ISSN: 0973-7510
E-ISSN: 2581-690X
The risk of zoonosis transmission when handling livestock or animal products is substantial, ‘One Health’ interventions should be an effective strategy for the control of many zoonotic bacteria. In this study, 26 fresh fecal samples from 2 clinically healthy goats were collected at different day ages to survey goat-borne zoonotic bacterial infection, and 19 fresh fecal samples from diarrhetic goats were tested to evaluate the possible role of zoonotic pathogens in goat diarrhea. Following all samples were analyzed by Metagenomic Sequencing, a total of 20 kinds of zoonotic bacteria were screened from healthy goats, and 11 (55%) of them were infection mainly during the preweaned period. Of the 19 fresh fecal samples from diarrhetic goats, all were confirmed to be zoonotic bacterial infection positive (range from 11 to 12 species). After comparison with healthy samples of the same or similar day-age goats, it was found that Lactococcus garvieae, Helicobacter pylori, Klebsiella pneumoniae, Shigella sonnei, Shigella boydii, Campylobacter coli, Salmonella enterica, Acinetobacter baumannii, Shigella flexneri, Shigella dysenteriae and Clostridium perfringens and Campylobacter fetus were highly increased incases in some diarrheic cases, while the remains had no significant change. The results suggest that goats may act as a reservoir for many zoonotic bacteria, and some of them may be associated with goat intestinal inflammation.
Goat, Zoonotic, Bacterium, Infection, Reservoir
Zoonoses, defined by WHO and FAO, refer to “those diseases and infections which are naturally transmitted between vertebrate animals and man”.1 Many organisms can cause zoonotic disease, as long as it can pass from other animals to humans. The numbers of microbial species that can infect human beings are shown to be 1415, of which 868 species (61%) are zoonotic.2 The transmission may occur directly or indirectly by means of vectors. The severity of zoonotic diseases in humans varies from mild symptoms to life-threatening conditions.
Zoonoses have a major societal impact on global health security, and timely diagnosis and appropriate management of clinical cases are often challenging. Preventive ‘One Health’ interventions should be an effective strategy to overcome some of these challenges.3 Both veterinary medicine and public health have roles to play in the surveillance, prevention, and control of this on-going issue.
Meat from goats is very popular in many populations and areas. The exposure in goat raising, slaughtering, and raw meat processing, as well as consumption of meat products has a zoonotic risk. Regarding sheep and goats, the main pathogens are: Brucella melitensis, Campylobacter spp., Listeria spp., Salmonella spp., Yersinia spp., Shiga-toxin producing Escherichia coli, Staphylococcus aureus, tick-borne encephalitis virus, and Toxoplasma gondii.4,5
It is important to have zoonotic bacteria epidemiological data to evaluate the impact on public health and animal diseases, so this work investigated the pathogens in goat intestine and analyzed the possible role of zoonotic pathogens in goat diarrhea.
Sample collection
The samples for use in this study were allowed to be collected in a goat farm (the number of animals raised was around 1,800) in Jiangsu Province, China. House-feeding mode was used in the farm. All lambs were treated with Tutreuli 1 ml by oral within 24 hours of birth, and vaccinated against infectious pleural pneumonia, clostridial disease, peste des petits ruminants, foot-and-mouth disease and goat pox at the age of 15, 20, 30, 40 and 50 days old respectively. Supplementary feeding was started at 15 days old, and weaning was performed at 44 to 45 days old followed by feeding total mixed rations.
To understand the impact of zoonotic pathogens in the intestine of goats, two goats (A and B) were randomly selected from the farm and fresh fecal samples were collected at the age of 1, 3, 5, 10, 20, 30, 40, 45, 50, 60, 70, 80 and 90 days old respectively. To analyze the possible role of zoonotic pathogens in goat diarrhea, fresh fecal samples were collected from nineteen diarrheic goats from the same farm (Table 1). To minimize environmental microbial contamination, approximately 2~3 grams fecal specimen (new excreted and no touching the ground) of each goat was collected by using an alcohol-sterilized scoop and placed in a 5 ml sterile EP tube. Following the EP tubes were stored in the biosafety transport box (UN3373), all samples were sent to Suzhou Genomics Biotech Co., Ltd. (China) for metagenomics sequencing and bioinformatics analysis. The sample collection in the farm occurred under the direction of the farm’s usual vet as part of their routine veterinary herd health programme from August to October, 2021.
Table (1):
Number and days old of diarrheal goats (%).
Goat number |
C1 |
C2 |
C3 |
C4 |
C5 |
C6 |
C7 |
C8 |
C9 |
C10 |
C11 |
C12 |
C13 |
C14 |
C15 |
C16 |
C17 |
C18 |
C19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Day of age (d) |
19 |
20 |
25 |
25 |
30 |
30 |
35 |
40 |
45 |
60 |
75 |
90 |
90 |
90 |
105 |
105 |
105 |
105 |
210 |
DNA extraction
To investigate the microbial species contained in the subcutaneous abscess, genomic DNA from each sample was extracted by using HiPure Stool DNA Kit (Shanghai Magen Biotech Co., Ltd, China). The extracted DNA concentration was tested by using Thermo NanoDrop 2000 Spectrophotometer (America), and the ratio of OD260/280 as well as that of OD260/230 was used to determine the purity of DNA.
Library Preparation and Metagenomic Sequencing
Following 200 μg genomic DNA was randomly fragmented by Covaris S220 Focused ultrasonicator (America) to an average size of 300-350bp, the sheared fragments were treated with End Prep Enzyme Mix (Beijing Biolab Technology Co., Ltd, China) for end repairing, 5’ phosphorylation and 3’ adenylation, to add adaptors to both ends. Size selection of Adaptor-ligated DNA was then performed by DNA Cleanup beads. Each sample was then amplified by PCR for 8 cycles using P5 and P7 primers, with both primers carrying sequences which can anneal to Flowcell to perform bridge PCR and P7 primer carrying a six-base index allowing for multiplexing. The PCR products were cleaned up and validated using an Agilent 2100 Bioanalyzer. The qualified libraries were sequenced pair end PE150 on the illumina HiseqXten/Novaseq/MGI2000 System.
Data Analysis
Raw shotgun sequencing reads were trimmed using Cutadapt (v1.9.1). Low-quality reads, N-rich reads and adapter-polluted reads were removed. Then host contamination reads were removed. Samples were each assembled de novo to obtain separate assemblies. Whole genome de novo assemblies were performed using Megahit (v1.1.3) with different k-mer. The best assembly result of Scaffold, which has the largest N50, was selected for the subsequent analysis.
Prodigal (v3.02) software was to predict coding genes and then integrated the gene sequences of all samples. Use the sequence clustering software MMseq2 for further de-redundancy processing. By default, identity 95% and coverage 95% were used for clustering. Using the alignment software SoapAligner (v2.21), we aligned the clean reads to construct nonredundant gene sets and obtained the number of reads aligned by unigene in each sample. Then, based on the number of reads and gene length in each unigene alignment, the abundance information of unigene in each sample was calculated.
Diamond (v0.8.15.77) was used to search the protein sequences of the unigenes with the NR database, CAZy database, eggNOG database, CARD database, and KEGG database with
E<10-5. The matched result with the best score was selected for annotation. To explore the microbial composition of the samples, we used Diamond to align the unigene sequences with the NR database and obtained the species annotation results of each sequence through the taxonomic annotation information corresponding to each sequence in the NR database. The abundance of a species in one sample equals the sum of the gene abundance annotated for the species.
The samples for this research were analyzed without removal of the host genome, so some genes could be annotated to the host animal or closely related species. The figures of the relative abundance comparison as well as the developmental trends of the 20 zoonotic pathogens are generated by Microsoft Excel.
Zoonotic pathogens in clinical healthy goats
After removing the host animal and closely related species, about 2,800 kinds of microorganisms were detected in each goat intestine. Out of all microorganisms, 20 kinds of zoonotic bacteria were screened (Table 2), including Lactococcus garvieae, Helicobacter pylori, Klebsiella pneumoniae, Shigella sonnei, Shigella boydii, Campylobacter coli, Salmonella enterica, Acinetobacter baumannii, Shigella flexneri, Shigella dysenteriae, Clostridium perfringens, Campylobacter jejuni, Campylobacter lanienae, Salmonella bongori, Campylobacter fetus, Listeria monocytogenes, Acinetobacter haemolyticus, Mycobacterium tuberculosis, Mycobacterium neglectum and Burkholderia pseudomallei, and each of them is a tiny part of the total community. No Brucelella was detected and potential zoonotic agents were not screened.
Table (2):
The relative abundance of 20 zoonotic pathogens in clinical healthy goats (%).
| Bacterium | goat | 1 d | 3 d | 5 d | 10 d | 20 d | 30 d | 40 d | 45 d | 50 d | 60 d | 70 d | 80 d | 90 d | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactococcus garvieae | A | 0.77 | 1.77 | 1.22 | 3.25 | 3.73 | 0.22 | 1.08 | 0.35 | 0.26 | 0.02 | 0.01 | 0 | 0.03 | 0.978 |
| B | 8.27 | 2.48 | 5.06 | 4.5 | 0.56 | 1.62 | 0.66 | 0.04 | 0 | 0.11 | 0 | 0.01 | 0.06 | 1.798 | |
| Helicobacter pylori | A | 0.58 | 1.28 | 1.41 | 2.39 | 2.74 | 0.17 | 0.81 | 0.27 | 0.21 | 0.02 | 0 | 0 | 0.03 | 0.762 |
| B | 1.49 | 1.86 | 3.86 | 3.39 | 0.45 | 1.23 | 0.51 | 0.03 | 0 | 0.09 | 0 | 0.01 | 0.05 | 0.998 | |
| Klebsiella pneumoniae | A | 0.17 | 0.63 | 1.49 | 1.38 | 1.31 | 0.06 | 0.30 | 0.11 | 0.08 | 0.03 | 0.04 | 0.04 | 0.05 | 0.438 |
| B | 0.19 | 1.11 | 1.92 | 1.51 | 0.19 | 0.55 | 0.20 | 0.04 | 0.02 | 0.05 | 0.03 | 0.03 | 0.04 | 0.452 | |
| Shigella sonnei | A | 0.15 | 0.43 | 0.36 | 0.73 | 0.73 | 0.06 | 0.20 | 0.07 | 0.06 | 0.01 | 0.02 | 0 | 0.01 | 0.218 |
| B | 1.48 | 0.7 | 1.2 | 0.91 | 0.14 | 0.31 | 0.14 | 0.02 | 0 | 0.03 | 0.01 | 0.01 | 0.02 | 0.382 | |
| Shigella boydii | A | 0.10 | 0.26 | 0.21 | 0.46 | 0.49 | 0.04 | 0.14 | 0.05 | 0.04 | 0 | 0.01 | 0 | 0 | 0.138 |
| B | 1.07 | 0.40 | 0.72 | 0.58 | 0.08 | 0.21 | 0.08 | 0.01 | 0 | 0.02 | 0 | 0 | 0.01 | 0.245 | |
| Campylobacter coli | A | 0.04 | 0.12 | 0.08 | 0.28 | 0.29 | 0.03 | 0.09 | 0.04 | 0.03 | 0.01 | 0.01 | 0.01 | 0.01 | 0.080 |
| B | 0.72 | 0.18 | 0.34 | 0.33 | 0.03 | 0.13 | 0.06 | 0.01 | 0 | 0.02 | 0.01 | 0.01 | 0.01 | 0.142 | |
| Salmonella enterica | A | 0.01 | 0.2 | 0.24 | 0.23 | 0.09 | 0.03 | 0.01 | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 0.02 | 0.073 |
| B | 0.01 | 0.45 | 0.39 | 0.12 | 0.05 | 0.02 | 0.02 | 0.03 | 0.01 | 0.01 | 0.07 | 0.01 | 0.01 | 0.092 | |
| Acinetobacter baumannii | A | 0.03 | 0.07 | 0.04 | 0.16 | 0.17 | 0.01 | 0.04 | 0.02 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0.045 |
| B | 0.43 | 0.11 | 0.20 | 0.19 | 0.01 | 0.06 | 0.02 | 0.02 | 0 | 0.01 | 0.03 | 0.01 | 0.02 | 0.085 | |
| Shigella flexneri | A | 0 | 0.10 | 0.11 | 0.10 | 0.03 | 0.01 | 0 | 0 | 0.01 | 0 | 0.01 | 0 | 0 | 0.028 |
| B | 0 | 0.22 | 0.19 | 0.05 | 0.02 | 0.01 | 0 | 0.01 | 0 | 0 | 0.01 | 0 | 0 | 0.039 | |
| Shigella dysenteriae | A | 0 | 0.07 | 0.08 | 0.08 | 0.03 | 0.01 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0.022 |
| B | 0 | 0.12 | 0.16 | 0.04 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.026 | |
| Clostridium perfringens | A | 0.02 | 0.12 | 0 | 0 | 0.01 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 | 0.020 |
| B | 0 | 0.09 | 0.02 | 0 | 0.01 | 0 | 0.02 | 0.02 | 0 | 0.01 | 0.02 | 0.03 | 0.01 | 0.018 | |
| Campylobacter jejuni | A | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0 | 0 | 0 | 0.01 | 0.008 |
| B | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.03 | 0.01 | 0 | 0 | 0 | 0.01 | 0 | 0.008 | |
| Campylobacter lanienae | A | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0 | 0.03 | 0.009 |
| B | 0 | 0 | 0 | 0 | 0 | 0 | 0.21 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0.017 | |
| Salmonella bongori | A | 0 | 0.02 | 0.02 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.005 |
| B | 0 | 0.04 | 0.03 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.006 | |
| Campylobacter fetus | A | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 0 | 0 | 0.01 | 0.01 | 0.01 | 0.006 |
| B | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0.005 | |
| Listeria monocytogenes | A | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.01 | 0 | 0.01 | 0.02 | 0.006 |
| B | 0 | 0.01 | 0.01 | 0.01 | 0.01 | 0 | 0 | 0.01 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0.007 | |
| Acinetobacter haemolyticus | A | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0.001 |
| B | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.001 | |
| Mycobacterium tuberculosis | A | 0.01 | 0.01 | 0 | 0.01 | 0.02 | 0 | 0 | 0.01 | 0 | 0 | 0.01 | 0.02 | 0.01 | 0.008 |
| B | 0.04 | 0.01 | 0.02 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0.012 | |
| Mycobacterium neglectum | A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.000 |
| B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0.001 | |
| Burkholderia pseudomallei | A | 0.01 | 0 | 0 | 0.01 | 0.01 | 0 | 0.01 | 0.01 | 0 | 0 | 0 | 0.01 | 0 | 0.005 |
| B | 0.02 | 0.02 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0.006 |
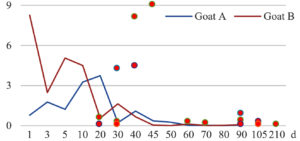
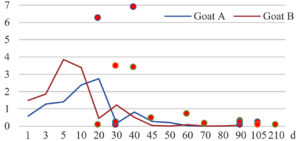
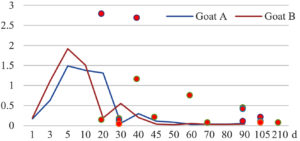
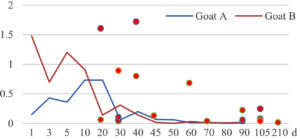
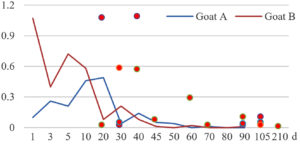
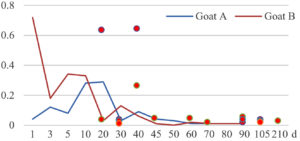
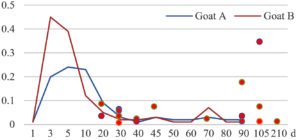
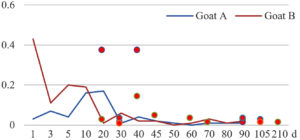
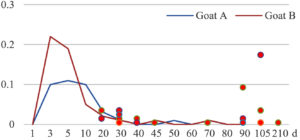
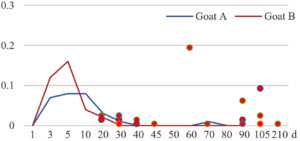
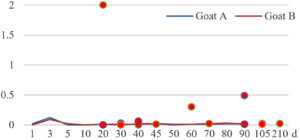
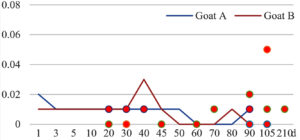
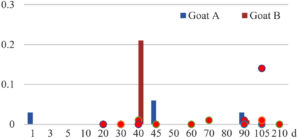
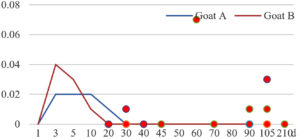
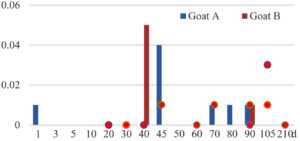
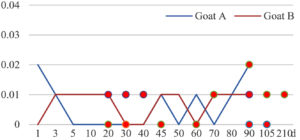
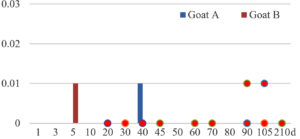
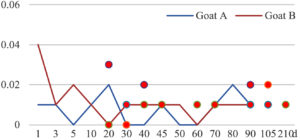
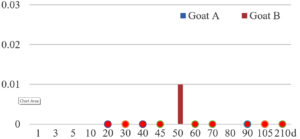
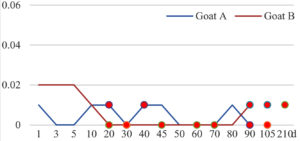
Among the 20 bacterial species, the top five are Lactococcus garvieae, Helicobacter pylori, Klebsiella pneumoniae, Shigella sonnei and Shigella boydii (Figure 1). By analyzing the abundance at different day ages, it was found that 11 (55%) of them, including Lactococcus garvieae, Helicobacter pylori, Klebsiella pneumoniae, Shigella sonnei, Shigella boydii, Campylobacter coli, Salmonella enterica, Acinetobacter baumannii, Shigella flexneri, Shigella dysenteriae and Salmonella bongori, were infectioned mainly during preweaned period (Figure 2 to 11, and Figure 15, line), while the remaining ones have no obvious regularity(Figure 12 to 14, and Figure 16 to 21, line or column). The abundant presence of 20 bacterial species in the goat intestine suggests an important role of these bacteria as foodborne or waterborne agents and possibly as zoonotic agents.
Zoonotic pathogens in clinical diarrheic goats
To reveal the possible role of zoonotic pathogens in goat diarrhea, the relative abundance of 20 zoonotic bacteria were analyzed in the samples from in clinical diarrheic cases (Table 3). Of the 19 diarrhetic goats, all were confirmed to be zoonotic bacterial infection positive (range from 11 to 12 species). After comparison with healthy samples of the same or similar day-age goats (Table 3, Figures 2 to 21, dot), it was found that Lactococcus garvieae was highly increased in 3 preweaned, 1 weaning and 1 postweaned cases, Helicobacter pylori in 4 preweaned and 1 postweaned cases, Klebsiella pneumoniae in 4 preweaned and 2 postweaned cases, Shigella sonnei in 4 preweaned and 3 postweaned cases, Shigella boydii in 4 preweaned and 1 postweaned cases, Campylobacter coli in 4 preweaned cases, Salmonella enterica in 3 postweaned cases, Acinetobacter baumannii in 3 preweaned cases, Shigella flexneri in 2 postweaned cases, Shigella dysenteriae in 1 postweaned cases, Clostridium perfringens in 1 pretweaned and 2 postweaned cases, and Campylobacter fetus in 1 pretweaned cases, while the remains no significant change.
Table (3):
The relative abundance of 20 zoonotic pathogens in clinical diarrheic goats (%).
Goat number Bacterium |
C1 |
C2 |
C3 |
C4 |
C5 |
C6 |
C7 |
C8 |
C9 |
C10 |
C11 |
C12 |
C13 |
C14 |
C15 |
C16 |
C17 |
C18 |
C19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Lactococcus garvieae |
0.51 |
0.02 |
0.27 |
0.06 |
4.23↑ |
0 |
8.03↑ |
4.45↑ |
8.97↑ |
0.23 |
0.10 |
0.39 |
0.08 |
0.82↑ |
0.10 |
0.27 |
0.03 |
0.06 |
0.01 |
Helicobacter pylori |
0.05 |
6.21↑ |
0.02 |
0 |
0.22 |
3.44↑ |
3.32↑ |
6.86↑ |
0.40 |
0.63↑ |
0.09 |
0.28 |
0.03 |
0.15 |
0.08 |
0.07 |
0.21 |
0.06 |
0.01 |
Klebsiella pneumoniae |
0.12 |
2.75↑ |
0.14 |
0.04 |
0.12 |
1.73↑ |
1.12↑ |
2.65↑ |
0.18 |
0.73 |
0.04 |
0.43↑ |
0.07 |
0.38↑ |
0.08 |
0.17 |
0.08 |
0.05 |
0.03 |
Shigella sonnei |
0.05 |
1.58↑ |
0.02 |
0.04 |
0.08 |
0.87↑ |
0.77↑ |
1.71↑ |
0.10 |
0.67↑ |
0.02 |
0.20↑ |
0.02 |
0.04 |
0.07 |
0.23↑ |
0.05 |
0.02 |
0 |
Shigella boydii |
0.02 |
1.07↑ |
0.01 |
0.02 |
0.04 |
0.57↑ |
0.56↑ |
1.08↑ |
0.07 |
0.28↑ |
0.01 |
0.09 |
0.01 |
0.03 |
0.03 |
0.10 |
0.04 |
0.01 |
0 |
Campylobacter coli |
0.03 |
0.63↑ |
0.03 |
0.03 |
0.02 |
0.41↑ |
0.26↑ |
0.64↑ |
0.04 |
0.04 |
0.01 |
0.05 |
0.01 |
0.03 |
0.02 |
0.03 |
0.02 |
0.01 |
0.02 |
Salmonella enterica |
0.08 |
0.03 |
0.03 |
0.06 |
0.05 |
0.01 |
0.02 |
0.01 |
0.07 |
0.90↑ |
0.02 |
0.17↑ |
0.03 |
0.01 |
0.07 |
0.34↑ |
0.01 |
0.01 |
0.01 |
Acinetobacter baumannii |
0.02 |
0.37↑ |
0 |
0.01 |
0.03 |
0.23↑ |
0.14 |
0.37↑ |
0.04 |
0.03 |
0.01 |
0.02 |
0.01 |
0.03 |
0.01 |
0.01 |
0.02 |
0.01 |
0.01 |
Shigella flexneri |
0.03 |
0.01 |
0.01 |
0.03 |
0.02 |
0 |
0.01 |
0 |
0 |
0.42↑ |
0 |
0.09 |
0.01 |
0 |
0.03 |
0.17↑ |
0 |
0 |
0 |
Shigella dysenteriae |
0.02 |
0.01 |
0.01 |
0.01 |
0.02 |
0 |
0.01 |
0 |
0 |
0.19↑ |
0 |
0.06 |
0.01 |
0 |
0.02 |
0.09 |
0 |
0 |
0 |
Clostridium perfringens |
2.00↑ |
0 |
0.01 |
0 |
0.03 |
0 |
0 |
0.06 |
0.01 |
0.30↑ |
0.02 |
0.01 |
0.01 |
0.49↑ |
0.02 |
0.01 |
0.02 |
0.01 |
0.02 |
Campylobacter jejuni |
0 |
0.01 |
0.01 |
0.01 |
0 |
0.01 |
0.01 |
0.01 |
0 |
0 |
0.01 |
0.02 |
0.01 |
0 |
0.01 |
0 |
0 |
0.05 |
0.01 |
Campylobacter lanienae |
0 |
0 |
0 |
0 |
0 |
0 |
0.01 |
0 |
0 |
0 |
0.01 |
0 |
0.01 |
0 |
0.01 |
0.14↑ |
0 |
0.01 |
0 |
Salmonella bongori |
0 |
0 |
0 |
0.01 |
0 |
0 |
0 |
0 |
0 |
0.07 |
0 |
0.01 |
0 |
0 |
0.01 |
0.03 |
0 |
0 |
0 |
Campylobacter fetus |
0 |
0 |
0 |
0 |
0 |
0.02 |
0.12↑ |
0 |
0.01 |
0 |
0.01 |
0.01 |
0 |
0 |
0.01 |
0.03 |
0.01 |
0.01 |
0 |
Listeria monocytogenes |
0.02 |
0.01 |
0 |
0 |
0.01 |
0 |
0 |
0 |
0.02 |
0.01 |
0.02 |
0.01 |
0.01 |
0.02 |
0.02 |
0.01 |
0.02 |
0.01 |
0.01 |
Acinetobacter haemolyticus |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.01 |
0 |
0 |
0 |
0 |
0.01 |
0 |
0 |
Mycobacterium tuberculosis |
0 |
0.03 |
0 |
0 |
0.01 |
0.02 |
0.01 |
0.02 |
0.01 |
0.01 |
0.01 |
0.01 |
0.02 |
0.01 |
0.01 |
0.01 |
0.01 |
0.02 |
0.01 |
Mycobacterium neglectum |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Burkholderia pseudomallei |
0 |
0.01 |
0 |
0 |
0 |
0.01 |
0.01 |
0.01 |
0 |
0 |
0 |
0.01 |
0 |
0.01 |
0.01 |
0 |
0.01 |
0 |
0.01 |
With the continuous global threat of zoonotic pathogens, effective strategies must be found to control these diseases from the root. Due to the complexity of these pathogens’ infection in animals and humans, it should be detected as early as possible and prevent and control the infection of these pathogens in the process of human-animal contact, to achieve the ultimate goal of maintaining human health and ecological environment.6,7 The results of this study show us that identifying threat factors from animal sources remains the most effective and economical way to protect human public health, which fits exactly with ‘One Health’ interventions.3
The bacteriological analysis of healthy goats in this study revealed that goats could harbor a variety of zoonotic pathogens in their intestines during their growth, especially in the pre-weaning stage. These zoonotic bacteria mainly include the previous reported Lactococcus garvieae,8 Helicobacter pylori,8,9 Klebsiella pneumoniae,10 Shigella sonnei,11 Shigella boydii,12 Campylobacter coli,13 Salmonella enterica,14 Acinetobacter baumannii,15 Shigella flexneri,12 Shigella dysenteriae,12 Clostridium perfringens,16 Campylobacter jejuni,13 Campylobacter lanienae,17 Salmonella bongori,18 Campylobacter fetus,19,20 Listeria monocytogenes,21 Acinetobacter haemolyticus,22 Mycobacterium tuberculosis,23 Mycobacterium neglectum,24 and Burkholderia pseudomallei.25 The results suggest that goats may act as a reservoir for many zoonotic bacteria.
The analysis of diarrheal goats revealed that Lactococcus garvieae, Helicobacter pylori, Klebsiella pneumoniae, Shigella sonnei, Shigella boydii, and Campylobacter coli were highly increased incases in some lambs with diarrhea before weaning. Salmonella enterica, Acinetobacter baumannii, Shigella flexneri, and Shigella dysenteriae were highly increased incases in some with diarrhea after weaning. Clostridium perfringens and Campylobacter fetus were highly increased incases in few cases, while the remains had no significant change. The results suggest that some of the zoonotic bacteria may be associated with goat intestinal inflammation.
Whether in healthy or diarrheic goats, the abundance of each zoonotic bacteria in the goat intestine is a tiny part of the total community, but they can spread through shedding in the faeces and subsequent faecal contamination of raw food.26 Therefore, in the process of raising, transportation, slaughter, and processing, it will inevitably bring public health risks to human beings. To take action to prevent this risk, OIE proposed that the best way to protect human health is to eliminate the pathogens at its source, which requires efforts to establish a multiparty participation and cooperation mechanism around the concept of “one health”. Therefore, it is important to have epidemiological data collecting systems and information systems that allow complete diagnostic tracing from herd to slaughterhouse and vice versa. All sides, including research and surveillance, as well as producers are called upon to actively share in protecting the health of consumers as far as it is threatened by latent infections in domestic stock.27
The deficiency of this research is only investigated in the healthy and diarrheic goats of one farm. More farms need further investigation, which is more consistent with the ‘One Health’ perspective.
A total of 20 kinds of zoonotic bacteria were screened from healthy goats, and some of them were highly increased incases in some diarrheic cases. The results suggest that goats may act as a reservoir for many zoonotic bacteria, and some of them may be associated with goat intestinal inflammation.
ACKNOWLEDGMENTS
The authors would like to thank the five farms that allowed sample collection for use in this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
CC and DC conceived and designed the study, and jointly executed the experiments with GW, ML, SZ and XC. CC, DC and JT co-wrote the paper. All authors read and approved the final manuscript.
FUNDING
This study was funded by the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS[2021]492), and partially funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (2018).
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study did not require official or institutional ethical approval. The animals were handled according to high ethical standards and national legislation. The sample collection in these farms occurred under the direction of the farms’ usual vets as part of their routine veterinary herd health programme; therefore, no changes in animal treatment or handling occurred as a result of the sample collection required for this study.
- Rantsios AT. Zoonoses. In: Encyclopedia of Food and Health. Elsevier; 2016:645-653.
Crossref - Yamada A. [Zoonoses]. Uirusu. 2004;54(1):17-22.
Crossref - Cleaveland S, Sharp J, Abela-Ridder B, et al. One Health contributions towards more effective and equitable approaches to health in low- and middle-income countries. Philos Trans R Soc Lond B Biol Sci. 2017;372(1725):20160168.
Crossref - Corry JE, Hinton MH. Zoonoses in the meat industry: a review. Acta Vet Hung. 1997;45:457-479.
- van den Brom R, de Jong A, van Engelen E, Heuvelink A, Vellema P. Zoonotic risks of pathogens from sheep and their milk borne transmission. Small Rumin Res. 2020;189:106123.
Crossref - Zinsstag J, Crump L, Winkler M. Biological threats from a “One Health” perspective. Rev Sci Tech. 2017;36(2):671-680.
Crossref - Jemmi T, Danuser J, Griot C. [Zoonoses as a risk when associating with livestock or animal products]. Schweizer Archiv fur Tierheilkunde. 2000;142:665-671.
- Rodrigues MX, Lima SF, Higgins CH, Canniatti-Brazaca SG, Bicalho RC. The Lactococcus genus as a potential emerging mastitis pathogen group: A report on an outbreak investigation. J Dairy Sci. 2016;99(12):9864-9874.
Crossref - Wroblewski LE, Peek RM. Helicobacter pylori, Cancer, and the Gastric Microbiota. Adv Exp Med Biol. 2016;908:393-408.
Crossref - Marques C, Menezes J, Belas A, et al. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: population structure, antimicrobial resistance and virulence genes. J Antimicrob Chemother. 2019;74(3):594-602.
Crossref - Torraca V, Holt K, Mostowy S. Shigella sonnei. Trends Microbiol. 2020;28(8):696-697.
Crossref - Peng JP, Yang J, Jin Q. Research progress in Shigella in the postgenomic era. Sci China Life Sci. 2010;53(11):1284-1290.
Crossref - Sheppard SK, Maiden MCJ. The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb Perspect Biol. 2015;7:a018119.
Crossref - Lamas A, Miranda JM, Regal P, Vazquez B, Franco CM, Cepeda A. A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiol Res. 2018;206:60-73.
Crossref - Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292-301.
Crossref - Cheng C, Zhang T, Cheng D, Cao S, Xin H, Tao J. Surveillance of Clostridium perfringens infection in Clinical Healthy Meat Sheep in Jiangsu Province.
J Yangzhou Univ. 2021; 42: 34–38. http://qikan.cqvip.com/Qikan/Article/Detail?id=7106026698 (in Chinese) - Fornefett J, Busch A, Dopping S, Hotzel H, Rimek D. Bacterial gastroenteritis caused by the putative zoonotic pathogen Campylobacter lanienae: First reported case in Germany. Access Microbiol. 2021;3(3):000199.
Crossref - Giammanco GM, Pignato S, Mammina C, et al. Persistent endemicity of Salmonella bongori 48:z35:-in Southern Italy: molecular characterization of human, animal, and environmental isolates. J Clin Microbiol. 2002;40(9):3502-3505.
Crossref - Fujimoto S. [Pathogenesis of Campylobacter fetus infection]. Fukuoka Igaku Zasshi. 2003;94:225-234.
- Costa D, Betancor L, Gadea P, et al. Polyclonal Campylobacter fetus Infections Among Unrelated Patients, Montevideo, Uruguay, 2013-2018. Clin Infect Dis. 2020;70(6):1236-1239.
Crossref - Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55(3):476-511.
Crossref - Al Bshabshe A, Joseph MRP, Al Hussein A, Haimour W, Hamid ME. Multidrug resistance Acinetobacter species at the intensive care unit, Aseer Central Hospital, Saudi Arabia: A one year analysis. Asian Pac J Trop Med. 2016;9(9):903-908.
Crossref - Koch A, Mizrahi V. Mycobacterium tuberculosis. Trends Microbiol. 2018;26(6):555-556.
Crossref - Nouioui I, Carro L, Sangal V, et al. Formal description of Mycobacterium neglectum sp. nov. and Mycobacterium palauense sp. nov., rapidly growing actinobacteria. Antonie Van Leeuwenhoek. 2018;111(6):1209-1223.
Crossref - vander Broek CW, Stevens JM. Type III Secretion in the Melioidosis Pathogen Burkholderia pseudomallei. Front Cell Infect Microbiol. 2017;7:255.
Crossref - Carol McWilliam Leitch E, Duncan SH, Stanley KN, Stewart CS. Dietary effects on the microbiological safety of food. Proc Nutr Soc. 2001;60:247-255.
Crossref - Grossklaus D. [Zoonoses control–new challenges in health protection of consumers]. Berliner und Munchener tierarztliche Wochenschrift. 114:420-427.
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.