Ally C. Antony1,2*, Mini K. Paul2, Reshma Silvester1, Aneesa P.A.1, Suresh K.1, Divya P.S.1, Simmy Paul3, Fathima P.A.4 and Mohamed Hatha Abdulla1
1Department of Marine Biology, Microbiology and Biochemistry, School of Marine Sciences,
Cochin University of Science and Technology, Lakeside Campus, Cochin – 682 016, India.
2 M.E.S. College, Marampally, Ernakulam – 683 107, India.
3School of Medical Education, Mahatma Gandhi University, Kottayam, India.
4State Council of Educational Research and Training, Trivandrum, India.
ABSTRACT
The performance of Hicrome E. coli agar (Himedia) versus EMB agar (Himedia) was compared for the detection of E. coli from food and environmental sources. Four hundred bacterial isolates producing typical E. coli like green metallic sheen on EMB agar were subjected to biochemical analysis by IMViC tests and they exhibited 6 different types of reactions. Isolates exhibiting only two distinct reaction types were proved to be typical E. coli, which were reconfirmed by molecular detection of species-specific uid A gene as well as 16S rRNA sequencing. Isolates exhibiting the remaining four reaction types comprised of Enterobacter asburiae, Kluyvera georgiana, Pantoea agglomerans and Phytobacter diazotrophicus respectively, all belonging to the family Enterobacteriaceae. The E. coli strains recovered from Hicrome E. coli agar comprised of 20 different serogroups including glucuronidase positive atypical O157: H7. Hicrome E. coli agar had better sensitivity (100%), specificity (99%) and efficiency (99.5%) than EMB agar in differentiating the false positives and false negatives and does not require any further confirmatory tests.
Keywords: Escherichia coli; EMB agar; Hicrome E. coli agar, metallic sheen.
INTRODUCTION
- coli is the widely accepted faecal indicator, since its presence does not occur in other niches, unless and until there is faecal contamination. It is easily detectable and its presence in food, water and other sources is indicative of recent faecal contamination and possible presence of other pathogens (1, 2). Even though most E. coli are non-pathogenic, there are several potentially pathogenic strains with virulent factors capable of causing diarrheal disease in humans (3, 4). The concentration of faecal coliforms or E. coli is the universally accepted microbiological quality parameter, determining the sanitary quality of food commodities meant for domestic as well as export purposes. In order to ensure public safety, it is strictly monitored in case of water bodies used for recreation, drinking water purposes or shellfish growing. Certification of shellfish harvesting areas based on E. coli levels has been made mandatory (5, 6), failing of which stringent actions like closing of the harvesting areas are implemented until the required standards are met with. For all these evaluations, isolation and identification of E. coli is a prerequisite which is often complicated in practice, due to the co-occurence of other interfering enteric bacteria such as Citrobacter, Klebsiella and Enterobacter, which also ferment lactose and exhibit similar colony morphology on Levine’s Eosin Methylene Blue (EMB) agar selective for E. coli. (7, 8, 9). These coliforms, unlike faecal coliforms, may also be present in other environmental sources and hence their presence need not essentially indicate faecal contamination. Incubation at elevated temperatures (44.5 °C) and use of Brilliant Green Lactose Bile Broth (BGLB) medium are employed, to distinguish these competing microbiota, but is not always possible, since they also exhibit similar reactions.
The problem is more evident especially when these competing flora are present in relatively high numbers, increasing the number of false positives, thus interfering with the isolation of the actual required isolates and reducing the efficiency of this selective medium (7). Even though many researchers often encounter this problem, there are very few reports suggesting an alternative to this medium and still less on the identification of the interfering microbiota, which may be helpful in avoiding them. There are many reports related to use of different chromogenic media for isolation of enterohaemorhagic E. coli O157:H7, from different sources; but it is not so with other pathogenic or non-pathogenic E. coli serotypes (10, 11). Hence, newer media or methods should be attempted for the rapid and precise isolation of this sanitary indicator which may be helpful in avoiding erroneous interpretations.
The current study compares the performance of a chromogenic medium, Hicrome E. coli agar (Himedia, India), over Eosin Methylene Blue (EMB, Himedia, India) agar, for isolation of E. coli from various sources. The interfering microbiota was also identified and a phylogenetic comparison of the same with E. coli was being undertaken.
Materials and Methods
Bacterial cultures and standard strains
Four hundred presumptive E. coli isolates obtained from various sources such as, live clam (n=140), bivalve shellfish meat available in market (n=45), water (n=100) and sediments from a tropical estuary (n=100) and drinking water (n=15) were used for the study.
- coli ATCC 25922 and 4 other strains CUSMBES1, CUSMBES 9, CUSMBES10 and CUSMBES 11a maintained in our laboratory with GenBank accession numbers JX183939.1, JX183940.1, JX183941.1, and JX183942.1 respectively were used as the standard reference strains.
Sample processing, isolation and biochemical analysis
Samples were aseptically collected and processed as per standard guidelines (12, 13 14). Approximately 25 g of shellfish meat was aseptically transferred to a sterile stomacher bag and blended with 225 ml of sterile peptone water in a stomacher (IUL Instruments, Spain). Five tube MPN test using lactose broth was performed for the isolation of E. coli. Similarly, sediment and water samples were also diluted (1:9 ratio) and examined for the presence of E. coli by the same method. The tubes were examined for evidence of lactose fermentation after incubation at 44.5 °C for 24 – 48 hours. From the tubes showing growth and gas production, loopful was streaked on to conventional Levine’s Eosin-methylene blue agar (L-EMB, Himedia, India) and alternatively to Hicrome E. coli agar (14). The plates were observed following 24 hours of incubation at 37 °C. On EMB, strong acid producers like E. coli usually form colonies with green metallic sheen, showing a dark nucleated centre. Under acidic conditions, the dye eosin Y is precipitated, producing dark coloured colonies usually with a green metallic sheen due to the formation of an amide bond between eosin Y and methylene blue in the medium (15,9). On the other hand, Hicrome E. coli agar detects E. coli based on the presence of the glucuronidase enzyme. The chromogenic substrate p-nitrophenyl-p-D-glucopyranosiduronic acid (PNPG) in the medium is absorbed by E. coli and the intracellular b-Glucuronidase enzyme present in most serotypes other than O157:H7 cleaves the bond between chromophore and the glucuronide, releasing the former, producing a blue-green coloured colony (16). Prior to testing samples, the efficiency of the two media was assessed, under controlled conditions using five standard strains mentioned above and was found to be 100% for both EMB agar and Hicrome E. coli agar alike. Typical colonies were subjected to biochemical confirmation by IMViC tests, which included Indole production test, Methyl red test, Voges-Proskauer and Citrate utilization tests (17,14).
Molecular confirmation by detection of uid A gene
Genomic DNA was extracted by boiling method and molecular confirmation was done by PCR amplification of uid A gene which codes for
b-D-Glucuronidase enzyme of E. coli using the primer set UAL- 754 (5′-AAAACGGCAAGAAA AAGCAG-3′) and UAR- 900 (5’ACGCGTGG TTACAGTCTTGCG-3′) (18,19). The PCR products were resolved by agarose (1.5% w/v) gel electrophoresis in 1X TBE buffer, containing 0.5 µg/ml of ethidium bromide and visualized by Gel Documentation System (Bio-Rad Gel Doc EZ Imager, USA). Amplicon sizes were compared with 100 bp DNA ladder; 147 bp PCR products confirmed the presence of E. coli.
Serotyping of E. coli
The confirmed E. coli strains were serotyped at the National Salmonella and Escherichia centre, Central Research Institute (Kasauli, Himachal Pradesh, India).
16S rRNA amplification and sequencing
Representative isolates from the confirmed and suspected groups were subjected to 16S rRNA gene amplification using the universal primer set (27F: 5′ -AGA GTT TGA TCC TGG CTC AG-3′ , 1492R: 5′ -GGT TAC CTT GTTACG ACTT-3′) as described elsewhere (20). PCR products were sequenced by Sangers dideoxy method (Scigenome, India) and the sequences edited using Gene Tool lite version 1.0 software. The percentage of homology was determined against known sequences in the NCBI GenBank database, using the Basic Local Alignment Search Tool algorithm (BLAST;http://www.ncbi.nlm.nih.gov/BLAST) and the sequences were submitted in the GenBank and allotted with an accession number.
Phylogenetic analysis
The partial 16S rRNA gene sequences of the E. coli and other false positive isolates obtained in this study, together with 16S rRNA gene sequences of similar isolates from previous reports and other related genera obtained from GenBank, were aligned using the CLUSTAL W program and the phylogenetic tree was constructed using Neighbour-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test with 1000 replicates (Boot strap value) is shown below the branches. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA6 software.
Statistical analysis
Statistical comparison of the performances of the two media, EMB agar and Hicrome E. coli agar was done by calculating percentages of sensitivity, specificity and efficiency using the equations below21.
% Sensitivity = True positives x 100/ (True positives + False Negatives)
% Specificity = True negatives x 100/ (True negatives +False positives)
% Efficiency = (True positives + True negatives) x 100/ Total
Results and Discussion
EMB agar and Hicrome E. coli agar showed 100% efficiency in identification of the five standard strains used in the study (data not shown). However, problem arose when both metallic sheen producing non E. coli and true E. coli were present together, as in the natural samples tested. The results of the isolation and identification of E. coli from the sources mentioned above using the two media under comparison are summarised in Tables 1 and 2. False positive results due to competing flora were found to be very high on EMB agar (40 %) whereas on Hicrome it was found to be very low (0.5%). False negatives were also high on EMB agar (15.75%), due to the sheen of true E. coli masked by the highly mucoid background flora. No false negatives were detected on Hicrome E. coli agar (Table 1). Similarly the sensitivity, specificity and efficiency of Hicrome E. coli agar were far above those of the Levine’s EMB agar (Table 2).
Table 1. Characteristic appearance of true positive, false positive, true negative and false negative colonies on EMB and Hicrome E. coli agar respectively.
Media |
True positive (n %) |
True negative (n %) |
False positive (n %) |
False negative (n %) |
EMB |
Green metallic sheen (34.25) |
No sheen (10) |
Green metallic sheen (40) |
No sheen (15.75) |
Hicrome E. coli agar |
Blue green (50) |
Colourless (49.5) |
Blue green (0.5) |
Colourless (0) |
Table 2. Comparison of EMB agar and Hicrome E. coli agar in terms of percentages of sensitivity, specificity and efficiency.
Appearance of E. coli and Non-E. coli on Hicrome E. coli agar vs EMB agar
True E. coli produced colonies with typical high metallic sheen on EMB agar and blue-green colonies on Hicrome E. coli agar respectively (Fig. 1). On the contrary, a large number of typical high metallic sheen E. coli like colonies isolated from EMB agar, produced white colonies on Hicrome E. coli agar and when tested biochemically, were proved to be Non E. coli (Fig. 2). It was also observed that, if the E. coli concentration is comparatively less in the sample, the sheen is often masked by the highly mucoid interfering background flora such as Klebsiella (Fig. 3), resulting in false negative results, which could also be effectively differentiated by the use of Hicrome E. coli agar.
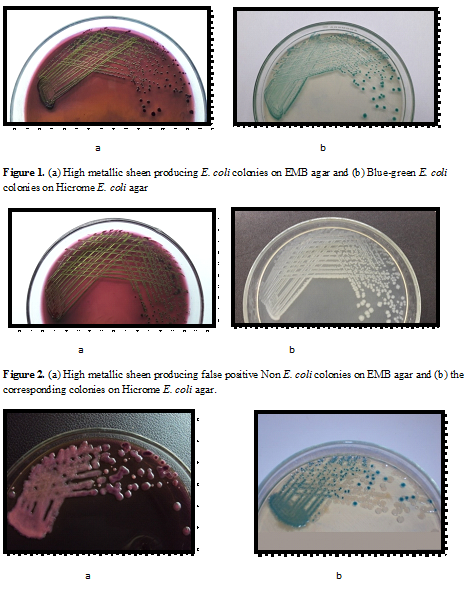
Figure 3. Interfering microbiota on EMB agar (a) and corresponding Hicrome E. coli agar plate (b). On Hicrome E. coli agar, true E. coli producing blue-green colonies are clearly differentiated from the interfering microbiota which formed white colonies.
Biochemical characterisation of the isolates
The isolates with high metallic sheen isolated from EMB agar exhibited 6 different types of IMViC reactions (Table 3). The first two reaction types comprised true E. coli which belonged to biotype I and II respectively (14), whereas all others were proved to be non-E. coli.
Table 3: Summary of the biochemical, molecular identifications and appearance of the isolates on EMB as well as Hicrome E. coli agar.
IMViC reaction |
Appearance on EMB agar |
Appearance on Hicrome E. coli agar |
uid A gene |
Organism |
+ + – – |
Green metallic sheen |
Blue green |
+ |
E. coli |
– + – – |
Green metallic sheen |
Blue green |
+ |
E. coli |
– + – + |
Green metallic sheen |
Colour less |
– |
Enterobacter asburiae |
+ + – + |
Green metallic sheen |
Colour less |
– |
Kluyvera georgiana |
– + + + |
Green metallic sheen |
Colour less |
– |
Pantoea agglomerans |
+ + – – (atypical reaction, yellow colour for Citrate utilization test) |
Green metallic sheen |
Colour less |
– |
Phytobacter diazotrophicus |
Molecular confirmation of the isolates
All the biochemically confirmed E. coli strains were positive for uid A gene, yielding 147 bp specific PCR amplicons(not shown ). The remaining four biochemical groups yielded either non specific PCR products or no products at all when tested for uid A gene.
16S rRNA amplification and sequencing
When subjected to 16S rRNA sequencing, isolates exhibiting the first two reaction types were reconfirmed to be E. coli (EF2p4) and the sequence was submitted in the GenBank with accession number KT804408. Non E. coli isolates exhibiting the remaining four reaction types were identified to be Enterobacter asburiae (E19), Kluyvera georgiana (E3), Pantoea agglomerans (E 30) and Phytobacter diazotrophicus (2ss1B3) all belonginng to the Enterobacteriaceae family and their 16S rRNA sequences were submitted in the GenBank with accession numbers KT804411, KT804412, KT804413 and KU240006 respectively.
Phylogenetic analysis
The phylogenetic analysis showed that the E. coli (EF2p4) and P. diazotrophicus (2ssiB3) belong to the same cluster with a boot strap value of 37. E. asburiae (E19) and P. agglomerans (E30) form a cluster with a boot strap value of 100. K. georgiana (E3) does not belong to any of these clusters but form a cluster with K. ascorbata strain ATCC 33433 with a boot strap value of 82 (Fig. 4).
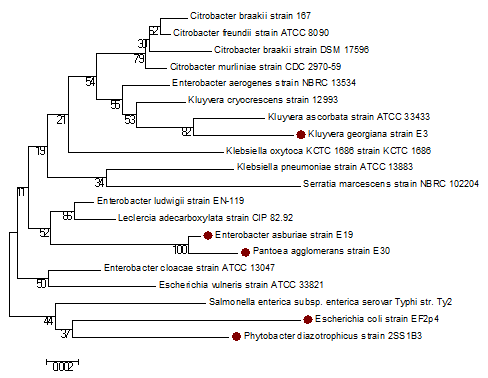
Figure 4: Phylogenetic tree of 16S rRNA gene sequences, constructed using Neighbour-Joining method, showing relationships between the false positive E. coli strains, and typical E. coli identified in this study (marked with bullets) and similar false positive isolates in other published reports and other closely related genera is shown. The scale on bottom measures evolutionary distance in substitutions per nucleotide. Boot strap values are shown below the branches.
Serotyping of E. coli
The confirmed E. coli isolates when serotyped constituted 20 different serogroups, including enterohaemorrhaegic O157:H7 (3.5%), enterotoxigenic O8 (8%), shigatoxigenic O121 (4.5%), enteroinvasive O124 (0.5%) and enteropathogenic O91 (0.5%) serogroups. Thus Hicrome E. coli agar enabled the isolation of b-D- Glucuronidase positive atypical E. coli O157:H7 strains also, which would have been otherwise missed on media specific for the same, since they specifically detects, glucuronidase negative typical isolates only (21). The other serogroups identified included O2, O4, O9, O17, O27, O33, O34, O35, O84, O118, O119, O120, O126, O141, and O156.
The study has proved the diagnostic efficiency of Hicrome E. coli agar (99.5%) over EMB agar (44.25%). Similarly, the sensitivity and specificity of Hicrome E. coli agar were also far higher than those of EMB agar (Table 2). Increased incidence of false positives on EMB agar make the isolation of this otherwise simple organism cumbersome, involving too many colony picks, finally yielding very few positives or no positives at all, while missing the true ones (Fig. 2 and Fig. 3).
Often it was also observed that E. coli isolated from EMB agar, existed in strong association with other flora, which on prolonged storage outgrew the E. coli, leading to the loss of the original required isolate. This phenomenon also could be overcome or rather reduced considerably by the use of chromogenic medium. Most of the false positives isolated from EMB belonged to the family Enterobacteriaceae, which reveals the inefficiency of EMB agar in differentiating E. coli from other Enterobacteriaceae members exhibiting similar phenotypic and biochemical characteristics. There have been previous reports regarding Citrobacter spp. Enterobacter spp., Escherichia vulneris, Kluyvera cryocrescens, Hafnia, Klebsiella, Rahnella and Serratia forming E. coli like false positive colonies on EMB agar (14,7,8). However to our knowledge, this is the first report of Enterobacter asburiae, Kluyvera georgiana and Phytobacter diazotrophicus forming false positive colonies on EMB agar.
Conclusion
Even though Hicrome E. coli agar is more expensive than EMB agar as any other chromogenic medium, when all the factors are considered, it is much more efficient, rapid and cost effective method and could save time and money. In short, this medium is highly recommended for the analysis of large number of samples, due to the accuracy and simplicity of which, even less experienced lab personnel can confirm the identification without further confirmatory biochemical or molecular tests.
Acknowledgements
The authors are thankful to the Head, Department of Marine Biology, Microbiology and Biochemistry, Cochin university of Science and Technology, Cochin, India, for providing the facilities to carry out the research work. The FIP-teacher fellowship granted to Mrs Ally C. Antony, by the University Grants Commission, Government of India is also gratefully acknowledged.
References
- Edberg, S.C., Rice, E.W. Karlin, R.J. Allen, M.J. Escherichia coli: the best biological drinking water indicator for public health protection. J. Appl. Microbiol. Symposium Supplement; 2000; 88: 106S–116S.
- Costa, R.A. Escherichia coli in seafood/ : A brief overview. Adv. Biosci. Biotechnol., 2013; 4, 450–454.
- Kaper, J.B., Nataro, J.P., Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol., 2004; 2: 123-140.
- Bacteriological Analytical Manual: Chapter 4A, Diarrheagenic Escherichia coli, 2011. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070080.htm
- European Commission (EC), Council Directive 91/492/EEC. Official Journal of the European Communities No. L268, 24/09/1991; 1-14.
- NSSP (National Shellfish Sanitation Program). Guide for the control of molluscan shellfish. Chapter IV. 2013; U.S. Food and Drug Administration, Centre for Food Safety and Applied Nutrition, Washington, D.C, USA, (availablehttp:// www.fda.gov/Food/GuidanceRegulation/FederalStateFoodPrograms/ucm2006754.htm)
- Kim, H.S., Kim, Y.J., Chon, J.W., Kim, D.H. Kim, K.Y., Seo, K.H. Citrobacter braakii: A Major Cause of False-Positive Results on MacConkey and Levine’s Eosin Methylene Blue Selective Agars Used for the Isolation of Escherichia coli from Fresh Vegetable Samples. J. Food Saf., 2016; 36: 33–37.
- Venkateswaran, K., Murakoshi, A., Satake, M. Comparison of commercially available kits with standard methods for the detection of coliforms and Escherichia coli in foods. Appl. Environ. Microbiol., 1996; 62, 2236–2243.
- Leininger, D.J., Roberson, J.R., Elvinger, F. Use of eosin methylene blue agar to differentiate Escherichia coli from other gram-negative mastitis pathogens. J. Vet. Diagn. Invest., 2001; 13: 273–275.
- Hirvonen, J.J., Siitonen, A., Kaukoranta, S. Usability and performance of CHROMagar STEC in detection of shiga toxin-producing Escherichia coli strains. J. Clin. Microbiol., 2012; 50: 3586-3590.
- Gill, A., Huszczynski, G., Gauthier, M., Blais B. Evaluation of eight agar media for the isolation of shiga toxin—producing Escherichia coli. J. Microbiol. Meth., 2014; 96, 6-11.
- American Public Health Association: Recommended Procedures for the Examination of Seawater and Shellfish, 4th ed. APHA, Washington, DC, 1970.
- American Public Health Association: Compendium of Methods for the Microbiological Examination of Foods, 3rd ed. APHA, Washington, DC, 1992.
- Bacteriological Analytical Manual: Chapter 4, Enumeration of Escherichia coli and the Coliform Bacteria, 2002. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm064948.htm
- Horvath, R.S., Ropp, M.E. Mechanism of action of Eosin-Methylene Blue Agar in the differentiation of Escherichia coli and Enterobacter aerogenes. Int. J. Syst. Bacteriol., 1974; 24: 221–224.
- Hansen, W., Yourassowsky, E. Detection of ²-glucuronidase in lactose-fermenting members of the family Enterobacteriaceae and its presence in bacterial urine cultures. J. Clin. Microbiol., 1984; 20: 1177–1179.
- Barrow, G.I., Feltham, R.K.A. Cowan and Steel’s manual for the identification of medical bacteria. Cambridge: Cambridge University Press, 1993; p 317.
- Bej, A.K., DiCesare, J.L., Haff, L. Atlas, R.M. Detection of Escherichia coli and Shigella spp in water by using the Polymerase Chain-Reaction and gene probes for Uid. Appl. Environ. Microbiol., 1991; 57: 1013–1017.
- Devi, R., Surendran, P., Chakraborty, K. Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the southwest coast of India. World J. Microbiol.Biotechnol., 2009; 25: 2005–2012.
- Silvester, R., Alexander, D., Ammanamveetil, M.H.A. Prevalence, antibiotic resistance, virulence and plasmid profiles of Vibrio parahaemolyticus from a tropical estuary and adjoining traditional prawn farm along the southwest coast of India. Ann. Microbiol., 2015; 65: 2141-2149.
- Manafi, M., Kremsmaier, B. Comparative evaluation of different chromogenic/fluorogenic media for detecting Escherichia coli O157:H7 in food. Int. J. Food Microbiol., 2001; 71: 257–262.
- Manal A. H., Saad, S. F., Zahraa, A.J., Saba, T.H. Chromogenic agar media for rapid detection of Enterobacteriaceae in food samples. Afr. J. Microbiol. Res., 2015; 9, 2354–2357.

