ISSN: 0973-7510
E-ISSN: 2581-690X
Streptomyces sp. VITGV38 (MCC8469) was obtained from the VIT University Microbiology Laboratory where it was isolated from tomato plants. This strain was mass-cultured for 15 days and its extracellular metabolites were extracted in ethyl acetate using a liquid–liquid extraction method. The antibacterial test was performed on the ethyl acetate crude extract against selected urinary tract pathogens, Proteus mirabilis (MTCC-442), Enterococcus faecalis (MTCC-439), Klebsiella pneumoniae (MTCC-109), and Escherichia coli (MTCC-1687), The extract developed a clear inhibition zone that measured between 17–21 mm. The minimum inhibition concentration was observed from a concentration of 25 μg/ml against all selected uropathogens. GC-MS analysis revealed 35 diverse compounds in the ethyl acetate crude extract, which includes 1,2 benzenedicarboxylic acid bis(2methylpropyl) ester, dibutyl phthalate, bis (2-ethylhexyl) phthalate, didodecyl phthalate, octadecanoic acid dodecyl ester, and dodecane. These six compounds are the major antimicrobial compounds present in the ethyl acetate extract. VITGV38 showed greyish aerial mycelium. Scanning electron microscopy revealed elliptical spores with a chain-like smooth orientation.
Antibacterial Activity, Streptomyces sp. VITGV38 (MCC4869), Gram-negative Bacteria, Minimum Inhibition Concentration, Uropathogens
Microbes occupy the entire human body. Some organisms are harmful and may cause diseases (e.g. Escherichia coli). These pathogens may also be referred to as germs or infectious agents. Every living organism on Earth is susceptible to illnesses. There are approximately 1013 human cells in the human body, but approximately 1014 microbes are considered to be part of the normal flora of the body.1 These normal microbes can change if the immunity of an individual is low.2 The human body is rich in nutrients and has warm and uniform temperatures, making it the best host for pathogens to live comfortably.
Among all body parts, the urinary system is the most prone to infection. UTIs are primarily caused by gram-negative bacteria; however, gram-positive bacteria have also begun to become causative agents. Every year, 150 million people worldwide suffer from UTIs, resulting in global economic losses of more than 1.6 billion USD.3 Approximately 10.5 million people are affected in India.
A wide range of pathogenic bacteria and fungi causes this UTI. Urinary system infections can affect any part of the kidney, bladder or urethra. A variety of symptoms are associated with bladder infection, including pelvic pain, an increased urge to urinate, difficulty in urinating, and even blood in the urine.4
The spectrum of predominant pathogens UTIs is quite diverse and includes well-known causative agents, such as Proteus mirabilis, Enterococcus faecalis, Staphylococcus saprophyticus, Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, various Candida species, and ubiquitous E. coli.5 Repeated occurrences of UTIs have the potential to inflict lasting harm on the kidneys, culminating in irreversible damage, renal hypertension, and ultimately, renal failure, particularly in severe cases.6 In the realm of treatment, a variety of antibiotics are commonly employed to combat straightforward UTI infections. No tablet among these are nitrofurantoins, which are marketed under various names such as Furadantin, Macrobid, and Macrodantin. Interestingly, this antibiotic is often used as a first-line treatment option; however, its efficacy is limited to certain strains of pathogens that develop resistance.7 Augmentin, a combination of amoxicillin and clavulanate, is an essential therapeutic option. Furthermore, specific cephalosporins carve their niches for managing UTIs. These choices of antibiotics reflect the ongoing evolution of treatment strategies aimed at addressing the dynamic landscape of microbial resistance.8
An additional class of antibiotics, fluoroquinolones, is reserved for more severe infections and circumstances in which alternatives have been proven to be inadequate. However, the FDA has issued strong safety recommendations advocating their use only in cases of genuine clinical necessity, owing to concerns about potential adverse effects and the development of antibiotic resistance.9
Therefore, it is imperative to emphasize the importance of judicious antibiotic use. The alarming rise in antibiotic resistance underscores the urgent need for responsible prescription practices and the development of alternative treatment modalities to counter the escalating challenge of antimicrobial resistance in UTI.10 The irrational use of antibiotics causes pathogens to become resistant to these antibiotics (e.g., Ofloxacin and norfloxacin), and everyone in the world is dealing with this issue. Recurrent infections can lead to antimicrobial resistance development. Bacterial resistance to various antimicrobials has increased dramatically, leaving doctors with limited options.
In the pursuit of novel therapeutic agents, the imperative to explore uncharted territories in the biological realm has never been more pressing. Over the last five decades, scientists have witnessed the discovery of an impressive array of approximately 30,000 natural drugs. Yet, there is an alarming trend: the pace of unearthing novel compounds has notably dwindled in recent years, necessitating a paradigm shift in our approach to drug discovery.11
Microorganisms with remarkable diversity and adaptability are promising sources of untapped bioactive compounds with immense therapeutic potential. These microorganisms, when cultivated under controlled conditions, can yield a valuable trove of bioactive molecules that can target various diseases and ailments.12
Among these microorganisms, Streptomyces is an invaluable contributor to the search for novel bioactive compounds. Owing to their remarkable capacity to produce an astonishing array of secondary metabolites, Streptomyces species are potential drug candidates. These metabolites, which often exhibit intricate structures and multifaceted biological activities, are promising candidates for treating diverse medical conditions.13
Recent advances in genetic and biotechnological techniques have enabled researchers to delve deeper into the biosynthetic potential of Streptomyces and other microorganisms. By harnessing the power of genetic manipulation and metabolic engineering, scientists can unlock pathways that lead to the production of entirely novel compounds or the enhancement of existing ones.14 As the challenges of antibiotic resistance and the quest for innovative treatments intensify, it becomes increasingly evident that the exploration of natural sources for novel drug candidates is far from exhausted. Vigorous research combined with cutting-edge technologies has the potential to unveil a wealth of bioactive molecules that can revolutionize the pharmaceutical landscape and address pressing global health concerns. The objective of this research was to investigate the antibiotic-producing potential of a newly discovered species, Streptomyces sp. VITGV38, against specific uropathogens.
Streptomyces sp. VITGV38
A pure culture of Streptomyces strain VITGV38 was obtained from the Microbiology Lab, SBST, VIT University, Vellore (stored at -20°C). To reactivate the strain, a series of streaks was performed on fresh International Streptomyces Project 2 (ISP2) agar.
Morphological characteristics
The aerial mass, color, and substrate mycelia were recorded and categorized as morphological parameters. For scanning electron microscopic studies, Streptomyces sp. VITGV38 cells were fixed on a 14 mm × 14 mm agar block using a standard protocol.15 Scanning electron microscopy was then used to analyze the samples (EVO-50 CarlZeiss).
Molecular characterization
The 16S rRNA was analyzed using both forward (5′-GAGTTTGATCCTGGCTCA-3′) and reverse (5′-ACGGCTACCTTGTTACGACTT-3′) primers. The resulting PCR products were then amplified and sequenced. Nucleotide sequences were compared using BLAST to find matches. A phylogenetic tree was constructed using the neighbor-joining method, and the sequences were deposited in GenBank.
Primary screening
Streptomyces sp. VITGV38 was initially screened against uropathogens by cross-streaking ISP2 agar plates.16 Double culture methods were used to screen the isolated Streptomyces strains for antagonistic properties against the selected uropathogens. Inhibition zones (mm) were recorded. Lawn cultures were performed using sterile cotton swabs and allowed to remain for one minute. All experiments were performed in duplicates.
Secondary metabolite production and extraction
Streptomyces sp. VITGV38 cells were mass-cultured for 21 days in an ISP2 agar medium. Ethyl acetate (1:1) was used as the solvent to extract the secondary metabolites. The solvent and filtrate were vigorously shaken for 20 mins. The ethyl acetate phase was separated from the aqueous phase by using separation funnels.17 This was done twice. Concentrating ethyl acetate was accomplished using a rotary evaporator operating at 50°C, and the extract was dried completely without any solvent.
Screening against uropathogens
A sterile steel cork borer was used to create wells on the MHA agar plates measuring approximately 6 mm in diameter. The air-dried crude extracts of Streptomyces sp. VITGV38 was evaluated for its antibacterial effects in comparison to those of tetracycline (positive control) and 5% DMSO (negative control). Four different concentrations (10, 25, 50, and 100 µg/ml) of the extract were tested against four distinct uropathogens; P. mirabilis, E. faecalis, K. pneumonia, and E. coli. After incubating the plates for 24 hrs at 37°C, the diameter of the inhibition zone was measured to calculate the activity index. The measurements were performed in three distinct fixed directions, and the average values were noted according to the methodology of Zorkey et al.18
GC-MS analysis
The chemical components of the crude ethyl acetate extract were identified using an Agilent GC-MS device consisting of a GC 5890 series II and MSD 5972. The MS was connected to a fused silica HP-5 capillary column with dimensions of 30 m × 0.25 mm ID and 0.25 m film thickness.19 Helium was used as a carrier gas at a flow rate of 1.2 ml per minute. The oven temperature was programmed to start at 50°C for 1 min and then ramped up to a range of 50 to 280°C at a rate of 5°C/min, remaining isothermal for 20 min.12 The injector port was set at 250°C, and the detector was set at 280°C. Mass spectrometry was used to analyze the peaks, and the spectra were examined using library data from the NIST-MS search (version 2.0), including the NIST’02 mass-spectral library, Agilent p/n G1033 A.20
Morphological characteristics
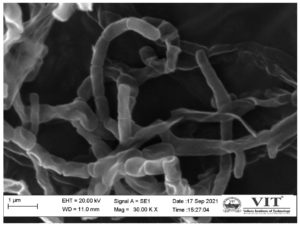
Streptomyces sp. VITGV38 was found to be a potential strain. This strain produces white-greyish aerial and substrate mycelia (Figure 1) on ISP2 agar. However, after ten days, they produced ethyl acetate-soluble pigments ranging from pink to maroon red. They had a chain-like spore morphology (straight and curved chains) and smooth spore ornamentation. There were 10–25 spores per mycelial segment. The SEM morphology (Figure 2) was similar to that of many other Streptomyces spp.
Figure 2. Spore chain morphology of Streptomyces sp. VITGV38 observed under SEM. Molecular characterization of VITGV38
Molecular characterization of VITGV38
To identify the species similarity of strain VITGV38, its 16S rRNA sequence consisting of 1040 nucleotides was used and compared with other strains. This analysis revealed a high degree of similarity between VITGV38 and the other members of the Streptomyces genus. The phylogenetic tree of strain VITGV38 formed a branch distinct from that of a neighboring Streptomyces strain (Figure 3). The 16S rRNA sequence of VITGV38 shared 99% similarity with Streptomyces ardesiacus, its nearest neighbor. A phylogenetic tree was constructed using sequences aligned with the System Software aligner, and the results confirmed that strain VITGV38 clustered within the genus Streptomyces, exhibiting a close phylogenetic relationship and high sequence similarity to Streptomyces ardesiacus NRRL B-1773 (99%), Streptomyces spoangioformans (99%), Streptomyces xiangluensis (98%), and Streptomyces tricolor NBRC 15461 (98%). This study reports the first identification of this particular Streptomyces species using GenBank (accession number MT792902).
Figure 3. Phylogenetic analysis of the 16s ribosomal sequence of Streptomyces sp. The tree was constructed using BLAST. Streptomyces sp. VITVGV38 is highlighted
Primary Screening
A dual-culture assay was performed using Streptomyces sp. The VITGV38 strain showed favorable antibacterial activity against the gram-positive bacteria P. mirabilis (MTCC 442) and E. coli (MTCC 1687) (Figure 4).
Figure 4. Primary screening of Streptomyces sp. VITGV38 for its antibacterial activity (cross-streak method)
Secondary Screening
The selected strain (VITGV38) was grown in liquid broth, and metabolites were extracted in ethyl acetate. Using the well-diffusion method, the crude extract was evaluated for its antibacterial activity against the same four test microorganisms. Significant antibacterial activities were observed against P. mirabilis, E. faecalis, E. coli, and K. pneumoniae (Figure 5). Table 1 shows the zones of inhibition observed in antibacterial assays. According to the findings, the crude extract (100 µg/ml) displayed a higher inhibition zone against K. pneumoniae (21 mm) and E. coli (19 mm). In the positive control experiment with tetracycline, the maximum zone size was 24 mm for E. coli. and 20 mm for K. pneumoniae.
Table (1):
Secondary screening of crude extract from Streptomyces sp. VITGV38 against E. coli, K. pneumoniae, P. mirabilis and E. faecalis with its zone of inhibition (mm)
| Streptomyces sp. VITGV38 ethyl acetate extract (µg/ml) | Zone of Inhibition (mm) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae | E. coli | P. mirabilis | E. faecalis | |||||||||||||
| 25 | 50 | 75 | 100 | 25 | 50 | 75 | 100 | 25 | 50 | 75 | 100 | 25 | 50 | 75 | 100 | |
| Extract dissolved in 5% DMSO | 17 | 17 | 19 | 21 | 11 | 14 | 15 | 19 | 10 | 14 | 15 | 17 | 11 | 13 | 15 | 17 |
| 5% DMSO | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Tetracycline | 19 | 20 | 20 | 24 | 18 | 18 | 19 | 20 | 13 | 18 | 19 | 19 | 16 | 17 | 20 | 21 |
Figure 5. Secondary screening of the culture extract from Streptomyces sp. VITGV38 against E.coli, K. pneumoniae, S. aureus, and B. subtilis at different concentrations (25, 50, 75 and 100 µg) by the well-diffusion method
Antimicrobial metabolites in GC–MS
GC-MS was used to identify the metabolites in the crude extract. The GC-MS chromatogram displayed 35 peaks (Figure 6), which were compared with the NIST database. Table S1 lists the antimicrobial compounds present in 35 peaks and their retention times, mol. Wt., mol. formula, and percentage area. Table S1 provides a list of the compounds corresponding to all 35 peaks. Six compounds were found to occupy a significant area in the chromatogram. These compounds had a previous record of antibacterial activity according to the NCBI database. The six compounds tested were 1,2 benzenedicarboxylic acid bis(2methylpropyl) ester, dibutyl phthalate, bis (2-ethylhexyl) phthalate, didodecyl phthalate, octadecanoic acid dodecyl ester, and dodecane. All basic information from the chromatogram, such as molecular weights, % area, structure, and spectrum of activity, are shown in Table 2. The mass spectrum is shown in the Figure S1.
Table (2):
Antimicrobial chemicals identified in the crude extract of Streptomyces sp. VITGV38 by GC-MS using the NIST database
No. |
Chemical Compound |
RT |
Mol. weight |
Mol. Formula |
Area % |
Compound |
Activity |
Spectrum of action & Ref. |
|---|---|---|---|---|---|---|---|---|
1 |
1 2 benzenedi – carboxylic acid bis(2methylpropyl) ester |
18.458 |
278.3 |
C16H22O4 |
11.6 |
Carboxylic acid |
Antimicrobial |
Broad |
2 |
Dibutyl phthalate |
19.004 |
278.0 |
C6H4(COOC4H9)2 |
18.85 |
Methylene |
Antimicrobial |
Broad |
3 |
Bis(2-ethylhexyl) phthalate |
24.138 |
278.5 |
C17H30OSi |
13.28 |
Phenolic |
Antimicrobial |
Narrow |
4 |
Didodecyl phthalate |
18.651 |
256.4 |
C16H32O2 |
12.92 |
carboxylic |
Antibacterial |
Narrow |
5 |
Octadecanoic acid, dodecyl ester |
25.396 |
452.8 |
C30H60O2 |
5.27 |
Ester |
Antimicrobial |
Broad |
6 |
Dodecane |
23.643 |
150.2 |
C9H10O2 |
2.80 |
Phenolic |
Antimicrobial |
Broad |
Streptomyces have been explored for their potential to produce antimicrobial compounds that are effective against pathogens infecting the urinary tract. Several studies have investigated the antimicrobial activity of Streptomyces-derived secondary metabolites against pathogens commonly associated with UTIs. UTIs are often caused by bacteria, such as Escherichia coli, Enterococcus spp., Proteus mirabilis, and Staphylococcus saprophyticus. Streptomyces bacteria produce a wide range of secondary metabolites, including antibiotics and other bioactive compounds. Some of these compounds exert inhibitory effects against UTI-causing pathogens. Researchers have isolated and tested various Streptomyces strains to identify potential antimicrobial agents for the treatment of UTIs. In this study, the novel isolate, Streptomyces sp. VITGV38 (MCC 4869) was used to treat infections caused by infectious pathogens. Spore morphology is another essential characteristic of the life cycle and is closely associated with survival and reproductive strategies. The spore mother cells undergo several rounds of division to form a chain of smaller spore cells. These cells are packed with reserves that help them survive harsh conditions. Spore cells mature and become more resistant to environmental stresses. They often develop pigmentation, resulting in unique colors. The spore chain of VITGV38 matches that of VMS-A10, as reported by Shaik et al.21
16S rRNA sequencing is a powerful and widely used molecular technique for identifying and classifying bacterial species, including Streptomyces. The 16S rRNA sequence of the Streptomyces isolates did not match any existing sequences, which may indicate the discovery of a novel species. This finding contributes to our understanding of bacterial diversity. A similar conformation for Streptomyces was reported by Duddu et al.22
The antagonistic activity of Streptomyces is of immense importance in various fields, including medicine, agriculture, and environmental management. However, it is worth noting that not all Streptomyces strains exhibit strong antagonistic properties, and the specific mechanisms and compounds involved can vary widely among different strains. Primary antimicrobial screening was reported against P. mirabilis, B. subtilis, E. aerogenes, P. aeruginosa, and E. coli using Streptomyces spp. ES2.23 The antagonistic activity of Streptomyces refers to its ability to inhibit the growth of microorganisms, particularly pathogenic or harmful bacteria.
This current result coincides with the antibacterial activity of Streptomyces sp. NLKPB45 against E. coli.24 A similar antimicrobial study was reported against E. coli (19.0 ± 0.6 mm) and K. pneumoniae (12.0 ± 0 mm) using the isolate Streptomyces spp. RVE002.25 Analysis of the GC-MS results showed that the NIST database contained six antimicrobial compounds from the extract. Six compounds were identified in the extracts of Streptomyces sp. VITGV38. The compounds with a higher retention time and % area in the crude extract were 1,2 benzenedicarboxylic acid bis(2methylpropyl) ester (11.6), dibutyl phthalate (18.85%), bis (2-ethylhexyl) phthalate (13.28%), didodecyl phthalate (12.92), octadecanoic acid dodecyl ester (5.27), and dodecane (2.80). Their molecular structures are shown in Figure 6. A complete list of compounds, area (%), retention time (%), molecular weight, and formulas is compiled in Supplementary Table 1. The major compound, dibutyl phthalate (area, 18.85%; molecular structure, Figure 6A), produced by Streptomyces albidoflavus 321.2, showed strong activity against Gram-positive and Gram-negative bacteria, as well as unicellular and filamentous fungi.26 Furthermore, its mechanism of inhibitory action against human GSK-3β (Hs GSK-3β) and Plasmodium falciparum 3D7 (Pf 3D7) malaria parasites was displayed by dibutyl phthalate generated by Streptomyces sp. H11809.27 The second major compound was bis (2-ethylhexyl) phthalate (13.28%; molecular structure, Figure 6B), which showed antimicrobial and cytotoxic activity against the Nile-derived fungus Aspergillus awamori.28 The third major bioactive compound was n-didodecyl phthalate (12.92; molecular structure, Figure 6C), which showed antibacterial activity against the essential oils from Launaea resedifolia L.29 The third major bioactive compound is 1,2 benzenedicarboxylic acid bis(2methylpropyl) ester (area 11.6%; Figure 6D), which previously revealed antibacterial molecular activities in poplar wood extractives.30 Another antimicrobial compound, octadecanoic acid dodecyl ester (area 5.27%; Figure 6E), has been reported to exhibit the antibacterial activity of medicinal plants against biofilm-forming methicillin-resistant Staphylococcus aureus.31 Octadecanoic acid acts as a bactericide against both bacterial strains by damaging bacterial cell wall integrity, permeating cells, and then interacting with DNA, as well as disturbing the activity of the respiratory electron transport chain to induce high-level toxic ROS (hydroxyl radicals) generation and the upregulation of ROS genes.32 The sixth major compound, dodecane (area 2.80%; Figure 6F), broad-spectrum antimicrobial and antioxidant activities against the flower essential oil of Halimodendron halodendron.33 Dodecane inhibits minor concentrations and limits the functionality of enzymes participating in cytokine interactions or less significant cellular activities, whereas, at higher concentrations, it inhibits ATPase.34
The ethyl acetate extracts obtained from Streptomyces sp. VITGV38 exhibited significant antibacterial activity against the selected uropathogens, indicating its potential to produce novel antibiotic compounds. Further studies are required to separate the individual compounds based on their specific activities.
Additional file: Additional Table S1- S3. Additional Figure S1.
ACKNOWLEDGMENTS
The authors acknowledge the technical support rendered by Mr. P. Veilumuthu.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was funded by the VIT-SEED grant no. SG20220080 from the Vellore Institute of Technology, India.
DATA AVAILABILITY
The 16s rRNA sequence of VITGV38 was submitted to GenBank (accession no: MT792902). The draft genome of Streptomyces sp. VITGV38 was submitted to the NCBI SRA portal under the BioProject accession number PRJNA750621, BioSample accession number SAMN20475137, and SRA accession number SRR15293244.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Davis CP. Normal Flora. In: Baron S, Ed. Medical Microbiology. 4th Edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996.
- Charles A Janeway J, Travers P, Walport M, Shlomchik MJ. Immunobiol Immunobiol. 2001;(14102):1-10.
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269.
Crossref - Scionti A, Rossi P, Gulino P, Semeraro A, Defilippi C, Tonerini M. Acute Pyelonephritis. Imaging Non-Traumatic Abdominal Emergencies in Pediatric Patients. 2021:255-268.
Crossref - Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7(12):653-660.
Crossref - Aggarwal N, Lotfollahzadeh S. Recurrent Urinary Tract Infections. StatPearls;2022.
- Vogel T, Verreault R, Gourdeau M, Morin M, Grenier-Gosselin L, Rochette L. Optimal duration of antibiotic therapy for uncomplicated urinary tract infection in older women: a double-blind randomized controlled trial. CMAJ. 2004;170(4):469-473.
- Gupta K, Hooton TM, Naber KG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52(5):e103-e120.
Crossref - Shah HA, Syed A, Bhat MA, et al. Genomic Determinants and Antimicrobial Resistance Pattern of Clinical Isolates of Extended Spectrum Beta Lactamase (ESBL) Producing Escherichia coli. J Pure Appl Microbiol. 2023;17(3):1679-1690.
Crossref - Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318-327.
Crossref - Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770-803.
Crossref - Berdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. The Journal of Antibiotics. 2012;65(8):385-395.
Crossref - Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1-26.
Crossref - Velasquez JE, Van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15(1):11-21.
Crossref - Nandhakumar P, Vasantha VS, Veilumuthu P, Christopher G. 7, 8- Dihydroxyflavone, An Effective Natural Product Reduce Ralstonia solanacearum Populations and Control Tomato Bacterial Wilt. Indian J Agric Res. 2020;54(6):731-737.
Crossref - Akshatha SJ, Ishwara KM. Evaluation of Mangrove Soil Streptomyces spp. exhibiting Culture and Biochemical Variation for Determination of Antibacterial Activity. J Pure Appl Microbiol. 2022;16(4):2458-2476.
Crossref - Ahmed AA. Production of Antimicrobial Agent by Streptomyces violachromogenes. Saudi J Biol Sci. 2007;14(1):7-16.
- Al-Zoreky NS. Antimicrobial activity of pomegranate (Punica granatum L.) fruit peels. Int J Food Microbiol. 2009;134(3):244-248.
Crossref - Sanjotha G, Shivasharana CT, Shettar AK, Manawadi S, Devendra BN. GC-MS Analysis and In-vitro Apoptosis Induction and Anticancer Activity of Methanol Extract of Aspergillus terreus against Lung Cancer. J Pure Appl Microbiol. 2022;16(4):2934-2948.
Crossref - P V, J GC. Characterization of Secondary Metabolites Derived from Tomato Endophyte – Streptomyces sp. Shanivit. Curr Trends Biotechnol Pharm. 2022;16(Supplement 1):141-152.
Crossref - Shaik M, Girija Sankar G, Iswarya M, Rajitha P. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. J Genet Eng Biotechnol. 2017;15(1):87-94.
Crossref - Duddu MK, Guntuku G. Isolation, screening and characterization of antibiotic producing actinomycetes from kapuluppada plastic waste dumping yard, visakhapatnam. Int J Pharm Pharm Sci. 2016;8(11):221-229.
Crossref - Al-Ansari M, Alkubaisi N, Vijayaragavan P, Murugan K. Antimicrobial potential of Streptomyces sp. to the Gram positive and Gram negative pathogens. J Infect Public Health. 2019;12(6):861-866.
Crossref - Kalyani BS, Krishna PS, Sreenivasulu K. Screening and identification of novel isolate Streptomyces sp., NLKPB45 from Nellore costal region for its biomedical applications. Saudi J Biol Sci. 2019;26(7):1655-1660.
Crossref - Elias F, Muddada S, Muleta D, Tefera B. Antimicrobial Potential of Streptomyces spp. Isolated from the Rift Valley Regions of Ethiopia. Adv Pharmacol Pharm Sci. 2022;2022.
Crossref - Roy RN, Laskar S, Sen SK. Dibutyl phthalate, the bioactive compound produced by Streptomyces albidoflavus 321.2. Microbiol Res. 2006;161(2):121-126.
Crossref - Mahmud F, Lai NS, How SE, et al. Bioactivities and Mode of Actions of Dibutyl Phthalates and Nocardamine from Streptomyces sp. H11809. Molecules. 2022;27(7):2292.
Crossref - Lotfy MM, Hassan HM, Hetta MH, El-Gendy AO, Mohammed R. Di-(2-ethylhexyl) Phthalate, a major bioactive metabolite with antimicrobial and cytotoxic activity isolated from River Nile derived fungus Aspergillus awamori. Beni Suef Univ J Basic Appl Sci. 2018;7(3):263-269.
Crossref - Zellagui A, Gherraf N, Ladjel S, Hameurlaine S. Chemical composition and antibacterial activity of the essential oils from Launaea resedifolia L. Org Med Chem Lett. 2012;2(1).
Crossref - Peng W, Li D, Zhang M, et al. Characteristics of antibacterial molecular activities in poplar wood extractives. Saudi J Biol Sci. 2017;24(2):399-404.
Crossref - Manilal A, Sabu KR, Shewangizaw M, et al. In vitro antibacterial activity of medicinal plants against biofilm-forming methicillin-resistant Staphylococcus aureus: efficacy of Moringa stenopetala and Rosmarinus officinalis extracts. Heliyon. 2020;6(1):e03303.
Crossref - Shi Y-g, Zhang R-r, Zhu C-m, et al. Antimicrobial mechanism of alkyl gallates against Escherichia coli and Staphylococcus aureus and its combined effect with electrospun nanofibers on Chinese Taihu Icefish preservation. Food Chem. 2021;346:128949.
Crossref - Wang J, Liu H, Gao H, et al. Antimicrobial and Antioxidant Activities of the Flower Essential Oil of Halimodendron halodendron. Natural Product Communication. 2011;6(11):1749-1753.
Crossref - Nazzaro F, Fratianni F, De Martino L, Coppola R, De Feo V. Effect of Essential Oils on Pathogenic Bacteria. Pharmaceuticals. 2013;6(12):1451-1474.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.