection for healthcare workers. At the disease onset, SARS-CoV-2 was found in posterior oropharyngeal saliva samples, with a high viral load.13,24 An early morning saliva sample was collected after coughing up and cleaning the throat in this investigation. Although the collected sample may contain both saliva and sputum, the likelihood of the patient coughing up sputum is minimal, as studies have shown that dry cough is the most prevalent symptom, occurring in roughly 80% of patients at the onset of the disease.26
Our study revealed that for all suspects whose NP swabs tested negative for SARS-CoV-2 (n = 44), all saliva swabs for them tested negative. This finding was consistent with the results of a previous study.24 However, the positive detection rate in the saliva sample in our study (31/56) was much lower than in the study by To et al. (11/12).24 This study was performed on hospitalized patients, and the coughing-out was used to collect the saliva samples. During our study, we performed RT-PCR on COVID-19-positive suspects and collected saliva samples on our own. Many studies have been conducted to investigate the possibility of using saliva as an alternative specimen in the detection of SARS-CoV-2.3,5,20 There are different results among various studies regarding the comparison of the RT-PCR test of the NP and saliva specimens.20,27,28 Some investigations reported the superiority of the NP specimens over saliva samples, while the others showed the opposite finding, and other investigations revealed an equal sensitivity of both samples.20,28,29,30 The current study revealed that the sensitivity and specificity of the RT-PCR saliva samples in comparison with the NP swabs were 100% and 63.8%, respectively. Besides, the results showed that there was a mild agreement between the RT-PCR of the NP and saliva specimens (kappa coefficient = 0.522). Therefore, we don’t advise using the saliva samples as an alternative to the NP swabs to detect COVID-19 in the suspected cases. However, they can be used during the screening process or in cases where NP swabs are difficult to obtain, such as with children, the elderly or handicapped individuals.
A previous study suggested that the RT-PCR for NP and saliva in the detection of SARS-CoV-2 had high sensitivity (84.2% for saliva and 79.5% for NP swabs) in the early time period from the onset of the COVID-19 (1-5 days).16 A similar finding of the high detection rate concerning the timing of presentation was also reported by other researchers.31 On the other hand, Dogan et al. reported a low rate of positivity (55%-63%) at the early onset of symptoms.32 However, the detection rate decreased gradually from 71.4% at 0 time to 33.3% at 5 days from the onset of symptoms. Our study found that there was no significant association between the detection rate of SARS-CoV-2 by RT-PCR saliva of 2 durations (2-5 days, vs 6-8 days). Furthermore, the detection rate remained the same in the first and second weeks from the time of presentation but reduced to half in the third week, and to 0 detection rate in the fourth week (Table 6). Our results are of benefit for the decision-makers to regulate the use of saliva samples as a good option for detection of SARS-CoV-2 in the early stages of the onset of the symptoms when in need of mass detection.
Our findings showed that there was no significant association between the detection rate of the SARS-CoV-2 by RT-PCR saliva and the severity of the COVID-19 suspects, and clinical symptoms (P-value > 0.05). However, the study by Uddin et al. reported a significant association between the detection rate and the following symptoms; fever, cough, chills, altered smell, muscle aches, and loss of appetite.16 It is crucial to consider certain symptoms like fever, cough, and olfactory dysfunction as a possible presentation of the COVID-19 as well as send the patients for laboratory tests to exclude or confirm the diagnosis of the disease.
This study’s approach ensures that only pure saliva is collected, with no contamination from other respiratory secretions. Saliva and NP matched samples acquired from the same suspects reduce other background factors. The quick turnaround time (less than 24 hours) used in this study ensures that the positive difference isn’t due to overlapping factors like transportation or cold chain issues. COVID-19 suspects were used to collect these matched samples, which contained both positive and negative cases. As a result, when the intended use is for mass self-testing, it may reflect reality and minimize the need for healthcare personnel to collect samples. However, the study didn’t measure the viral load, which can be considered a shortcoming of the current study. Owing to the study design, the study was performed on COVID-19 suspects who presented within 2-8 days from the onset of the symptoms. Therefore, the current study did not test those who were asymptomatic or those who presented outside of the range of 2-8 days. However, the disease spectrum in the affected individuals ranged from asymptomatic to critically ill patients. This was considered another shortcoming of the present study.
Owing to the mild agreement of the saliva RT-PCR in comparison with the NP swabs, we recommend not using this test in clinical practice. Examination of the saliva by RT-PCR can provide a useful tool for follow-up of the COVID-19 patients. Moreover, saliva samples can be used in the screening process, patients with an extreme age (children and old age), and those with handicaps.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All the authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, University Of Anbar, Iraq with reference number 20, dated 8-5-2022.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- WHO, “Guidelines for the collection of clinical specimens during field investigation of outbreaks,” World Health Organization, 2000.
- WHO, “Use of laboratory methods for SARS diagnosis.” 2003.
- Bidkar V, Mishra M, Gade N, Selvaraj K. Conventional Naso-Oropharyngeal Sampling Versus Self-Collected Saliva Samples in COVID-19 Testing. Indian J Otolaryngol Head Neck Surg. 2021:1-7.
Crossref - Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843-1844.
Crossref - Pasomsub E, Watcharananan SP, Boonyawat K, et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2021;27(2):285-e1.
Crossref - Sapkota D, Soland TM, Galtung HK, et al. COVID-19 salivary signature: diagnostic and research opportunities. J Clin Pathol. 2021;74(6):344-349.
Crossref - Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457-460.
Crossref - Shirato K, Nao N, Matsuyama S, Kageyama T. An ultra-rapid real-time RT-PCR method for detecting Middle East respiratory syndrome coronavirus using a mobile PCR device, PCR1100. Jpn J Infect Dis. 2020;73(3):181-186.
Crossref - Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.
Crossref - Xu J, Li Y, Gan F, Du Y, Yao Y. Salivary glands: potential reservoirs for COVID-19 asymptomatic infection. J Dent Res. 2020;99(8):989.
Crossref - Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85(8):4025-4030.
Crossref - Wang W-K, Chen S-Y, Liu I-J, et al. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 2004;10(7):1213-1219.
Crossref - To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565-574.
- Castro T, Sabalza M, Barber C, et al. Rapid diagnosis of Zika virus through saliva and urine by Loop-mediated isothermal amplification (LAMP). J Oral Microbiol. 2018;10(1):1510712.
Crossref - Martelli F, Mencarini J, Rocca A, Malva ND, Bartolozzi D, Giannecchini S. Polyomavirus microRNA in saliva reveals persistent infectious status in the oral cavity. Virus Res. 2018;249:1-7.
Crossref - Uddin MKM, Shirin T, Hossain ME, et al. Diagnostic Performance of Self-Collected Saliva Versus Nasopharyngeal Swab for the Molecular Detection of SARS-CoV-2 in the Clinical Setting. Microbiol Spectr. 2021;9(3):e00468-21.
Crossref - Kamel M, Maher S, El-Baz H, Salah F, Sayyouh O, Demerdash Z. Non-Invasive Detection of SARS-CoV-2 Antigen in Saliva versus Nasopharyngeal Swabs Using Nanobodies Conjugated Gold Nanoparticles. Trop Med Infect Dis. 2022;7(6):102.
Crossref - Abdollahi A, Salarvand S, Ghalehtaki R, et al. The role of saliva PCR assay in the diagnosis of COVID-19. J Infect Dev Ctries. 2022;16(01):5-9.
Crossref - Bulfoni M, Sozio E, Marcon B, et al. Validation of a saliva-based test for the molecular diagnosis of SARS-CoV-2 infection. Dis Markers. 2021;2022:6478434.
Crossref - Tsujimoto Y, Terada J, Kimura M, et al. Diagnostic accuracy of nasopharyngeal swab, nasal swab and saliva swab samples for the detection of SARS-CoV-2 using RT-PCR. Infect Dis (Lond). 2021;53(8):581-589.
Crossref - Takeuchi Y, Furuchi M, Kamimoto A, Honda K, Matsumura H, Kobayashi R. Saliva-based PCR tests for SARS-CoV-2 detection. J Oral Sci. 2020;62(3):350-351.
Crossref - Czumbel LM, Kiss CS, Farkas N, et al. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. 2020;465.
Crossref - Altawalah H, AlHuraish F, Alkandari WA, Ezzikouri S. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: A cross-sectional study. J Clin Virol. 2020;132:104652.
Crossref - To KK-W, Tsang OT-Y, Yip CC-Y, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841-843.
Crossref - Matic N, Stefanovic A, Leung V, et al. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur J Clin Microbiol Infect Dis. 2021;40(2):447-450.
Crossref - Chen Q, Zheng Z, Zhang C, et al. Clinical characteristics of 145 patients with corona virus disease 2019 (COVID-19) in Taizhou, Zhejiang, China. Infection. 2020;48(4):543-551.
Crossref - Senok A, Alsuwaidi H, Atrah Y, et al. Saliva as an alternative specimen for molecular COVID-19 testing in community settings and population-based screening. Infect Drug Resist. 2020;13:3393.
Crossref - Jamal AID, Mozafarihashjin AJ, CoomesM, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. 2021;72(6):1064-1066.
Crossref - Skolimowska K, Rayment M, Jones R, Madona P, Moore LSP, Randell P. Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect. 2020;26(12):1711-1713.
Crossref - Kerneis S, Elie C, Fourgeaud J, et al. Accuracy of antigen and nucleic acid amplification testing on saliva and naopharyngeal samples for detection of SARS-CoV-2 in ambulatory care. MedRxiv. 2021.
Crossref - Nagura-Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58(9):e01438-20.
Crossref - Dogan OA, Kose B, Agaoglu NB, et al. Does sampling saliva increase detection of SARS-CoV-2 by RT-PCR? Comparing saliva with oro-nasopharyngeal swabs. J Virol Methods. 2021;290:114049.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.