ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcal biofilms are the prominent cause of chronic wound infection and antibiotic resistance. It acts as a reservoir for bacteria, making wound healing difficult. Biofilm infections increase the hospital stays and cost to the patients. The current study explores the phenotypic and genotypic detection of S. aureus biofilm from chronic traumatic wounds and their association with antibiotic resistance. A prospective observational study was conducted from April 2020 to March 2021. S. aureus isolates were identified by the MALDI-TOF. Antibiotic susceptibility was determined by VITEK 2. Biofilm production detected by tissue culture plate method and associated ica genes were diagnosed through multiplex PCR. Overall, 67 isolates were investigated. The frequency of biofilm production in S. aureus was 73.1%, and most of the isolates were moderate biofilm producers (38.8%). The presence of intracellular adhesion (ica) operon among these isolates was 85.7% and also significantly associated with the strength of biofilm mass formation. Ica A was the predominant biofilm-producing gene (42.9%). Biofilm producing Methicillin-resistant S. aureus were as high as 75%, and multidrug resistant strains were significantly associated with biofilm formation. But frequency of ica genes were noted more in Methicillin sensitive S. aureus (65.2%). High frequency of biofilm in S. aureus of isolates was responsible for the development of chronic non-healing traumatic wounds. Biofilm-producing isolates showed greater multidrug-resistance. Phenotypically MRSA expressed more biofilm, but ica operon was documented in MSSA. It emphasized the further need for ica independent pathway exploration for MRSA biofilm synthesis.
MRSA, Biofilm, Ica operon, traumatic wound, Multidrug resistance
Staphylococcus aureus, together with methicillin resistant S. aureus (MRSA), is a predominant pathogen identified in chronic traumatic wound infection. It possesses several virulence factors. Biofilm formation is one of the important virulence factors this bacteria retains and is considered a severe problem, specifically in chronic wound infection. S. aureus attaches to the highly proteinaceous surfaces of wounds and forms multi layered exopolysaccharide matrix surrounding the bacterial cells. It protects the bacteria from adverse environments by shielding them from various external and internal factors. This virulence factor delays the drug penetration, promotes antimicrobial resistance, impair wound healing & increases the chronicity of the wound.1
S. aureus biofilm production is chiefly regulated by intercellular adhesion operon (ica operon).2 Several ica independent factors also contribute to the manufacturing and maintenance of biofilm. These are biofilm-associated protein (Bap), clumping factor A and B (ClfA and ClfB), fibronectin binding proteins A and B (FnBpA and FnBpB), and etc.3
MRSA shows resistance to various antibiotics and it is inevitable in biofilm-producing strains. Some antibiotics induce biofilm in these MRSA isolates on exposure to sub MIC concentrations.4
Traumatic wounds smeared with MRSA biofilm require prolonged antibiotic therapy and repeated debridement, which increase the hospital stay and add over the cost to the patients. Significant progress in the development of remedies against these biofilm infection producing strains is still lacking. Therefore, this study was focused on ascertaining the phenotypic and genotypic characteristics of S. aureus biofilm from chronic traumatic wounds and their relation with antibiotic resistance.
Identification and Antimicrobial Susceptibility testing
A total of 67 strains of S. aureus isolated from the chronic traumatic wound infections were analysed for biofilm. Deep wound swabs (48), tissues (11), and pus samples (19) were collected for the analysis from the patients admitted to our tertiary care hospital from March 2020 through March 2021. Biological samples were cultivated on 5% sheep blood agar and MacConkey agar. S. aureus strains identified by an automated mass spectrometry microbial identification system that uses Matrix Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) technology. It examines the patterns of proteins detected directly from intact bacteria. The sample to be analyzed is mixed with another compound, called a matrix to generate charged molecules and to measure their mass-to-charge ratio. Such molecular signatures used for rapid bacterial identification (ID) from isolated colonies. Antibiotic sensitivity testing was carried out by Vitek®2 (BioMerieux, Inc, Durham, NC) for the following antibiotics: Cefoxitin, oxacillin, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, erythromycin, doxycycline, clindamycin, vancomycin, linezolid, teicoplanin.
Phenotypic detection of Biofilm
The Tissue culture plate (TCP) method was used for the phenotypic analysis of the biofilm of S. aureus strains.5
An enriched medium Trypticase soya broth contains pancreatic digest of casein and papaic digest of soyabean meal supplemented with additional 1% Glucose was used for detecting biofilm production in tissue culture plates. The bacterial suspension was added to this medium. After inoculation, the tissue culture plates were incubated at 37°C for 24hours. Each well of the TCP was washed with phosphate-buffered saline of pH 7.2. Microtiter plates with the attached bacteria were fixed by adding methanol into each well of the microtiter plate and kept at room temperature for 20 minutes. The biofilm layer formed by the bacteria in the microtiter plate was stained by using Hucker’s 2% crystal violet for 15 minutes at room temperature. The optical density of each stained well was measured at 570nm using a microtiter plate reader (Thermo Scientific, Zürich, Switzerland). Each assay was performed three times, and the results were averaged. S. aureus ATCC 25923 was used as positive control, and three wells with TSB + 1 % glucose alone were kept as a negative control. Interpretation of biofilm production was executed according to the criteria described by Stepanovic et al.5 The bacteria were categorised into non-producers, weak, moderate, or strong biofilm producers.
Detection of intracellular adhesion genes (ica)
S. aureus ica genes ica A, and ica C were detected by multiplex PCR. Qiagen (Germany) DNA extraction kit was used to extract S. aureus genomic DNA. The amplification cycle consisted of one cycle of denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 30 s, and elongation at 72°C for 1 min, with a final extension at 72°C for 5 min. The amplified products were visualized by electrophoresis in an agarose gel stained with ethidium bromide. The primers specific for the ica A, and ica C, are listed in Table 1.
Table (1):
Primer sequence of ica genes.
Gene Name |
Primer sequence |
Amplified Size |
|---|---|---|
ica A |
F- TCGTTGATCAAGATGCACCA R- CCAACATCGACAACTGCACT |
238 |
ica C |
F- TTATTTCGTCGTCCCACCCC R- AGTAAAACATTGGGCGAGGC |
199 |
Statistical analysis
Descriptive statistics, such as frequencies, and percentages were used. Comparisons between categorical variables were performed using the chi-square test. Univariate logistic regression analysis followed by multivariate analysis was performed to analyse the association between biofilm production and ica gene carriage. A p-value less than 0.05 was considered statistically significant.
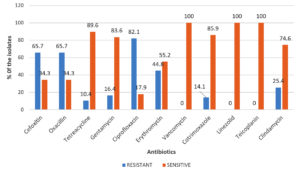
A total of 67 S. aureus strains were explored. Among them, MRSA was 65.7% (44/67). Out of 44 MRSA isolates, 15 (34.1%) isolates were found to be multidrug-resistant. Entire S. aureus isolates were sensitive to teicoplanin, vancomycin, and linezolid. Other than these drugs, they showed overall high susceptibility to doxycycline (89.6%) followed by cotrimoxazole (85.9%). The least susceptibility was seen for ciprofloxacin (17.9%), and it was also documented more in biofilm-producing isolates. (Fig. 1) (Table 3).
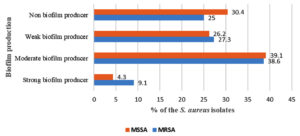
A total of 49 (73.1%) isolates produced the biofilm and 18 (26.9%) isolates were non-biofilm producers. Based on absorbance values at 570nm, 5 (7.5%) out of 67 strains were found to be strong biofilm producers, 26 (38.8%) were moderate, and 18 (26.8%) were weak biofilm producers.
Biofilm producing isolates were analysed for carriage of ica genes. The presence of the ica operon was noted in 85.7% (42/49) of S. aureus. The isolates harboured ica AC gene was 38.8% (19/49). Only ica A and ica C genes were present in 42.9% (21/49) and 4% (2/49) of the isolates, respectively. The comparison of the strength of biofilm formation by the isolates with ica genes divulged that only ica A was significantly associated with weak to strong biofilm production. In contrast, ica AC was associated with moderate to weak biofilm production. (Table 2)
Table (2):
Association between biofilm production and ica genes.
| Ica genes | Strength of biofilm production | ||
|---|---|---|---|
| Strong (5) | Moderate (26) | Weak (18) | |
| Ica A (%) | 4 (80) | 10 (38.5) | 7(38.9) |
| p=.00 | p=.004 | p=.007 | |
| Ica C (%) | – | 1 (3.8%) | 1 (5.6) |
| Ica AC (%) | – | 10 (38.5) | 9 (50) |
| p=.003 | p=.001 | ||
Among 67 isolates, 44 (65.7%) isolates were MRSA. Biofilm producing MRSA and MSSA were 75% (33/44) and 69.6% (16/23), respectively. A large number of MRSA (38.6%; 17/44) and MSSA (39.1%, 9/23) produced biofilm at moderate strength (Fig. 2).
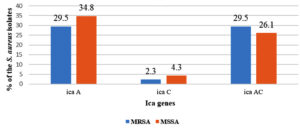
The majority of the MSSA (65.2%; 15/23) possessed biofilm forming ica genes compared to MRSA (61.4%, 27/44). Most of the MSSA was only ica A gene (34.8%; 8/23) positive, whereas MRSA isolates had both ica A and C (29.5%; 13/44) (Fig. 3).
Out of 44 MRSA isolates, 15 were MDR MRSA. Among 15 MDR MRSA strains, 10 (66.7%) were biofilm producers, and 5 (33.3%) were non-biofilm producers. All MDR MRSA strains significantly produced the biofilm with a p-value of .020.
Table (3):
Correlation of antibiotic resistance with the biofilm production.
No. |
Antimicrobials |
Biofilm producers N=44 |
Non-biofilm producers N=23 |
P-value |
|---|---|---|---|---|
1. |
Oxacillin |
33 (75 %) |
11 (47.8 %) |
0.634 |
2. |
Gentamicin |
8 (18.2 %) |
3 (13 %) |
0.973 |
3. |
Ciprofloxacin |
40 (90.9 %) |
15 (65.2 %) |
0.03 |
4. |
Erythromycin |
19 (43.2 %) |
10 (43.4 %) |
0.419 |
5. |
Doxycycline |
4 (9 %) |
2 (8.7 %) |
0.780 |
6. |
Clindamycin |
12 (27.3 %) |
5 (21.7 %) |
0.784 |
7. |
Co-trimoxazole |
9 (20.5 %) |
1 (4.3 %) |
0.02 |
Chronic wound infections are an important cause of a significant healthcare burden. Delayed wound healing increases hospitalisation due to prolonged treatment and leads to amputation. S. aureus is one of the important microorganisms isolated from these chronic wounds. They express an important virulence factor like biofilm that impairs the healing of chronic wounds. The frequency of biofilm production by S. aureus was 73.13%, and maximum isolates were moderate biofilm producers (38.8%). Only 7.5% were strong biofilm producers. Earlier high occurrence of biofilms was well documented. Malone et al. observed a prevalence of 78.2% biofilm in chronic wounds, while Asati et al. found in 68.9% of burn wounds examined.6,7 A study by Piechota et al. also noted moderate biofilm production (48%) by most S. aureus strains isolated from the wound.8 The ica ADBC locus mediates this biofilm synthesis of S. aureus strains. The presence of the ica operon was noted in 85.7% of S. aureus. Bimanand L et al. also indicated that ica is positively associated with biofilm production.9 In our study, few strong to moderate biofilm-producing isolates were found to be ica negative. The PIA-independent mechanism of biofilm formation must account for this. Strains with ica AC had moderate to weak biofilm, whereas strains possessed only ica A significantly strong biofilm producers. The presence ica locus was also significantly associated with the strength of biofilm mass formation, and there was no significant difference in the distribution of the ica genes in strong and weak strains. It is similar to the findings of others.10
The relationship between biofilm production and resistance to antibiotics was also assessed. The preponderance of MRSA ranges from 13 to 74.0% in different parts of the world.11 In this study, 75% of isolates were MRSA. It was found that resistance levels to antibiotics significantly for ciprofloxacin and co-trimoxazole were more for biofilm-producing S. aureus strains. A similar observation was noted by Bhattacharya et al. in their study also.12 Biofilm is one of the significant factors in the foreground for the emergence of multidrug resistance S. aureus. MRSA produced more biofilm compare to MSSA. Significantly MDR strains were frequent producers, which is in accordance with the observations obtained by Neopane et al.13 Earlier studies conducted by S Jain et al. and Neetu Peedikayil et al. showed a high prevalence of biofilm production among MRSA isolates.14,15 Ica genes were found more among MSSA compare to MRSA. This finding of our study is similar to others where authors mentioned PIA plays a significant role in biofilm development of MSSA rather than MRSA.16 In MRSA, it is mediated by accessory gene regulator (agr) and Staphylococcal accessory regulator (SAR). So ica independent mechanism is essential for MRSA biofilm-producing strains.
The current study revealed that the frequency of biofilm-producing MRSA strains in chronic traumatic wound infection is relatively high, and multidrug resistance was more with biofilm-producing strains. Phenotypically most of them produced biofilm at moderate strength. But possession of ica operon that encodes the synthesis of biofilm was more observed in MSSA. It highlights that further analysis is required on ica independent pathway for MRSA biofilm synthesis. Worrisome increases of multidrug resistance among biofilm-producing isolates also deserve future attention.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
BB and PS conceived and designed the experiments. PS and KTA conducted experiments, collected data and prepared first draft of the manuscript. BB and KTA revised the manuscript and structured the work order. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Kasturba Medical College and Kasturba Hospital, Manipal, Karnataka, India (IEC:189/2020).
AVAILABILITY OF DATA
All datasets generated or analyzed during the study are included in the manuscript.
- Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547-569.
Crossref - Tahaei SAS, Stajer A, Barrak I, Ostorhazi E, Szabo D, Gajdacs M. Correlation between Biofilm-Formation and the Antibiotic Resistant Phenotype in Staphylococcus aureus Isolates: A Laboratory-Based Study in Hungary and a Review of the Literature. Infect Drug Resist. 2021;14:1155-1168.
Crossref - Nguyen HTT, Nguyen TH, Otto M. The staphylococcal exopolysaccharide PIA – Biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol. 2020;18:3324-3334.
Crossref - Haddadin RNS, Saleh S, Al-Adham ISI, Buultjens TEJ, Collier PJ. The effect of subminimal inhibitory concentrations of antibiotics on virulence factors expressed by Staphylococcus aureus biofilms. J Appl Microbiol. 2010;108(4):1281-1291.
Crossref - Stepanovic S, Vukovic D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis. 2007;115(8):891-899.
Crossref - Malone M, Bjarnsholt T, McBain AJ, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care. 2017;26(1):20-25.
Crossref - Asati S, Chaudhary U. Prevalence of biofilm producing aerobic bacterial isolates in burn wound infections at a tertiary care hospital in northern India. Ann Burns Fire Disasters. 2017;30(1):39-42.
- Piechota M, Kot B, Frankowska-Maciejewska A, Gruzewska A, Wozniak-Kosek A. Biofilm Formation by Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus Strains from Hospitalized Patients in Poland. Biomed Res Int. 2018;2018:4657396.
Crossref - Bimanand L, Taherikalani M, Jalilian FA, et al. Association between biofilm production, adhesion genes and drugs resistance in different SCCmec types of methicillin resistant Staphylococcus aureus strains isolated from several major hospitals of Iran. Iran J Basic Med Sci. 2018;21(4):400-403.
Crossref - Azmi K, Qrei W, Abdeen Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genomics. 2019;20(1):578.
Crossref - Prakash PH, Rajan V, Gopal S. Predominance of SCCmec types IV and V among biofilm producing device-associated Staphylococcus aureus strains isolated from tertiary care hospitals in Mysuru, India. Enferm Infecc Microbiol Clin. 2017;35(4):229-235.
Crossref - Bhattacharya S, Bir R, Majumdar T. Evaluation of multidrug resistant Staphylococcus aureus and their association with biofilm production in a Tertiary Care Hospital, Tripura, Northeast India. J Clin Diagn Microbiol. 2015;9:DC01-DC04.
Crossref - Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25-32.
Crossref - Jain S, Chowdhury R, Datta M, Chowdhury G, Mukhopadhyay AK. Characterization of the clonal profile of methicillin resistant Staphylococcus aureus isolated from patients with early post-operative orthopedic implant based infections. Ann Clin Microbiol Antimicrob. 2019;18(1):8.
Crossref - Neetu TJ, Murugan S. Genotyping of Methicillin Resistant Staphylococcus aureus from Tertiary Care Hospitals in Coimbatore, South India. J Glob Infect Dis. 2016;8(2):68-74.
Crossref - Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesion in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.