ISSN: 0973-7510
E-ISSN: 2581-690X
The World Health Organization (WHO) has recognized antimicrobial resistance to be one of the top 10 threats to mankind in the coming future. Therefore, it requires solutions that are targeted, sustainable, and economically effective. Carbapenem-resistant Pseudomonas aeruginosa is associated with nosocomial infections affecting mostly patients with chronic lung disease. The goal of the current investigation was to gain insight into significant P. aeruginosa genes responsible for carbapenems, beta-lactams, and other antimicrobials resistance through a systems biology approach. To proceed with the methodology, 866 genes were retrieved from the NDARO database and a gene interaction network of 45 genes and 195 functional partners was constructed using STRING v9.0 with high confidence and analyzed using Cytoscape 3.10.0. Using clustering analysis, four closely linked clusters (C1-C4) associated with mechanisms of multidrug-resistance were identified. The enrichment analysis revealed a substantial role for 43 genes in biological processes, 36 genes in molecular function, and 40 genes in cellular components. The gene interaction network analysis found that the genes oprD, oprM, oprN, mexR, nfxB, mexB, mexT, mexA, nalD, and nalC had the greatest number of gene interactions, which can be further used as potential drug targets for the development of novel therapeutics to manage the antimicrobial resistance associated with Pseudomonas aeruginosa.
Systems Biology Approach, Pseudomonas aeruginosa, Antimicrobial Resistance Genes, NDARO, Enrichment Analysis, Gene Interaction Network, Nosocomial Infections
Pseudomonas aeruginosa is an aerobic, Gram-negative bacterium that survives in habitats such as water, soil, and plants.1 According to the World Health Organisation (WHO), Carbapenem-resistant Pseudomonas aeruginosa has been identified to pose a severe threat to individuals with impaired immunity, especially diabetes and chronic lung disease. Along with the five other ESKAPE pathogens including Staphylococcus aureus, Enterococcus faecium, Klebsiella pneumonia, Enterobacter species and Acinetobacter baumannii, Pseudomonas aeruginosa have been identified as “critical” pathogens by WHO, which requires urgent attention for the development of sustainable therapeutics to tackle resistance mechanisms in them.2 Pseudomonas aeruginosa, being an opportunistic pathogen, infects immune-compromised patients causing cystic fibrosis,3 chronic obstructive pulmonary disorder2 along with aggravation of diabetic foot wounds in association with Staphylococcus aureus.4 This Gram-negative bacterium houses an extensive genome consisting of approximately 5.5 million to 7 million base pairs, which encodes regulatory enzymes crucial for the transportation, metabolism, and efflux of organic chemical compounds,5,6 responsible for its nutritional flexibility and adaptability to different environmental conditions.7 Several virulence factors, firstly, lipopolysaccharides in the outer cell membrane responsible for host cell attachment, recognition of antibiotics,2,8 secondly secretion systems most importantly T3SS secretion system which participates in escaping the host immune system,9 and third polysaccharides namely alginate, Psl, and Pel associated with biofilm formation, allows the bacteria to adapt and survive in the adverse environment in presence of antibiotics and other stresses.10 As a part of the intrinsic resistance mechanisms, the bacteria also possess several families of multi-drug efflux pump systems such as mexAB-oprM, mexXY-oprM, mexCD-oprJ and mexEF-oprN belonging to resistance nodulation division (RND), PmpM belonging to multidrug and toxic compound extrusion (MATE) family, Mfs1 and Mfs2 of major facilitator superfamily (MFS) and others which contribute to resistance to antimicrobial compounds such as aminoglycosides, macrolides, carbapenems, antimicrobial peptides and beta-lactams.11-14
In addition to these intrinsic resistance mechanisms, the bacteria has also evolved to acquire mutations that enhance existing resistance mechanisms such as mutations in oprD gene leading to decreased expression or defect in oprD protein, thereby enhancing resistance to carbapenems antibiotics,15 gyrA, gyrB, parC, and parE mutations resulting to fluoroquinolone resistance,16 nfxB mutations leading to overexpression of mexCD-oprJ efflux pump thereby contributing to Ciprofloxacin resistance17 or involving mexXY induction and biofilm formation, resulting in modifications in the target gene or protein expression systems.5,18,19
Currently, anti-quorum sensing, anti-biofilm, and immuno-therapeutics are being investigated as alternative approaches to control Pseudomonas infections.20 In our previous study, a combination therapy containing recombinant Lactonase and antibiotics (fluoroquinolone and carbapenem) was developed targeting the quorum-sensing pathway.21 The main issue that exists while designing new antibacterial drugs is improving their accumulation within bacterial cells and enhancing the antibiotic efficacy in controlling the infections. This can be accomplished by enhancing antibiotic flow across the outer membrane, interfering with the regulatory genes, creating compounds that circumvent the efflux process effectively repurposing existing drugs as efflux pump inhibitors, or designing stable antimicrobial peptides. Previously, a group of researchers,22 had performed the systems biology approach to identify the potential genes responsible for antimicrobial resistance in 2019 where they had identified oprJ, oprM, oprN, ampC, gyrA, mexA, oprD, mexB and nfxB genes to have shown maximum number of interactions in Pseudomonas aeruginosa PAO1 strain. In the present study, we focused on identifying the hub genes responsible for the antimicrobial resistance of P. aeruginosa, specifically not restricting to any strain to recognizing the target genes for the development of specific therapeutic targets to enhance the susceptibility of multidrug-resistance P. aeruginosa strains was done using clustering analysis.
Collection of raw data from the NDARO database
The National Database of Antibiotic Resistant Organisms (NDARO) is maintained by NCBI and serves as a central site for gathering and disseminating information about bacterial strains and their genes resistant and sensitive to antibiotics. After the removal of the duplicates, a total of 866 antimicrobial resistance genes of Pseudomonas aeruginosa were retrieved from the NDARO database.
Protein-protein interaction using STRING v9.0 database
A pre-computed database STRING v9.0 was used to predict the physical and functional gene/protein relationships. STRING employs data selected from sources like experimental and published data, data mining, and co-expression study analysis. A confidence score or combination score has been assigned to each interaction. The total scores indicate the likelihood of the correlation based on the sources that the interaction selected. The total scores fall between 0 and 1, with the lowest values denoting a lower likelihood of that specific relationship occurring and the highest values denoting a higher likelihood. Based on the confidence scores, interactions are characterized as low (0.15-0.39), medium (0.40-0.69), high (0.70-0.89), and highest (0.90-1.0) to obtain an optimum number of gene interactions and the network was visualized using Cytoscape 3.10.0.
Network analysis and clustering analysis using Cytoscape
Cytoscape 3.10.0 is a versatile software that provides multiple plugins to carry out detailed network analysis. One of the finest plugins of Cytoscape 3.10.0 is Network Analyser which helps in evaluating multiple topological variables such as nodes, edges, average shortest path length, betweenness centrality, and clustering coefficient. Whereas Cytoscape program’s Molecular Complex Detection (MCODE) software was used to detect the network’s highly interacting nodes. The method relies heavily on topology to establish clusters. The clusters were formed by identifying protein groupings with similar activities. It depended on vertex weighting, complicated prediction, and optimal post-processing by assigning weight to the vertex in local neighbourhood density from dense regions depending on a specific parameter. The collected clusters were scored based on their size and density.23 Numerous metrics, including the number of nodes, edges, average shortest path length, betweenness centrality, the average number of neighbours, and clustering coefficient of the individual network as well as the clusters were effectively calculated using the Network Analyser tool of Cytoscape 3.10.0.
Retrieval of antimicrobial resistance genes of P. aeruginosa from NDARO database and STRING analysis
National Database of Antimicrobial Resistance Organisms (NDARO) was used to retrieve the antimicrobial resistance (AMR) genes of Pseudomonas aeruginosa and a total of 866 genes were obtained after the removal of the duplicates and AMR genes belonging to another Pseudomonas sp. from the list. These genes were queried for their protein sequences in the NCBI Protein database and UNIPROT database. The protein sequences were searched in the STRING v7.0 database to obtain the protein-protein interaction against Pseudomonas aeruginosa. Out of the 866 genes queried in the STRING database, we obtained interaction with 46 genes with high confidence (Confidence score = 0.70-0.89). A list of the top 10 genes with a maximum number of interactions along with their interaction partners is given in Table 1.
Table (1):
List of top 10 genes with their interacting partners at high confidence scores
No. |
Target Genes |
Interacting Partners (Confidence Score = 0.70-0.89) |
|---|---|---|
1 |
oprM |
parE, parR, parC |
2 |
mexA |
mexR, mexT, nfxB, nalC, oprD, nalD, oprN, oprJ, mexF, mexD, mexB, oprM |
3 |
mexB |
mexR, mexT, nfxB, nalC, oprD, nalD, oprN, oprJ, mexE, mexC, oprM |
4 |
mexR |
parE, oprD, mexT, oprN, oprJ, nalD, nfxB, nalC, parC, oprM |
5 |
mexT |
oprM, oprD, parS, parC, oprJ, nfxB, nalC, nalD, oprN |
6 |
nfxB |
oprM, oprD, oprN, oprJ, parE, parC |
7 |
oprN |
parC |
8 |
nalC |
oprM, oprD, oprN, nalD, oprJ, nfxB |
9 |
nalD |
oprM, oprD, oprN, parC, nfxB, oprJ |
10 |
oprD |
oprM, oprJ, parC, oprN |
Network analysis using Cytoscape
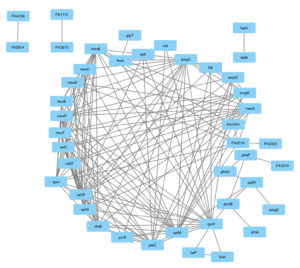
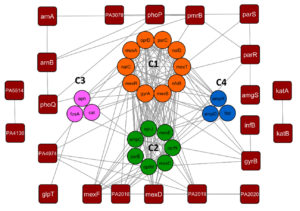
The protein-protein interactions obtained from the String database were visualized using Cytoscape 3.10.0 shown in Figure 1 and the network analysis was carried out using the Network analyzer tool of Cytoscape to obtain the topological parameters of the gene interaction which is given in Table 2. A dense interaction network with 45 nodes and 198 edges was obtained when we analyzed the interaction with high confidence followed by which clustering analysis was carried out using the MCODE tool of Cytoscape 3.10.0 given in Table 3. We have obtained 4 clusters, C1, C2, C3, and C4 which are shown in Figure 2. After carrying out the network analysis on the clusters, we have found that out of the 45 genes, 23 genes were clustered with 10 genes in Cluster 1 (score: 8.667), 7 genes in Cluster 2 (score: 4.333), 3 genes in Cluster 3 (score: 3) and 3 genes in Cluster 4 (score: 3). Network analysis of the entire network using the network analyzer resulted in 45 nodes, 198 edges with clustering coefficient 0.559 and network density 0.239. On the other hand, network analysis of the clusters revealed 10 nodes and 39 edges with an average number of neighbors of 7.8 for Cluster 1, 7 nodes, 13 edges, and the average number of neighbours of 3.714 for Cluster 2, 3 nodes, 3 edges for Cluster 3 and 4. Table 3 lists the topological parameters of the top 20 genes in the network obtained from network analysis based on their average shortest path length, closeness, betweenness centrality, degree, and clustering coefficient.
Table (2):
List of top 20 genes based on the topological parameters like degree, shortest path length, betweenness centrality, closeness centrality, and clustering coefficient
Gene Name |
Degree |
Average Shortest Path Length |
Betweenness Centrality |
Closeness Centrality |
Clustering Coefficient |
|---|---|---|---|---|---|
oprM |
24 |
1.55 |
0.218715 |
0.645161 |
0.391304 |
oprN |
17 |
1.875 |
0.026111 |
0.533333 |
0.566176 |
oprJ |
17 |
1.875 |
0.026111 |
0.533333 |
0.566176 |
mexA |
16 |
1.925 |
0.016607 |
0.519481 |
0.633333 |
mexB |
17 |
1.8 |
0.027839 |
0.555556 |
0.632353 |
ampC |
21 |
1.7 |
0.084808 |
0.588235 |
0.457143 |
mexR |
18 |
1.8 |
0.016233 |
0.555556 |
0.640523 |
oprD |
21 |
1.725 |
0.036374 |
0.57971 |
0.557143 |
gyrA |
18 |
1.65 |
0.220247 |
0.606061 |
0.431373 |
nfxB |
17 |
1.8 |
0.020869 |
0.555556 |
0.654412 |
parC |
16 |
1.825 |
0.036069 |
0.547945 |
0.541667 |
mexT |
16 |
1.8 |
0.078098 |
0.555556 |
0.625 |
nalD |
12 |
2.075 |
0.002051 |
0.481928 |
0.833333 |
nalC |
11 |
2.125 |
0.001506 |
0.470588 |
0.854545 |
PA2019 |
10 |
2.225 |
0.027126 |
0.449438 |
0.555556 |
mexF |
10 |
2.075 |
0.003942 |
0.481928 |
0.711111 |
mexD |
10 |
2.15 |
0.002886 |
0.465116 |
0.733333 |
PA2018 |
10 |
2.225 |
0.027302 |
0.449438 |
0.555556 |
mexR |
18 |
1.8 |
0.016233 |
0.555556 |
0.640523 |
ampR |
10 |
2.075 |
0.003766 |
0.481928 |
0.733333 |
Table (3):
Topological parameters of the whole network and the clusters obtained through network analysis
| No. | Topological parameters | Network | MCODE Cluster | |||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | |||
| 1 | Number of Nodes | 45 | 10 | 7 | 3 | 3 |
| 2 | Number of Edges | 198 | 39 | 13 | 3 | 3 |
| 3 | Average number of neighbours | 9.561 | 7.8 | 3.714 | ||
| 4 | Clustering co-efficient | 0.559 | 0.899 | 0.614 | ||
| 5 | Network density | 0.239 | 0.867 | 0.619 | ||
| 6 | Average Shortest path length | 2.17 | 1.133333 | 1.380952 | ||
| 7 | Average Betweenness Centrality | 0.03 | 0.016667 | 0.07619 | ||
| 8 | MCODE score | – | 8.667 | 4.333 | 3 | 3 |
| 9 | Genes in the cluster | – | nalC, mexA, mexT, mexR, nalD, nfxB, parC, gyrA, mexB, oprD | parE, oprN, mexC, prJ, oprM, ampC, mexE | ampR, ftsl, ampD | fosA, cat, Aph |
Figure 1. Gene interaction network of 46 genes of Pseudomonas aeruginosa as obtained from STRING v9.0 database and visualized using Cytoscape 3.10.0
Figure 2. Clustering analyses of antimicrobial resistance genes of Pseudomonas aeruginosa using Cytoscape-MCODE tool. Of the 46 genes in the network, 10 were grouped into Cluster 1, 7 were in Cluster 2, and 3 were in Cluster 3 and Cluster 4. The remaining 23 genes were ungrouped
Functional enrichment analysis
Gene Ontology (GO) terms and their annotations along with enriched KEGG pathways, UniProt keywords, and pFAM protein domains for the target genes and interaction partners were obtained from the STRING database with p-value ≤0.05 to understand the contribution of these AMR genes in molecular function (MF), biological processes (BP) or associating as a cellular component (CC) and others. Table 4 lists the genes enriched in biological process, molecular function, and cellular compartment, and Table 5 lists the genes enriched in the KEGG pathway, UniProt keywords, and Pfam Protein families. Among the 45 genes, 43 genes were enriched in biological processes, 36 genes were enriched in molecular function, and approximately 40 genes in the cellular compartment, 22 genes in the KEGG pathway, 45 in Uniprot keywords, and 11 in the Pfam protein family.
Table (4):
List of the genes enriched in biological processes, molecular function and cellular components with GO IDs
No. |
Functional Enrichment |
Genes Involved |
|---|---|---|
1 |
Biological Processes |
mexA, mexB, oprM, cat, fosA, pmpM, PA2018, PA2019, mexE, arr, PA3127, arnB, arnA, PA3676, aph, oprJ, mexC, phoP, phoQ, parS, parR, PA2019, soxR, PA3078, PA3127, ampC, PA4136, katA, katB, pmrB, PA4990, amgS, mexR, PA2020, mexT, nalD, nalC, nfxB, gyrB, gyrA, parC, parE, ftsI, ampR, soxR, infB |
2 |
Molecular Function |
mexB, oprM, PA2018, mexF, oprN, oprJ, mexD, PA4974, pmpM, PA3676, PA4136, gyrB, gyrA, parC, parE, PA4990, glpT, mexE, PA4974, mexC, mexT, nalD, nalC, nfxB, phoP, parR, soxR, ampR, gyrA, ksgA, infB, PA1290, parR, PA2020, soxR, ampR |
3 |
Cellular Compartment |
mexA, mexB, oprM, oprD, phoQ, pmpM, parS, PA2018, PA2019, mexE, mexF, oprN, arr, PA3676, PA4136, ftsI, ampD, oprJ, mexD, mexC, pmrB, parC, PA4990, amgS, glpT, PA5514, gyrB, gyrA, phoQ, infB, pmrB, fosA, phoP, ksgA, katA, ampR, ampC, pare, katB, nfxB |
Table (5):
List of KEGG pathway-enriched genes, UniProt keywords, and pFAM protein family
KEGG pathway-enriched genes |
Genes enriched in UniProt Keywords |
Genes enriched in the Pfam Protein family |
|---|---|---|
mexR, mexA, mexB, oprM, oprD, parS, parR, PA2018, PA2019, PA2020, nalD, nalC, ampR, ampC, ftsI, PA4974, PA5514, phoP, phoQ, arnB, pmrB, amgS |
mexA, mexB, oprM, cat, fosA, pmpM, arnB, arnA, ampC, aph, oprJ, PA2018, PA2019, mexE, mexF, PA3676, ftsI, mexD, mexC, gyrB, gyrA, parC, parE, phoP, phoQ, parS, parR, pmrB, amgS, PA4136, PA4990, oprN, katA, katB, glpT, arr, PA3078, PA3676, PA4136, gyrB, soxR, ampR, nfxB, nalC, nalD |
PA2018, mexF, PA3676, mexD, oprM, oprN, oprJ, PA4974, mexA, PA2019, mexC |
Pseudomonas aeruginosa is responsible for nosocomial infections in critical care patients or patients with low immunity. The prevalence of multidrug-resistant strains of P. aeruginosa has grown alarmingly in recent years. As per the Centres for Disease Control and Prevention’s statistics, 32,600 MDR P. aeruginosa infections were detected, resulting in 2,700 deaths in the United States in 2019.24 Antibiotic resistance-related genes or proteins, as well as the gene networks that support them, are extremely important because they offer important insights into biological and molecular complexes and the signaling pathways of the genes that are thought to be responsible for the resistance.25 The present study identified 45 antimicrobial resistance gene interactions in P. aeruginosa which were found to be involved in cellular processes (41 genes), molecular function (10 genes), and KEGG pathway (3 genes). The analysis of the network and the clusters showed that the average shortest path length of the whole network was 2.17 which was reduced to 1.13 in Cluster 1 and 1.38 in Cluster 2 along with average betweenness centrality ranging from 0.03 in the whole network to 0.01 in C1 but increased to 0.07 in C2. Betweenness centrality is calculated using the shortest path length whereas the average shortest path length calculates the shortest distance between the nodes and higher betweenness centrality and lower average shortest path length can impact the gene network and could be a therapeutic target.26-28
The C1 cluster has a high degree of connectivity, with an average neighbour count of 7.8, meaning that every node in the network is connected to 8 additional nodes. The cluster C1 has a closeness centrality of 8.936364 and an average shortest path length of 1.133333. C1 cluster consists of 10 genes (nalC, mexA, mexT, mexR, nalD, nfxB, parC, gyrA, mexB, oprD). mexB is a transmembrane protein with antiporter activity, whereas oprM is an outer membrane porin channel. The multidrug efflux system of P. aeruginosa is constitutively expressed by mexAB-oprM.29 mexB promotes the production of biofilms and its hyperexpression in conjunction with the increased efflux pump activity, enabling the bacteria to withstand antibiotic stresses and survive. Antibiotics such as aminoglycosides, beta-lactams, fluoroquinolones, macrolides, and carbapenems are consequently lodged out to the extracellular environment,30 which promotes bacterial growth and enhanced biofilm formation while impeding the would-healing process.31 The primary regulatory loci that control mexAB-oprM expression are mexR, nalC, and nalD. The gene mexR, which is present in the upstream region of the operon-mexAB-oprM, codes for a repressor protein. The repressor protein binds to the operon intergenic region and inhibit transcription by forming a stable homodimer. Overexpression mexAB-oprM is caused due to mexR gene mutations that impair the mexR protein’s capacity to dimerize and bind.32,33 The gene nalC produces the TetR family repressor protein nalC, which binds to armR operon. By blocking the mexR repressor, armR derepresses mexAB-oprM, acting as an anti-repressor.34,35 In addition, the secondary repressor, nalD, belonging to TetR family binds to a region close to the mexA promoter. Hence, nalD dysfunction leads to mexAB-oprM overexpression and leads to increased efflux of antibiotics.36 oprD is the second most prevalent and well-studied porin protein in P. aeruginosa and is responsible for carbapenem antibiotic penetration, particularly imipenem and meropenem. In biofilm outer membrane vesicles, oprD has been detected in large abundance. Carbapenem resistance-causing mutations have been linked to oprD expression downregulation.37,38 Pseudomonas pathogenicity is modulated by the transcriptional regulator mexT. It was discovered to regulate the mexEF-oprN operon in the wild-type strain, but in the nfxC type resistant strain, the effector molecules required to activate mexT are constitutively present, resulting in overexpression of mexT and the mexEF-oprN operon, which contributes to resistance to quinolones, beta-lactams, and chloramphenicol.39 mexT is upregulated and the mexEF-oprN efflux operon is overexpressed in nfxC-type bacteria, where oprD expression is downregulated. This reduces the entry of carbapenems and short basic peptides into the cell.40 Independent of the mexEF-oprN operon, mexT downregulates the expression of the genes coding for rhamnosyltransferase, elastase, and hydrogen cyanide (rhlA, lasB, and hcnB). It also adversely affects the production of homoserine lactone-dependent virulence factors such as pyocyanin and rhamnose.41 DNA gyrase of Pseudomonas contains two subunits, gyrA and gyrB. gyrB has ATPase activity whereas gyrA helps in binding to the double-stranded host DNA. gyrA proteins can be targeted by the quinolones, thereby killing the bacteria through inhibition of DNA replication.42 Further, studies suggest that gyrA possesses 67-106 amino acid motif called Quinolone-resistance determining region, where substitutions in the amino acids may lead to reduced affinity to quinolone drugs.43 In silico analysis of gyrA with amino acid substitutions like T83I and D87N reduced the affinity of gyrA towards ciprofloxacin.42 During functional enrichment analysis, mexA, mexB, oprD, and oprN were found to be enriched in biological processes (GO:0046677, GO:0050896, GO:0042221) being associated with response to antibiotics; mexR and mexT were enriched in biological process (GO:0051172, GO:0010605) associated with negative regulation of cellular and macromolecule metabolic process while nalC and nalD were enriched with molecular pathways (GO:0000976) having transcription regulation activity and biological processes (GO:0051051, GO:0032410, GO:0034763) mostly functioning as a negative regulator of cellular processes. Topological changes in DNA are also responsible for antimicrobial-resistance and gyrA gene was found to be enriched in molecular process (GO:0003918). Cluster 1 revealed the genes which upon mutation (either deletion/ base substitution/overexpression) could confer resistance to carbapenems, quinolones, and beta-lactams along with cationic peptide-like drug molecules in Pseudomonas aeruginosa.
Cluster 2 has 7 nodes and 13 edges with approximately one node connected to 4 neighbours. The clustering coefficient was found to be 0.634 which is comparatively less than cluster C1. The most important genes reported from these clusters are mexC, mexE, oprJ, oprN, and ampC which are enriched in biological processes (GO:0050896, GO:0051301, GO:0009987) and molecular functions (GO:0015562, GO:0042910). MexD is a transmembrane protein transporter, while oprJ is a porin channel. mexC is a periplasmic adaptor protein of the mexCD-oprJ efflux system.43 Unlike mexAB-oprM, mexCD-oprN is not constitutively produced in the cell and is present in a very small amount under normal conditions.44 mexCD-oprN expression is transcriptionally regulated by nfxB gene. Mutations in nfxB gene can lead to the overexpression of the efflux pump, contributing to resistance towards fluoroquinolones, tetracycline, chloramphenicol, and macrolides whereas increased susceptibility to beta-lactams, aminoglycosides, and complement-mediated killing.11 In nfxB mutants, the pump is also associated with the resistance towards Chlorhexidine. The pump is also associated with resistance towards Chlorhexidine, an antimicrobial agent in nfxB mutants.45 mexEF-oprN is also regulated by the lysR family protein mexT, which is located upstream to the MexEF-OprN operon.46 Studies have shown, insertional inactivation of mexT, overexpresses the efflux pump, thereby increasing the efflux of fluoroquinolones and chloramphenicol.46 Moreover, mexT inactivation is associated with the downregulation of OprD, which in turn confers imipenem resistance to the bacteria.47 mexEF-OprN overexpression also leads to the increased production of virulence molecules like rhamnolipids, elastase, and pyocyanin which also leads to quorum sensing mechanism in Pseudomonas aeruginosa.48 In addition to the genes encoding efflux pumps, cluster C2 consists of the ampC gene. Under normal circumstances, this gene is expressed at a low level; however, in cases of mutation, ampC overproduces ampC beta-lactamases, which hydrolyze beta-lactam antibiotics like cephalosporins and cephamycin.49,50
Clusters 3 and 4 revealed just 3 nodes with 3 edges, making them the least dense clusters found in the whole network. ampR, ampD, cat, aph, and fosA are a few of the crucial antimicrobial genes that were obtained from C3 and C4 play a significant role in Pseudomonas resistance. In P. aeruginosa, ampC beta-lactamase production is regulated by genes-ampD, ampR and ampG and ampD. ampR gene transcribes a LysR superfamily DNA-binding protein that has two regulatory properties. In case of β-lactam inducer absence, LysR binds to the pentapeptide protein (UDP-MurNAc) and inhibits ampC transcription. In the presence of β-lactam inducer, the UDP-MurNAc pentapeptide is competitively replaced by the 1,6-anhydro-MurNAc tripeptide, which turns ampR into an activator and causes ampC to produce β-lactamase.51 Thus, ampC mutations can lead to the upregulation of AmpC-β-lactamases production resulting in β-lactam antibiotics resistance50 ampD encodes N-acetyl-anhydromuramyl-L-alanine amidase that preferentially hydrolyzes the 1,6-anhydro-MurNAc peptide, hence suppressing ampC production.52 On the other side, ftsI encodes penicillin-binding protein 3 (PPB3), which is an important target for beta-lactam antibiotics, and also essential for bacterial survival.53 PPB3 exhibits structural changes as a result of ftsI gene mutations or horizontal gene transfer, and so resists the effect of beta-lactams by exporting them through efflux pumps or converting them into a functionally inactive form.54 Genes in Cluster 4 include fosA which is associated with the hydrolysis of Fosfomycin thereby hindering the treatment of P. aeruginosa-associated cystitis,55 cat gene encoding chloramphenicol acetyltransferase promotes chloramphenicol resistance in Pseudomonas,56 aph gene encodes for enzymes that result in acetylation, adenylation and phosphorylation of aminoglycosides.57
Based on cluster analysis, the genes oprD, oprM, oprN, mexR, nfxB, mexB, mexT, mexA, nalD and nalC had the greatest number of gene interactions and could be collectively known as hub genes which are almost like the ones predicted by Miryala et al.22 Because their expressed proteins are specific to the bacterial cell, they could be used as potential drug targets. mexAB-oprM plays a critical role in the development of both inherent and acquired resistance of P. aeruginosa. Overexpression of mexAB-oprM causes increased efflux of many antibiotics, including macrolides, β-lactams, quinolones, and tetracycline.12,58 Similarly, mutations in nalC can cause PA3720-PA3719 overexpression and subsequent upregulation of mexAB-oprM.59 A potentially effective strategy is the inhibition of the mexAB-oprM multidrug efflux operon, which could significantly reduce the carbapenem resistance as well as β-lactam resistance thereby enhancing the susceptibility of multidrug resistance P. aeruginosa. Deficient expression of oprD, an outer membrane porin can lead to baseline resistance to carbapenem antibiotics. Studies show upregulation of oprD expression can be used as a therapeutic strategy against carbapenem-resistant Pseudomonas strains.38,60
Multidrug-resistant (MDR) Gram-negative bacterial infections continue to pose problems for the medical community and complicate antibiotic options. Early commencement of adequate antipseudomonal medication is required for P. aeruginosa infections to avoid the negative consequences of delayed therapy. In the present study, a total of 46 genes were identified to be responsible for P. aeruginosa multidrug resistance. The study found out that the mexAB-oprM efflux pump system along with its regulator genes could be potential therapeutic targets. Repurposing already available clinically approved drugs, identifying plant-based therapeutics, or specifically targeting the exporter protein mexB with antimicrobial peptides could solve a part of the bigger problem of antibiotic resistance caused by Pseudomonas aeruginosa.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DC conceptualized the study, performed literature review and experiments. DC wrote the manuscript. KS critically reviewed and revised the manuscript. Both authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Diggle SP, Whiteley M. Microbe Profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology. 2020;166(1):30-33.
Crossref - Qin S, Xiao W, Zhou C, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7(1):199.
Crossref - Rossi E, La Rosa R, Bartell JA, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol. 2021;19(5):331-342.
Crossref - Banu A, Hassan MMN, Rajkumar J, Srinivasa S. Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: A prospective study. Australas Med J. 2015;8(9):280-285.
Crossref - Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39.
Crossref - Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646-D653.
Crossref - Abril D, Marquez-Ortiz RA, Castro-Cardozo B, et al. Genome plasticity favours double chromosomal Tn4401b-blaKPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019;19(1):45.
Crossref - Park WS, Lee J, Na G, et al. Benzyl Isothiocyanate Attenuates Inflammasome Activation in Pseudomonas aeruginosa LPS-Stimulated THP-1 Cells and Exerts Regulation through the MAPKs/NF-kB Pathway. Int J Mol Sci. 2022;23(3):1228.
Crossref - Yang JJ, Tsuei K-SC, Shen EP. The role of Type III secretion system in the pathogenesis of Pseudomonas aeruginosa microbial keratitis. Tzu Chi Med J. 2022;34(1):8-14.
Crossref - Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa Biofilms. Int J Mol Sci. 2020;21(22):8671.
Crossref - Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int J Mol Sci. 2022;23(24):15779.
Crossref - Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate Specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM Efflux Pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44(12):3322-3327.
Crossref - Okamoto K, Gotoh N, Nishino T. Pseudomonas aeruginosa Reveals High Intrinsic Resistance to Penem Antibiotics: Penem Resistance Mechanisms and Their Interplay. Antimicrob Agents Chemother. 2001;45(7):1964-1971.
Crossref - Pang Z, Raudonis R, Glick BR, Lin T-J, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Shen J, Pan Y, Fang Y. Role of the Outer Membrane Protein OprD2 in Carbapenem-Resistance Mechanisms of Pseudomonas aeruginosa. PLOS ONE. 2015;10(10):e0139995.
Crossref - Feng X, Zhang Z, Li X, et al. Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa. Infect Drug Resist. 2019;12:261-272.
Crossref - Monti MR, Morero NR, Miguel V, Argarana CE. nfxB as a Novel Target for Analysis of Mutation Spectra in Pseudomonas aeruginosa. PLoS ONE. 2013;8(6):e66236.
Crossref - Arzanlou M, Chai WC, Venter H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017;61(1):49-59.
Crossref - Langendonk RF, Neill DR, Fothergill JL. The Building Blocks of Antimicrobial Resistance in Pseudomonas aeruginosa: Implications for Current Resistance-Breaking Therapies. Front Cell Infect Microbiol. 2021;11:665759.
Crossref - Yin R, Cheng J, Wang J, Li P, Lin J. Treatment of Pseudomonas aeruginosa infectious biofilms: Challenges and strategies. Front Microbiol. 2022;13:955286.
Crossref - Srivastava P, Sivashanmugam K. Combinatorial Drug Therapy for Controlling Pseudomonas aeruginosa and Its Association With Chronic Condition of Diabetic Foot Ulcer. Int J Low Extrem Wounds. 2020;19(1):7-20.
Crossref - Miryala SK, Anbarasu A, Ramaiah S. Systems biology studies in Pseudomonas aeruginosa PA01 to understand their role in biofilm formation and multidrug efflux pumps. Microb Pathog. 2019;136:103668.
Crossref - Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2.
Crossref - Yang AF, Huang V, Samaroo-Campbell J, Augenbraun M. Multi-drug resistant Pseudomonas aeruginosa: a 2019-2020 single center retrospective case control study. Infect Prev Pract. 2023;5(3):100296.
Crossref - Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016;4(2).
Crossref - Anusha M, Tejaswini V, Kumar SU, Prashantha CN, Vasudevan K, Doss, CGP. Gene network interaction analysis to elucidate the antimicrobial resistance mechanisms in the Clostridium difficile. Microbial Pathogenesis. 2023;178:106083.
Crossref - Assenov Y, Ramirez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282-284.
Crossref - Dhasmana A, Uniyal S, Anukriti, et al. Topological and system-level protein interaction network (PIN) analyses to deduce molecular mechanism of curcumin. Sci Rep. 2020;10(1):12045.
Crossref - Kello E, Greenberg R, Li W, et al. The Effect of Antibiotic Treatment and Gene Expression of Mex B Efflux Transporters on the Resistance in Pseudomonas Aeruginosa Biofilms. Appl Microbiol. 2023;3(3):709-721.
Crossref - Hassuna NA, Darwish MK, Sayed M, Ibrahem RA. Molecular Epidemiology and Mechanisms of High-Level Resistance to Meropenem and Imipenem in Pseudomonas aeruginosa. Infect Drug Resist. 2020;13:285-293.
Crossref - Alav I, Sutton JM, Rahman KM. Role of bacterial efflux pumps in biofilm formation. J Antimicrob Chemother. 2018;73(8):2003-2020.
Crossref - Suresh M, Nithya N, Jayasree PR, Vimal KP, Kumar PRM. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J Microbiol Biotechnol. 2018;34(6):83.
Crossref - Braz VS, Furlan JPR, Fernandes AFT, Stehling EG. Mutations in NalC induce MexAB-OprM overexpression resulting in high level of aztreonam resistance in environmental isolates of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2016;363(16):fnw166.
Crossref - Tafti FA, Eslami G, Zandi H, Barzegar K. Mutations in nalc gene of Mex AB-OprM efflux pump in carbapenem resistant Pseudomonas aeruginosa isolated from burn wounds in Yazd, Iran. Iran J Microbiol. 2020;12(1):32-36.
- Sadeghifard N, Valizadeh A, Zolfaghary MR, et al. Relationship between the Presence of the nalC Mutation and Multidrug Resistance in Pseudomonas aeruginosa. Int J Microbiol. 2012;2012(1):575193.
Crossref - Morita Y, Cao L, Gould VC, Avison MB, Poole K. naID Encodes a Second Repressor of the mexAB-oprM Multidrug Efflux Operon of Pseudomonas aeruginosa. J Bacteriol. 2006;188(24):8649-8654.
Crossref - Shu J-C, Kuo A-J, Su L-H, et al. Development of carbapenem resistance in Pseudomonas aeruginosa is associated with OprD polymorphisms, particularly the amino acid substitution at codon 170. J Antimicrob Chemother. 2017;72(9):2489-2495.
Crossref - Li H, Luo Y-F, Williams BJ, Blackwell TS, Xie C-M. Structure and function of OprD protein in Pseudomonas aeruginosa: From antibiotic resistance to novel therapies. Int J Med Microbiol. 2012;302(2):63-68.
Crossref - Linares JF, Loipez JA, Camafeita E, Albar JP, Rojo F, Martiinez JL. Overexpression of the Multidrug Efflux Pumps MexCD-OprJ and MexEF-OprN Is Associated with a Reduction of Type III Secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187(4):1384-1391.
Crossref - Kohler T, Epp SF, Curty LK, Pechelre J-C. Characterization of MexT, the Regulator of the MexE-MexF-OprN Multidrug Efflux System of Pseudomonas aeruginosa. J Bacteriol. 1999;181(20):6300-6305.
Crossref - Tian Z-X, mac Aogain M, O’Connor HF, et al. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb Pathog. 2009;47(4):237-241.
Crossref - Sada M, Kimura H, Nagasawa N, et al. Molecular Evolution of the Pseudomonas aeruginosa DNA Gyrase gyrA Gene. Microorganisms. 2022;10(8):1660.
Crossref - Bialvaei AZ, Rahbar M, Hamidi-Farahani R, et al. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb Pathog. 2021;153:104789.
Crossref - Shigemura K, Osawa K, Kato A, et al. Association of overexpression of efflux pump genes with antibiotic resistance in Pseudomonas aeruginosa strains clinically isolated from urinary tract infection patients. J Antibiot. 2015;68(9):568-572.
Crossref - Fraud S, Campigotto AJ, Chen Z, Poole K. MexCD-OprJ Multidrug Efflux System of Pseudomonas aeruginosa: Involvement in Chlorhexidine Resistance and Induction by Membrane-Damaging Agents Dependent upon the AlgU Stress Response Sigma Factor. Antimicrob Agents Chemother. 2008;52(12):4478-4482.
Crossref - Maseda H, Saito K, Nakajima A, Nakae T. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2000;192(1):107-112.
Crossref - Sobel ML, Neshat S, Poole K. Mutations in PA2491 (mexS) Promote MexT-Dependent mexEF-oprN Expression and Multidrug Resistance in a Clinical Strain of Pseudomonas aeruginosa. J Bacteriol. 2005;187(4):1246-1253.
Crossref - Kohler T, van Delden C, Curty LK, Hamzehpour MM, Pechere J-C. Overexpression of the MexEF-OprN Multidrug Efflux System Affects Cell-to-Cell Signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183(18):5213-5222.
Crossref - Lister PD, Wolter DJ, Hanson ND. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin Microbiol Rev. 2009;22(4):582-610.
Crossref - Mirsalehian A, Kalantar-Neyestanaki D, Nourijelyani K, et al. Detection of AmpC-b-lactamases producing isolates among carbapenem resistant P. aeruginosa isolated from burn patient. Iran J Microbiol. 2014;6(5):306-310.
- Torrens G, Hernandez SB, Ayala JA, et al. Regulation of AmpC-Driven b-Lactam Resistance in Pseudomonas aeruginosa: Different Pathways, Different Signaling. MSystems. 20194(6).
Crossref - Schmidtke AJ, Hanson ND. Role of ampD Homologs in Overproduction of AmpC in Clinical Isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother.2008;52(11):3922-3927.
Crossref - Chen W, Zhang Y-M, Davies C. Penicillin-Binding Protein 3 Is Essential for Growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61(1):e01651.
Crossref - Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482-501.
Crossref - Yayan J, Ghebremedhin B, Rasche K. Antibiotic Resistance of Pseudomonas aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. PLOS ONE. 2015;10(10):e0139836.
Crossref - Nitzan Y, Rushansky NM. Chloramphenicol acetyltransferase fromPseudomonas aeruginosa-a new variant of the enzyme. Curr Microbiol. 1981;5(5):261-265.
Crossref - Zeng L, Jin S. aph(32 )-IIb, a Gene Encoding anAminoglycoside-Modifying Enzyme, Is under the Positive Control ofSurrogate RegulatorHpaA. Antimicrob Agents Chemother. 2003;47(12):3867-3876.
Crossref - Pesingi PV, Singh BR, Pesingi PK, et al. MexAB-OprM Efflux Pump of Pseudomonas aeruginosa Offers Resistance to Carvacrol: A Herbal Antimicrobial Agent. Front Microbiol. 2019;10.
Crossref - Cao L, Srikumar R, Poole K. MexAB OprM hyperexpression in NalC type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720 PA3719. Mol Microbiol. 2004;53(5):1423-1436.
Crossref - Kunz Coyne AJ, El Ghali A, Holger D, Rebold N, Rybak MJ. Therapeutic Strategies for Emerging Multidrug-Resistant Pseudomonas aeruginosa. Infect Dis Ther. 2022;11(2):661-682.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.