Tuberculosis (TB) is a significant public health challenge, especially in developing nations. Developing a TB eradication strategy is hampered by the global health concern of drug-resistant (DR) TB. Effective patient treatment, preventing TB transfer and avoiding the upsurge of DR strains depend primarily on the timely and accurate identification of DR TB. Due to inadequate sensitivity, the necessity of trained laboratory personnel, the sluggish growth pattern of Mycobacterium bacilli in culture, and the small number of bacilli that are usually found in extrapulmonary TB samples, TB diagnosis is still tricky in clinical practice. Although mycobacterial culture is the gold standard to identify TB and determine drug resistance, it takes 2 to 8 weeks to develop. Despite their high cost, nucleic acid amplification tests (NAATs) and whole-genome sequencing (WGS) are the commonly employed molecular-based methods for diagnosing and identifying TB. The WHO suggested the GeneXpert MTB/RIF to identify TB and detect resistance to rifampicin. In comparison, numerous molecular techniques were developed, including allele-specific PCR (MAS-PCR), solid-phase hybridization, real-time PCR (RT-PCR) and droplet digital PCR-based technique (DDPCR). This manuscript is intended to overview the current approaches for the phenotypic and genotypic diagnosis of TB disease and identifying resistance to antitubercular drugs depending on recently published articles, WHO and CDC reports, and commercially available diagnostic tools.
Mycobacterium tuberculosis, TB, Drug Resistance, Molecular Techniques, MDR-TB
Tuberculosis (TB) disease, caused by the bacterium Mycobacterium tuberculosis (MTB), is one of the world’s most fatal infectious illnesses. According to a recent estimation, MTB infects around one-fourth of the world’s population and kills 1.5 million individuals annually.1 TB was considered a primary reason for mortality from a single infectious agent until the pandemic coronavirus disease 2019 (COVID-19). Still, according to the World Health Organization (WHO) reports, TB is a worldwide prevalent health issue, with new cases rising from 7.1 million in 2019 to 10 million in 2020. MTB-affected people exhale germs into the air (e.g., by coughing and sneezing), which spreads the disease; therefore, air represents a primary source of illness in humans.2 Thus, the delay in the treatment of the active cases of TB leads to the further spread of the bacterium to others. Accordingly, early and accurate TB diagnosis and drug susceptibility determination are crucial. In the following sections, TB disease, its risk factors, and the available methods for diagnosing and detecting drug resistance, including the traditional and recent techniques, will be comprehensively reviewed.
TB disease, management and health risk factors
TB is a contagious disease caused by the infection with MTB. TB illness usually affects the respiratory system, causing pulmonary tuberculosis (PTB), but it can also affect other body organs causing extrapulmonary tuberculosis (EPTB). Adults are the most affected by TB, around 90% of patients, with more cases among men than women.2 The tubercle bacilli settle in the airways once the person inhales droplet nuclei formed from the evaporation of respiratory droplets. When the immune system cannot control the infection, it will spread locally within the lungs and nearby lymph nodes within three to eight weeks. Patients with PTB will likely experience dyspnea, cough, hemoptysis, irregular chest radiographs, night sweats, weight loss, anorexia and exhaustion. Over the past several years, more cases of EPTB have been reported. EPTB may affect several organs or parts of the body, including lymph nodes, pleura, central nervous system, eyes, musculoskeletal system and both genitourinary and gastrointestinal tracts. Therefore, the clinical manifestations of EPTB cases are determined according to the affected body site. EPTB patients may have stomach discomfort (most common), diarrhoea, infertility, monoarticular joint pain, headache, meningism and lymphadenopathy. However, EPTB patients are less likely to experience the typical clinical features of PTB.3
Macrophages are essential to the human host’s innate immune response to MTB. Still, the ability of MTB to live over extended periods within macrophages in a tubercle granuloma is a crucial pathogenic trait. Macrophages can kill mycobacteria by various mechanisms, including apoptosis, immune-inflammatory reactions and phagocytic activity.4 Conversely, the pathogen can evade and/or resist the host defences, ensuring survival and persistence by delaying phagosome-lysosome fusion, providing a favourable environment for tubercle bacilli survival and reproduction. Correspondingly, macrophages have developed other defence tactics against MTB, including activating autophagy to promote phagosome-lysosome fusion and bacilli clearance.5
The human immune responses to MTB infections may either disrupt bacterial development and eradicate the bacteria or, in most cases, cause latent tuberculosis infection (LTBI). Still, 5% to 15% of LTBI patients can develop active TB disease, including pulmonary or extrapulmonary illness. Active TB usually appears immediately after getting the organism, although it might occur years later in some instances due to debilitated immunity.6 WHO published worldwide programmatic management recommendations for drug-resistant TB in 2014. The recommended treatments included rifampicin (RIF), ethambutol (EMB), pyrazinamide (PZA), and levofloxacin for six months in RIF-susceptible and isoniazid (INH)-resistant TB infections. However, injectable medications, such as streptomycin (STR), are not recommended in patients with this RIF-susceptible and INH-resistant TB.7
Various predisposing factors for PTB, particularly for infection with multidrug-resistant TB (MDR TB), have been identified. These risk factors include two past bouts of PTB and illness that lasted more than 60 days, sputum acid-fast bacilli (AFB) smear score of 3+, and chest radiographs showing cavities or pleural effusion. However, international studies from different countries have identified variable records of these risk factors. Therefore, identifying risk variables and the possible impact of the geographic area are required to establish optimum measures for MDR TB control. In addition, to limit the MDR TB cases, physicians should ask for an early drug susceptibility test (DST) and give appropriate treatment in response to the clinical variables and radiographic chest abnormalities.8
Many reasons constrained the systematic reviews and meta-analyses of MDR TB risk factors. The reason for that is most investigations are a single-regional emphasis, making it impossible to evaluate the impact of the risk influences globally. In addition, the risk factors are studied from the perspective of either the host or the pathogen. Thus, a comprehensive evaluation to identify MDR TB risk factors should be carried out across all geographic regions to enhance international efforts to control MDR TB effectively.9 Many studies from different countries explored MDR TB risk factors, including Indonesia, Vietnam, Russia, and China.10-13 In China, MTB strains are likely to be MDR, linking the MTB Beijing genotype strain with past TB treatment or failure of remedy. In addition, MTB strains in Beijing showed more virulence in animal models, with more widespread tissue damage, faster expansion, and high mortality rates.13 The proposed possibilities for this link are the differences in cell wall construction and greater virulence, resulting in inadequate intracellular drug concentrations and more prolonged, persistent infections.10 Notably, higher incidence rates of TB have been documented in certain people groups than others. There are two categories concerning illness with MTB and getting TB disease: people at high risk for exposure to or getting infected with MTB and people at higher hazard for getting TB disease after infection with MTB (Table 1).14
Table (1):
High-risk groups for tuberculosis infection and disease14.
| People at high risk for infection with MTB [14] |
|---|
|
|
|
|
|
|
|
| People at increased risk for developing TB disease after infection with MTB [14] |
|
|
|
|
|
|
|
|
|
|
Transmission and epidemiology of TB
TB commonly affects the lungs, and it is an air-borne disease. PTB patients exhale the MTB bacteria when they cough, sneeze or spit. To get infected, a person only has to breathe in some MTB bacteria. When a person has active TB disease, the symptoms might be minor for months. Therefore, there might be delays in obtaining treatment, leading to the further spread of the bacterium to others. Consequently, over one year, a person with active TB can infect 5 to 15 persons through intimate contact.15
TB is one of the most widespread illnesses worldwide. Effective management and control of TB disease necessitate a broad knowledge of epidemiology and transmission risk factors.16 MTB is estimated to infect over two billion individuals (almost one-fourth of the world population). Nearly 10 million people have TB disease annually, with 1.6 million deaths. Accordingly, TB is the world’s most significant cause of mortality due to bacterial infection.14 Nearly 70% of TB cases worldwide exist in Southeast Asia and Africa. Even though the total number of cases in Southeast Asia was more prominent, the overall incidence in both areas was comparable (226 per 100,000 in Southeast Asia and 237 per 100,000 in Africa). In 2017, most high-incidence countries were in Southeast Asia and Africa; nevertheless, the percentage of TB cases among HIV-positive people in Africa (27%) was more significant than in Southeast Asia (3%). While the incidence of TB in Europe remains low, the proportion of TB patients with rifampicin-resistant TB (RR TB) or MDR TB was much higher than in the other areas (range = 3.6% – 6.3%).14
Globally, TB frequency decreased by 8.5% from 2015 to 2019. The decrease was from 142 cases of a newly developed active TB illness for every 100,000 people in 2015 to 130 cases per 100,000 in 2019, or an annual reduction rate of roughly 2.1%. The WHO End TB Strategy set its target of a 20% cumulative decrease by 2020. However, TB is still a significant infectious disease that causes death globally, which is higher than the mortality rate of acquired immunodeficiency syndrome (AIDS) caused by HIV infection, compared to a 14% decrease in the death rate from 2015. It fell off the WHO target of reducing TB-related deaths by 35% by 2020, with an estimated 1.4 million TB-related deaths, while 1.2 million among those who were HIV-negative and about 200,000 were positive for HIV).17
Diagnosis of TB disease and identification of MTB
An early, quick, and precise diagnosis of TB, or identification of MTB and determining the antimicrobial susceptibility of infecting strain, are essential for better treatment and management. This section discusses the conventional and advanced methods for TB diagnosis or identification of MTB and DST.
Conventional methods for diagnosis of TB
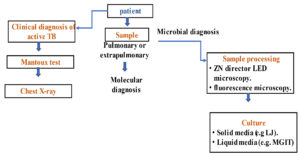
Three main approaches are utilized to diagnose TB: sputum smear microscopy, MTB culture and chest radiography. The smear microscopy sensitivity varies, especially in individuals with other infections like HIV patients. Due to MTB sluggish growth, the traditional culture procedures for MTB isolation, identification and drug susceptibility testing take several weeks. However, prolonged, inappropriate antituberculosis therapy contributes to developing resistance in MTB to available antitubercular drugs, which reduces the number of available treatments and increases the length and expense of TB therapy and morbidity and mortality rates. Therefore, innovative diagnostic techniques are necessary to quickly identify MTB and determine the drug susceptibility profile of the infecting strain. Accordingly, several molecular-based methods have been developed, which are more precise and have shorter turnaround times than the phenotypic methods. However, even though these techniques yield data a few hours after sample collection, MTB culturing and phenotypic susceptibility testing are still required to quantify a particular strain’s sensitivity to certain antibiotics and determine its drug susceptibility.18Acid-fast smear microscopy, nucleic acid amplification tests (NAATs), and culture-based procedures are currently used for the definitive laboratory diagnosis of TB (Figure 1).
Clinical diagnosis of active TB
The individual in contact with a PTB patient for at least two to three weeks is suspected of having the disease.19 However, MTB infection is frequently asymptomatic in healthy persons with no comorbidities affecting their immunity. This TB infection is LTBI, which implies the patient has no active TB infection but might get it at some point (known as TB reactivation). It is assessed that one in every three persons worldwide has LTBI, and 5 to 10% of these people are at risk of TB reactivation. In addition, most people with LTBI will get TB within the first five years after getting infected with MTB. However, when predisposing conditions are present, the likelihood of reactivation rises significantly. When an infected person has any disease threatening his health, such as HIV/AIDS or malnutrition, LTBI escalates to active TB infection.20 PTB can be diagnosed when a patient exhibits the following signs and symptoms: weakness, fever, evening or night sweats, a persistent cough with sputum (phlegm), and chest discomfort. Lung secretions are usually accompanied by blood or hemoptysis, the spitting of blood derived from the lungs or bronchial tubes hemorrhage, weight loss, pale complexion and bright sunken eyes in severe cases.21 A positive Mantoux test confirms the PTB diagnosis. In addition, an advanced PTB is indicated by the appearance of lung lesions (caverns) in a chest X-ray.22
Mantoux test
The Mantoux test (or the tuberculin skin test) assesses a patient’s tuberculin sensitivity. In this test, a tiny amount of a mycobacteria protein extract or purified protein derivative dissolved in glycerol is injected intradermally (5 units of tuberculin in 1 mL) on the forearm. The halo of erythema (skin redness) should be considered when measuring the diameter of the skin induration (thickened, hard skin) between 48 and 72 hours after injection. It is a positive response when the induration diameter is 5 to 15 mm. The positive test indicates that the individual has been exposed to MTB infection, but it does not necessarily mean he is sick. Even though the risk factors or medical history of TB, it is worth considering whether or not the person has the disease. Notably, persons allergic to tuberculin or vaccinated with the BCG TB vaccine may give false-positive results. In contrast, if a PTB person has comorbidities such as AIDS or has compromised immunity, false-negative results are probable.23
Chest X-ray
When a person shows a positive tuberculin test with no apparent symptoms, chest radiographs should be employed to exclude the potential of PTB. The chest X-ray is a critical diagnostic of TB or suspicion of TB patients. This X-ray shows the parenchymal infiltrates, hilar adenopathy, cavitation, nodules, and pleural effusion.24 Also, any aberration in HIV/AIDS patients or other immunocompromised people might suggest tuberculosis. On the other hand, the chest X-ray may seem completely normal. In addition, old cured tuberculosis lesions frequently manifest as pulmonary nodules in the hilar region or higher lobes, with or without fibrotic scarring, volume loss, and bronchiectasis (enlargement of parts of the airways within the lung). Scarring of pleura might also be evident. Notably, slowly growing tubercle bacilli may be seen in nodules and fibrotic scars, potentially leading to active TB. Regardless of age, people with these lesions should be considered high-priority candidates for latent infection therapy if they have positive tuberculin skin test results. However, calcified nodular lesions (calcified granuloma) have a minimal probability of evolving into active TB.25
Artificial intelligence-supported interpretation for chest X-ray radiography
The lack of experienced radiologists and the high inter- and intra-radiology reader interpretation variability have hampered the performance of chest radiography, particularly in areas with an increased incidence of TB and limited access to high-quality medical care.26,27 Lately, artificial intelligence (AI)-aided diagnostics systems have developed and evolved at an unprecedented rate, with many commercial systems available for clinical usage. These systems can potentially address existing limitations of chest X-ray radiography, including reducing human inter-reader variability and reproducibility and providing radiologic services where radiologists are unavailable.28 Many medical image-analyzing AI algorithms based on deep learning and deep convolutional neural networks were being used for radiograph reading at the same time.29AI evaluates radiographs and expresses anomaly scores in computer-aided detection products that suspect TB.30 The diagnostic performance of these computer-aided detection software was comparable to that of a human reader interpreting digital chest radiography, with a sensitivity ranging from 90-92% and a specificity ranging from 23-79%.28,30 WHO has just conditionally endorsed CAD as an alternative to human interpretation of digital chest radiography for TB screening and triage in 15-year-old adults and older.30 The most studied software is CAD4TB (Delft Imaging Systems, Netherlands) version 6. When compared with PCR amplification tests, CAD4TB has been shown to have from 90 to 100% sensitivity and from 23 to 45% specificity in detecting TB.31
Traditional laboratory-based methods to identify PTB
Traditional laboratory techniques for identifying MTB are still widely utilized because they are both feasible and trustworthy. Identifying MBT using AFB staining and fluorescence microscopy and the cultivation and isolation of mycobacteria are diagnostic approaches that allow for a more precise diagnosis of PTB than the clinical-based diagnosis. Because many of the former clinical features of PTB might be mistaken for other illnesses, such as coccidioidomycosis, in addition to a PTB clinical picture, detecting and characterizing MTB in patient sputum is necessary.32 In other words, for an accurate diagnosis, the clinical diagnosis of PTB is essential first, the confirmation or rejection by the laboratory-based methods.22
Identification of acid-fast bacilli (AFB)
Ziehl-Neelsen (ZN) staining technique
It is already long-established that the Gram staining approach does not work well with mycobacteria. Thus, Ziehl-Neelsen (ZN) staining was utilized for detecting AFB, which relies on direct observation of AFB by microscopy. Nonetheless, because certain bacterial species, such as Nocardia genus members, are also AFB, ZN is neither 100 % specific nor a sensitive diagnostic method of MTB.33 More than 120 AFB mycobacteria species are not PTB causal agents. Further, some mycobacteria other than MTB produce atypical pulmonary symptoms.34 The ZN staining process includes the following steps. The culture in suspension, or a liquid biological sample, is placed on a slide and dried, then fixed using a heat air flux. After submerging the fixed bacteria-containing slide in a phenol-carbol fuchsin solution, the smear is heated to allow the dye to penetrate the waxy cell wall of mycobacteria and binds to mycolic acids. After staining, the dyed preparation is washed with water and decolourized byan acid-decolorizing solution (1% hydrochloric acid in isopropyl alcohol or methanol) to remove the red dye from any non-AFB cells. Because of the characteristic waxy lipid coating of AFBs, only AFBs, including mycobacteria, will retain the phenol-carbol fuchsin dye. Malachite green or methylene blue counterstaining is used to stain non-AFB material that failed to hold the initial dye. Afterwards, a microscope will be used to observe the difference between the red AFB and the non-AFB material, green or blue hue. This process may differ owing to the variation in structure across mycobacteria genera. M. ulcerans, for example, has a robust AFB phenotype, but M. leprae has a mild AFB phenotype. M. ulcerans may be decolourized with 3% ethanol, but M. leprae requires 0.5 – 1% sulfuric acid. The staining and discolouration timeframes vary from one Mycobacterium genus to the other. In addition, this technique requires skilled persons.35
Fluorescence microscopy (FM) technique
The fluorescence microscopy (FM)technique is an alternative to ZN staining, employed where the required facilities are available. FM is more rapid than ZN because it is easier to see MTB bacilli. FM is also, as a minimum, 10% more sensitive than traditional light microscopy.36 That is important because the rapid turnaround time can be critical in identifying presumptive TB illness in high-volume laboratories. In addition, other test factors, such as the time length of staining and the fluorescence background, play a role in the effectiveness of laboratory reports.37 FM technique is a semi-automated approach, while ZN must be performed and investigated manually, which requires a skilled operator. However, the ZN method is available in low-resource laboratories than FM, particularly in developing countries, because fluorescence microscopes are significantly more expensive than light-field microscopes. Auramine and rhodamine are the fundamental stains used in FM to identify mycobacteria in biological materials. However, these dyes are non-specific fluorochromes that bind to the mycobacterial wall mycolic acids. The dyed mycolic acid resists discolouration by alcohol-acid solutions. The potassium permanganate as a counterstain avoids non-AFB fluorescence; thus, the test is specific and minimizes the artifacts. AFBs can be seen under an epifluorescence microscope once the staining is complete. When exposed to UV light, AFBs sparkle in yellow or brilliant orange on a dark background. An AFB smear is examined using a fluorescent microscope equipped with a 20 x or 40 x objective and a 100 x oil immersion objective lens to illustrate the morphology of fluorescing organisms.38
Currently, novel technologies for recognizing mycobacteria in clinical samples have been developed based on light-emitting diode (LED) technology. This LED technology is far less expensive than the original auramine and rhodamine systems.39 The LED-based approach involves using a ‘Royal Blue’ LuxeonTMLED to illuminate a standard fluorescent microscope, showing that this type of lighting is adequate for detecting auramine O-stained Mycobacterium spp. The developers argue that their approach is low-cost, low-power, and safe and that LEDs’ dependability makes them a viable replacement for mercury vapour lamps.40 Conclusively, despite using the AFB stain without a mycobacteria culture, AFB visualization has a weak negative predictive value when utilizing ZN staining or FM. Thus, AFB culture should be combined with an AFB stain since the latter has a significantly larger negative predictive value.38
MTB culture
Mycobacterial culture is still the gold standard for diagnosing TB, owing to its high sensitivity and specificity, mainly when molecular techniques are unavailable. MTB culturing enables species identification, drug susceptibility testing, monitoring the response to therapy, andstudying disease epidemiology. However, MTB cannot be grown on standard culture media and must be cultured on specialized media for a long incubation period. Thus, MTB culturing is a costly procedure.41,42 In addition, the sluggish growth rate of MTB prevents early diagnosis and management of the disease. Three types of culture media can isolate and support the growth of MTB from biological samples, including the Lowenstein-Jensen (LJ) medium and its adaption for the Ogawa-Kudoh technique and Egg-based medium.43 Agar-based media (Middlebrook 7H10 and 7H11) and liquid medium (Middlebrook 7H9 and MGITs) are also available. Although, the LJ medium is the commonly used solid medium for growing mycobacteria. The commercially available liquid-based culture systems may be manual, semi-automated, or automated using colourimetric or fluorometric detection techniques. Depending on the design and the source of the material, the Middlebrook 7H9 or the MGIT medium is frequently utilized. The Septi-Chek AFB systems (Becton Dickinson, Sparks, MD, USA), the ESP (Extra Sensing Power), Myco-ESP Culture System II (Trek Diagnostic Systems, USA), and the BacT/ALERT MB (bioMיrieux, Marcy-l’Etoile, France) are an example of these systems. Other systems include the BACTEC MB9000, BACTEC MGIT 960, and 320, and the most widely used is the BACTEC MGIT 960 system.18
Biomarker-based serodiagnosis
Serum biomarker-based technologies
Biomarker-based serodiagnosis might be beneficial for identifying TB in those hard-to-diagnose groups like pediatric TB patients, extrapulmonary TB and HIV co-infected patients. The WHO approved C-reactive protein (CRP) as a screening marker in HIV-infected TB patients. CRP is sensitive but not specific; it may be helpful when combined with other biomarkers.30 Several protein biomarkers have been discovered and could be efficient in TB diagnosis, such as interferon-γ, interferon-γ inducible protein-10, tumour necrosis factor-α, fibrinogen, α2-macroglobulin, matrix metalloproteinase-9, transthyretin and complement factor H. These combined protein biosignatures demonstrated 92% sensitivity and 72% specificity for detecting TB.44 Antibodies, cytokines, and ribonucleic acid (RNA) signatures are the most commonly evaluated host biomarkers for TB diagnosis. Host biomarkers seem sensitive but often have less specificity than pathogen biomarkers.45
Antibodies were the most studied host immune biomarkers. More recent studies showed promising results for antibodies as signatures for TB diagnosis.46 Antibody-based assays are preferred as they are low-cost and require limited operator training. TB-specific IgG4 has been shown to correlate with disease activity and decrease after treatment.47 Several companies are developing innovative antibody-based technologies, including MBio Diagnostics Inc. (Boulder, USA) and TB Biosciences (Bethlehem, USA).48
Another approach lies in the determination of host T cells as biomarkers such as cluster of differentiation (CD) 27, CD38, or CD153, as well as human leukocyte antigen (HLA)-DR and cell proliferation markers such as Kiel (Ki) using flow cytometry.49,50 These markers have repetitively shown sensitivity and specificity levels compatible with the targeted product profile (TPP) defined by the WHO, targets for a confirmatory TB test, particularly for children.51
Other biomarker-based technologies
Detection of TB biomarkers in urine is a noninvasive helpful approach to diagnose TB in patients unable to expectorate sputum as elderly, children and adults without a productive cough.52 The Alere Determine TB LAM Ag fast test (Alere Inc., Waltham, USA) detects LAM and assists in identifying TB in highly immunocompromised people with HIV. The LAMP test is a straightforward strip test that takes only a few minutes to complete. On the other hand, LAM tests have limited sensitivity in individuals with CD4 counts larger than 200 cells/µl, and the WHO does not recommend their use.53 The fact that the LAM test is not specific for MTBC presents a hurdle. Methods for detecting enzymes are also being developed. Global Bio Diagnostics Corp. in Texas, USA, has designed a biphotonic detection technology that uses a fluorescence reporter enzyme to identify live TB bacteria that produce b-lactamase.54
Early MTB infections can be detected using volatile organic compounds (VOCs) in the breath. Using VOCs may aid in diagnosing MTB in HIV patients and children. The possibility of employing VOCs for diagnosing TB was verified in gigantic African pouched cane rats practised to recognize VOCs in TB-infected sputum. Compared to smear microscopy, VOCs performed better in this proof-of-concept investigation.53 Commercial VOC-based solutions are being developed or evaluated, such as Aenose (eNose Company, Zutphen, The Netherlands), BreathLink (Menssana Research Inc, Newark, USA), and TB breathalyzer (Rapid Biosensor Systems, Cambridge, UK). However, a limited amount of data supports it.54
Phenotypic methods of determination of antimicrobial drug susceptibility of MTB
MTB has become resistant to the most clinically available antitubercular drugs. Drug-resistant MTB, particularly MDR MTB (showing resistance to as a minimum RIF and INH) and extensively drug-resistant MTB (XDR MTB), which is also resistant to fluoroquinolones and kanamycin or amikacin or capreomycin.55 The incidence of drug-resistant TB is increasing worldwide, making MDR TB treatment difficult and costly, with many adverse effects on patients. Furthermore, drug resistance is a significant barrier to efficient prevention and control of TB.56 Thus, TB is a global public health problem and one of the top ten mortality diseases worldwide.57
Previously, WHO recommended a universal policy for controlling TB called the directly observed treatment short-course (DOTS) approach.58 The strategy is effective if the MTB strain is not drug-resistant, i.e., sensitive to the first-line anti-TB drugs (INH, RIF, EMB, and PZA). However, the challenge arose when both MDR and XDR expanded globally. Accordingly, the DOTS-plus strategy for patients with MDR TB was established. It involves using the second-line anti-TB drugs (fluoroquinolone and kanamycin/amikacin/ capreomycin), although they have serious side effects.59 Therefore, determining drug resistance of MTB to first- and second-line anti-TB drugs is crucial to both TB therapy and control.60
Knowing each clinical MTB isolate’s entire drug susceptibility profile would allow for more individualized short-course effective therapy while minimizing exposure to unuseful and potentially harmful drug effects.61 Thus, the regular performance of DST for MTB-infecting strains might help prevent and treat drug-resistant TB; even DST for at least RIF is recommended. In addition, all concerned people must perform more actions and efforts to control MDR TB before it becomes a pandemic with terrible consequences.62 However, a significant challenge is facing developing countries to train and hire experienced laboratory technicians and implement suitable diagnostic laboratories with modern devices and a continuous supply of expensive diagnostic kits.38
Determination of drug resistance in MTB by conventional phenotyping methods
Phenotypic methods are the gold standard for the DST of MTB. However, these methods are time-consuming and require specialized laboratories and well-trained personnel to prepare media with various antitubercular drug concentrations, specimen processing and culturing. Three traditional methods use solid media: proportion, absolute concentration (minimal inhibitory concentration (MIC)), and resistance ratio. Phenotypic methods are efficiently employed to determine the sensitivity of MTB to the first-line anti-TB drugs, including RIF, INH, STR, and Ethambutol (EMB) (Figure 2).63,64
MTB proportional DST method
The proportion method compares growing a regulated inoculum of mycobacteria on a drug-free medium to growing on culture media with an anti-TB drug’s critical concentration.64,65 The concept of the MTB proportional DST technique is as follows. Suppose 1% of the inoculum on the drug-containing medium comprises resistant mutants. In that case, these mutants will only grow, and by dividing the number of colony-forming units (CFU) by those appearing on the drug-free medium, the microorganism can be identified as sensitive (≥ 1%). The number of susceptible CFUs on the drug-containing medium should be less than CFUs on the drug-free medium. This method takes around 4 to 6 weeks to get the results. It can be applied directly on the sputum or indirectly using mycobacterial culture. The proportion method using Middlebrook 7H10 agar has been the “gold standard” method for several decades.66
Absolute concentration method
In this method, an inoculum of MTB is cultured on LJ medium, or Middlebrook 7H10 agar has serial dilutions of anti-TB drug and is on drug-free media. Drug resistance is revealed by the lowest drug concentration inhibiting growth, i.e., minimal inhibitory concentration (MIC), which is indicated by obtaining less than 20 colonies by the end of incubation for four weeks.67 In this test, a standardized inoculum is cultured on media containing gradient concentrations of tested anti-TB drugs. The resistance is expressed in terms of MIC. However, the test is significantly affected by inoculum size and the capability of the MTB microorganisms to grow.64
Resistance ratio (RR) method
The resistance ratio (RR), an old method, includes determining the MIC of the tested drug to the MTB isolate in relation to the MIC against the drug-susceptible reference microorganism, such as M. tuberculosis strain H37Rv; both are tested simultaneously.68 Drug resistance is expressed as the resistance ratio of the MIC for the test MTB isolate divided by that for the standard strain. After incubation, growth is verified as the presence of 20 colonies or more, and MIC is the lowest drug concentration where less than 20 colonies are obtained. A ratio of 2 or less designates a sensitive strain, and a ratio of 8 or more indicates a resistant strain.69
Determination of drug resistance of MTB by rapid susceptibility test on solid media
Nitrate reductase assay (NRA)
In NRA, critical concentrations of the drug are tested. Changing the medium colour to pink and purple represents MTB ability to grow and, thus, resistance to the drug. The principle of this assay measures the reduction capability of nitrate to nitrite by MTB in LJ media with the same concentrations of anti-TB drugs used in the proportion method.70 The appearance of a pink or purple colour upon adding Griess reagent to the culture medium indicates the resistance to the drug.71 The NRA test is simple to set, reads the results and gives precise information on drug susceptibility. In addition, technicians get the results quicker than waiting for the visual detection of colonies.72
E-test drug susceptibility testing
E-test comprises strips containing concentration gradients of anti-TB drugs (AB BIODISK, Solna, Sweden). The antimicrobial agent disseminates from the strip into the medium, thus inhibiting the growth of susceptible strains. The MIC is determined, and the isolate is interpreted as resistant or susceptible.73 However, this method may give false results compared to conventional LJ proportion methods. That is because the diffused drugs degrade in the sluggishly growing mycobacteria, resulting in an unclear MIC cut-off point. Additionally, it needs a heavy inoculum (equivalent to McFarland 3 turbidity standards).65,74
Phage-based susceptibility testing
Phage-based susceptibility testing relies on the ability of live drug-resistant MTB to allow the growth of an infecting mycobacteriophage. The mycobacteriophage, a virus that infects mycobacteria, is pre-incubated with the test anti-TB drug before adding MTB, which forms plaques on the agar surface when MTB is resistant.75 Mycobacteriophage-based susceptibility assays are divided into two types; the first is the phage amplified biologically (PhaB) assay, which employs sensitive host bacteria such as M. smegmatis to multiply offspring phages capable of infecting M. tuberculosis.76 FASTPlaqueTBTM, a commercially available kit, offers greater sensitivity and specificity than standard MTB diagnostic methods and can more precisely represent MTB activity, metabolic features, and the proportion of drug-resistant bacteria.77 The second type is Bronx Box, which is based on fluorescent reporter phage and can decrease the time of DST testing to three days for INH, RIF, capreomycin and EMB. Mycobacteriophage-based assays have several advantages, including low cost, reduced detection time, ease of use, infecting only living bacteria and decreased false positive results. Conversely, it might be technically challenging, labour-intensive and have significant failure rates with uninterpretable results.65,75,76
Dio-TK culture system
Dio-TK Culture System, a rapidly automated colourimetric technique, is commercially available to isolate mycobacteria from clinical samples.78 This system contains dye as an indicator, and colour variations in the media reveal the metabolic activity due to mycobacterial growth. The culture media with different antitubercular drugs start red, change to yellow if MTB resists the specific drugs, and turn green if the sample is contaminated. Compared to solid media like LJ-medium, colour shifts appear sooner, thus reducing the time it takes to produce the results by around half (from six weeks to about three weeks).79
Determination of drug resistance of MTB by rapid tests in liquid media
Colourimetric oxidation-reduction methods
The colourimetric assays for detecting drug resistance of MTB are tests that employ oxidation-reduction reactions to produce a colour shift. These tests use the liquid medium 7H9 broth in 96-well microtiter plates.80 In addition, these tests use resazurin, an oxidation-reduction indicator dye, and tetrazolium bromide or 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2 H-tetrazolium bromide (MTT), a redox indicator reduced by dehydrogenases produced when cells are still alive.81 In these tests, MTB isolates are inoculated into 7H9 broth enriched with the test anti-TB drug and incubated at 37°C for seven days. Then, Alamar blue or resazurin reagent is added to wells. If MTB grows in the presence of the drug, it will reduce the blue reagent to a pink dye that may be seen with the naked eye or measured with a colourimeter. The blue-to-pink colour change in a drug-containing well suggests the existence of resistant MTB.82 The colourimetric methods have been evaluated in many studies with reported sensitivity and specificity ranges of 94 – 100%, with likely getting the results within eight days.83 However, colourimetric methods pose a significant bio-safety risk to laboratory staff.65
Microscopic-observation drug-susceptibility assay (MODS)
MODS assay is an approach for directly detecting MTB and MDR MTB in sputum or indirectly in mycobacterial broth culture. This assay uses an inverted microscope to observe MTB distinctive cord development in microscopically visible liquid media.84 Several studies have recommended this technique as relatively quick and cost-effective for early identification of MTB and drug resistance directly from sputum specimens.84,85 Some studies on MODS assay revealed a range between 92% to 100% of sensitivities and specificities for determining the sensitivity to each RIF, INH, and fluoroquinolone. The DST findings are obtained within seven days using this method.86
BACTEC radiometric method (BACTEC-460)
The BACTEC 460 (Becton Dickinson, Sparks, Maryland) is a radiometric method that detects 14CO2 for indicating mycobacterial growth. The Bactec vials contain Middlebrook 7H12 medium and 4C labelled palmitic acid substrates as the sole carbon source. When mycobacteria grow,14CO2 is produced as a metabolic end-product instead of CO2. The gas is removed and analyzed by the BACTEC 460 TB SYSTEM. The radioactive 14C released amount is converted to a numerical value of Growth Index (GI), which indicates a positive mycobacterial growth if it is ten or more. Although this test’s sensitivity and specificity are significantly high, it uses radioactive carbon, whose half-life is 5,000 years, requiring complicated and costly disposal. The BACTEC system can be used in level II laboratories and for particularly diagnosing the cases of extrapulmonary and smear-negative TB.71,87
BacT/alert 3D
The BacT/ALERT® 3D System (bioMe’rieux, Marcy Etoile France) is a nonradiometric, liquid-based automated test that is developed for mycobacteria primary isolation and DST. This test is performed in a tube using a liquid emulsion sensor and a complex computer algorithm that relies on detecting carbon dioxide decrease.88 Growing bacteria emit CO2, which interacts with the sensor, causing a colour shift monitored by a reflectometric detecting unit housed within each incubation drawer of the instrument. Some disadvantages of the BacT/alert 3D system include susceptibility to contamination, longer turnaround time, require expensive and non-robust machine, along with the complications and hazardousness of procedures.87
Mycobacterial Growth Indicator Tube (MGIT)
MGIT (Becton Dickinson, Sparks, Maryland, USA) detects mycobacterial growth by fluorescence. The test tube contains a modified Middlebrook 7H9 medium and a fluorescence quenching-based oxygen sensor at the bottom of the tube.89 The indicator fluoresces under UV light when the bacteria grow and use oxygen, and the proliferation in the presence of the test drug reveals resistance.90 When the growth unit (GU) in growth control hits 400, the GU values of the drugs containing tubes are observed immediately. If the GU of the drug tube is less than 100, the isolate is susceptible; if the GU of the drug tube is 100 or above, the isolate is resistant to the tested drug.91,92 The manual and automated MGIT 960 systems have demonstrated a great connection to traditional DST techniques for rapidly identifying resistance to first and second-line anti-TB drugs.93
Limitations of conventional methods for TB diagnosis and drug susceptibility testing
Sputum smear microscopy is the principal diagnostic technique for detecting PTB in many high-TB-burden countries.94 Traditional sputum smear microscopy is inexpensive and requires minimal laboratory infrastructure; competent technicians can read 20 – 30 slides daily. However, this diagnostic method has a low sensitivity (20 – 80%) and limited accuracy, particularly among HIV and immunocompromised patients.95 Unlike traditional staining, fluorescent dyes allow for easier detection at lower magnification powers, FM is around 10% more sensitive than light microscopy.96 However, conventional FM microscopes areexpensive and require special laboratory requirements, such as a dark room. Light-emitting diode (LED) microscopes, such as the Primo Star iLED and the CyScope® TB Fluorescence Microscope, have been developed to overcome these issues.94
Because various respiratory illnesses can induce X-ray deviations, chest X-rays have a high sensitivity (80 – 95%) but a low specificity (70 – 75%) for TB disease. As a result, a chest X-ray is an effective preliminary screening method that can identify suspected people who need further investigation to confirm TB. In chest radiography, automated reading algorithms have become increasingly significant. The utilization of digital radiography, as well as computer-assisted diagnostics, is revolutionizing radiographic instruments.97 Computed radiography uses a specific plate that produces an analogue signal, allowing the image to be digitized and stored. At the same time, direct digital radiography uses a digital detector.98 Although traditional radiography has lower costs, digital radiography is more cost-effective in the long run because it decreases recurrent expenditures and reduces reagent consumption and radiation exposure. Furthermore, digital radiography provides more convenient data preservation and transfer, but it should be emphasized that sufficient data transmission capability is required.94
Mycobacterial cultures are the gold standard for TB diagnosis and DST. MTB cultures are highly sensitive and can detect low numbers of bacterial cells. However, one of their significant problems is that culture-based TB diagnosis takes a long time. In addition, skilled personnel are required to process and monitor cultures. Further, biosafety level 3 settings are needed for culturing specimens suspected to have MTB.94 Compared to solid cultures, in which MTB takes a long time to grow, the liquid culture enables faster results, which may be produced within 10 to 14 days and up to 10% higher sensitivity. Moreover, the resistance to RIF, INH, PZA, EMB, and STR may all be identified using liquid cultures.99 Notably, the WHO has recommended automated liquid culture systems for DST, including the Bactec MGIT (BD Diagnostics, New Jersey, USA), mycobacterial growth indicator tube, scan-processed MGIT sputum bottles and BacT/Alert 3D (bioMיrieux, Marcy l’Etoile, France). However, liquid culture systems are more sophisticated and costly than solid culture systems and can be prone to contamination.94,100
Molecular-based approaches for diagnosis and detecting drug susceptibility of MTB
There are over 170 known mycobacterial species, most related to the nontuberculous mycobacteria (NTM) group, with some belonging to the MTB complex (MTBc), sharing 99.9% genomic similarity.66 Restriction endonuclease analysis and nucleic acid hybridization were employed for TB diagnosis. These approaches provided a strategy for rapidly diagnosing TB disease by identifying particular nucleotide sequences in the organism utilizing probes of labelled nucleic acid sequences.101,102 The FDA approved the Accuprobe MTBc test in 1990 for identifying MTBc isolated in culture. It was one of the most incredible accuracies ever reported of a molecular technique used for mycobacteria identification from the culture.103 The DNA-labeled probes hybridize with mycobacteria’s RNA, giving a stable DNA-RNA construction. These hybrids are detected by light emission on a luminometer after adding a selection component to distinguish hybridized probes from non-hybridized ones.104
Phenotypic drug susceptibility testing requires mycobacteria to be in an active growth phase; thus, results are obtained within at least two to three weeks if liquid culture media is utilized and up to four to eight weeks when grown on solid media. This process will delay the patient’s best therapy choice. Consequently, delayed results by conventional methods will likely contribute to acquiring further drug resistance and spreading drug-resistant MTB strains; thus, quick and dependable techniques for detecting drug-resistant TB are essential. The progress in molecular biology techniques and the knowledge of drug resistance on the molecular levels have provided advanced approaches for rapidly detecting resistance in MTB. The molecular-based approaches, detecting changes in genes coding for anti-TB drug resistance, could provide results within 24 to 48 hours.94,99,101
The clinical use of NAATs and whole-genome sequencing (WGS) has grown remarkably in recent years. However, owing to the high cost of WGS (Table 2), NAATs are the most commonly used. Indeed, several laboratories routinely employ NAAT-based methods for species and drug resistance identification from cultures or directly from specimens. The ribosomal 16S rRNA-targeted PCR followed by DNA sequencing is a gold standard for detecting MTB.105 The 16S rRNA includes species-specific hypervariable areas that benefit specific species identification. Other targets include the 16S-23S rRNA intragenic region and anti-TB drug resistance genes, including rpoB, gyrB, hsp65, recA and sodA.106,107
Table (2):
Principles and features of first-generation and next-generation sequencer
First-generation sequencer |
Next-generation sequencer |
|---|---|
· Essentially, automated electrophoresis devices that track the movement of labelled DNA fragments make up the first generation of DNA sequencers.134 |
· Rapid data production by parallel sequencing of large volumes of DNA utilizing many techniques.135 |
· Able to genotype genetic markers if just a DNA fragment’s length is known.136 |
· Create diverse tactics that combine genomic alignment and assembly approaches, template preparation, sequencing and imaging.137 |
Collectively, the sensitivity, specificity and capability to precisely identify MTB infections are all improved by molecular-based techniques. Several molecular-based methods have been developed for diagnosing TB and determining drug susceptibility, which have sped up turnaround times and allowed point-of-care testing.18 The following sections will outline the available molecular tests for TB diagnosis and determination of MTB drug susceptibility, addressing their benefits and limitations.
Molecular methods used for the identification of MTB
Nucleic acid amplification tests (NAATs)
The NAATs have been utilized as a practical alternative since the traditional phenotypic diagnosis of TB has several problems.108 Because of the NAAT systems’ quick turnaround times, testing and treatment can be started in the same visit, thus reducing the number of patients lost to follow-up.109 Most NAATs identify the MTB complex bacteria, which consists of a group of closely related species, by detecting the mycobacterial insertion element IS6110 (Table 3).110
Table (3):
Main characteristics of the molecular assays used for the detection of drug-resistance-associated mutations in M. tuberculosis Technology
| Technology: 1-Line probe assay (DNA•STRIP technology) | ||||||
|---|---|---|---|---|---|---|
| Test specification | Manufacturer | Detection Target | Mechanism | Time | Advantages and Limitations | Reference |
| INNO-LiPA Rif TB Assay | Innogenetics, Zwijndrecht, Belgium | MTBs and rpoB for RIF resistance | It has 10 probes identifies MTBc, 5 specific regions of the rpoB gene in wild types, and 4 detect mutations | 2 to 3 days | – It accurately detects resistance to RIF – Less sensitive for the detection of M. tuberculosis complex – Less sensitive when applied to clinical specimens |
[185] |
| GenoType MTBDRplus assay
VER 1.0 and VER 2.0 |
Hain LifeScience GmbH, Germany | MTBs rpoB for RIF resistance,
katG and inhA for INH resistance |
– 12 probes for rpoB gene (8 for wild type and 4 for mutant type)
– 3 probes for katG INH (1 for wild type and 2 for mutant type) – 6 probes for inhA promoter (2 for wild type and 4 for mutant type) |
2 to 3 days | – It is highly sensitive and specific for early detection of MDR-TB. However, – The diagnostic performance of this molecular assay in direct smear negative sputum sample is low and showed a high level of invalid results for the detection of M. tuberculosis and its resistance to RMP and/or INH |
[104, 186] |
| GenoType MTBDRsl assay VER 1.0 and 2.0 | Hain LifeScience GmbH, Germany | – MTBs
– rss gene for SLID, – gyrA gene and gyrB gene for FQs – embB gene for EMB (IN GenoType MTBDRsl assay VER 1.0 ONLY) – eis promoter for KAN SLID (second-line drugs) |
Each strip of GenoType MTBDRsl assay VER2.0 has 9 probes for gyrA,
3 probes for gyrB 4 probes for rss 4 probes for eis promotor |
48 h | – GenoType MTBDRsl VER2.0 had variations in the overall specificity and sensitivity for identifying XDR isolates. – Additionally, results for smear-negative sputum samples were difficult to interpret |
[106, 187, 188] |
| AID TB Resistance LPA | Aid Diagnostika GmbH | MTBs rpoB for RIF resistance, katG and inhA for INH resistance
rpsL and rrs 500 region for STR rrs 1400 region for SLID, gyrA for FQ. embB for EMB |
6 probes for rpoB
2 probes for katG 2 probes for inhA 5 probes for rpsL and rrs 500 region fo 5 probes for rrs 1400 region 7 probes for gyrA 5 probes for embB |
48 h | – All AID modules have moderate sensitivity and reasonable specificity for the detection of M. tuberculosis in sputum samples – However, the high proportion of invalid tests, especially concerning resistance testing, is problematic and hampers the evaluation |
[187, 189] |
| Nipro NTMC/MDRTB | Nipro Co., Osaka, Japan | MTBs rpoB for RIF resistance, katG, and inhA for INH resistance | 9 probes for rpoB
6 probes for katG 2 probes for inhA |
24 h | Good sensitivity and specificity for detection of MDR-TB in smear-positive samples only | [107] |
| 2- Real-time PCR | ||||||
| The Xpert MTB/RIF | Cepheid, Sunnyvale,
California, USA |
Semi-quantitative nested real-time PCR
(Molecular beacon) -MTBs –rpoB for RIF resistance |
Amplify the rpoB gene’s 81 bp hotspot region. | 2 h | – It is a completely automated technology -High sensitivity and specificity for smear-positive samples but – Moderate sensitivity and specificity for smear-negative samples – Inability to test for and detect INH resistance – High cost – False-positive findings owing to silent mutations |
[159] |
| Xpert MTB/RIF Ultra system | Cepheid, Sunnyvale,
California, USA |
Semi-quantitative nested real-time PCR
(Molecular beacon) -MTBs -rpoB for RIF resistance |
As a replacement for Xpert
MTB/RIF due to its greater sensitivity in detecting MTB and omitting the silent rpoB mutations Q513Q and F514F |
Less than 90 min | – Improve detection of MTB complex and rpoB silent mutations – Low specificity in patients from high-incidence countries – Inability to test for and detect INH resistance |
[163] |
| The Genedrive MTB/RIF ID Kit | Epistem, United Kingdom | Asymmetric real-time PCR (Highlighter Probes)
MTB and RIF-resistant TB |
Detects mutations in a rpoB 81-bp hotspot area of at codons 516, 526, and 531 | Less than 1 h | – Fast results and cheap cost – Accessible to low-income communities – Inability to test for and detect INH resistance – Do not accommodate all RIF mutations |
[39, 165, 191] |
| Abbott RealTime MTB RIF/INH | Abbott-RIF/INH | Real-time PCR | rpoB for RIF resistance and katG and inhA upper promoter region for INH | 2 h | Good assay for the diagnosis of TB and DR-TB | [166, 192] |
| Anyplex II MTB/MDR/XDR | (Seegene, South Korea | Multiplex real-time PCR dual-priming oligonucleotides and tagging oligonucleotide cleavage and extension for detection of RIF, INH, SL, FQ resistance | 18 mutations in rpoB
7 mutations in the katG and inhA promoters, 7 mutations in gyrA 3 mutations in the rrs and eis promoters |
2-3 h | – High specificity and moderate sensitivity for detection of MTB and drug resistance to TB | [67, 192] |
| FluoroType MTBDR | Hain Lifescience, Nehren, Germany | -Linear-after-the-exponential PCR (LATE-PCR) with probes using lights-on/lights-off detection technique.
– For the detection of RIF, INH resistance |
Can detect rpoB mutations, katG mutations, and inhA mutations in one single tube. | up to 96 samples in a closed system within 3-4 h from DNA extraction | – High sensitivity and specificity for the detection of rifampin and isoniazid resistance less hands-on hours, quicker outcomes, automated interpretation – The lower possibility of DNA contamination |
[169, 170] |
| BD MAX MDR-TB assay (BD MAX) | Becton, Dickinson and Company, New Jersey, USA) | Real-time PCR TaqMan probes for the detection of RIF, INH resistance |
rpoB and katG genes and the inhA promoter | 4 h for testing 24 samples | – High sensitivity and specificity for the detection of RIF and INH resistance in short time – It does not identify new mutations and the sample needs to be sequenced. |
[173] |
EMB, ethambutol; FQ, fluoroquinolones; INH, isoniazid; KAN, kanamycin; MDRTB, multidrug-resistant tuberculosis; MTB, M. tuberculosis; MUT, mutation; NTM, nontuberculous mycobacteria; RIF, rifampicin; SLID, second-line injectable drugs; STR, streptomycin; TB, tuberculosis; h, hour.
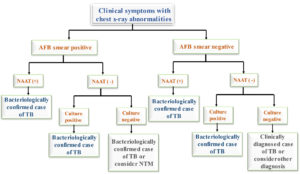
The AFB smear-positive and AFB smear-negative sputum specimens can both have MTB ribosomal DNA detected by NAATs.111 The NAATs demonstrated high sensitivity in patients with positive sputum smears and between 61% and 76% sensitivity in those with negative sputum smears.109 The GeneXpert/RIF MTB test is the NAAT system approved by the WHO.112 The other NAATs, Amplicor M. tuberculosis Test (Roche. Molecular Systems, Inc) and Amplified M. tuberculosis Direct (MTD; Gen-Probe, Inc), were approved by the FDA for evaluating respiratory AFB smear-positive samples.113-115 The available NAATs include the M. tuberculosis detection device based on loop-mediated isothermal amplification, the cross-priming amplification-based TB diagnostic system, and the Genedriv® Mycobacterium tuberculosis iD.116-118
A positive AFB smear with a positive NAAT would suggest active TB in areas with multibacillary illnesses with a high mycobacterial burden. Still, a positive AFB smear with a negative NAAT without inhibitors would indicate nontuberculous mycobacterial (NTM) disease.119 If the culture in this situation showed positive, the doctor might classify the patient as having TB.120 NAAT might be used for patients suspected of MTB infection with an AFB smear-positive to assess their TB stage.121 Significantly, starting anti-TB medication would depend on the clinical decision while awaiting culture findings if the NAAT test is positive, and the result of the AFB smear is negative.122 Centres for Disease Control (CDC) recommends testing other samples with NAAT if the sputum is smear-negative and the NAAT is also negative.123 However, if the culture shows MTB growth, the patient might be considered to have PTB. The NAAT method for diagnosing TB is shown in Figure 3 and follows the WHO and the CDC recommendations.124 Limiting the NAAT to cases with positive smear findings may be more effective in areas with low rates of TB cultures. On the other hand, a NAAT should be employed in patients with negative smear results in high TB rates areas.125 According to a meta-analysis of 125 trials, NAATs should not replace standard tests for diagnosing PTB.126
Figure 3. The role of nucleic acid amplification tests (NAATs) in the diagnosis algorithm of TB. TB: tuberculosis, NAAT: nucleic acid amplification test, NTM: nontuberculous mycobacterium.109
NAAT-based TB diagnostic device: GeneXpert MTB/RIF assay
The real-time PCR (RT-PCR)technology generally uses fluorescent TaqMan or Molecular Beacons probes for concurrent target amplification and detection. The probes have a fluorochrome and an inhibitor that is closely spaced from one another. As a result, separation occurs when a link forms with the amplified target and the emitted light is detected, equivalent to the quantity of DNA in the sample.66 This approach has the advantage of quantification of the microorganism in the clinical sample, in addition to a lower risk of contamination and a quicker turnaround time compared to Line probe assays (LPAs) due to the removal of the hybridization phase.127 The sensitivity and specificity of RT-PCR for smear-positive TB cases are 100% and 99 %, respectively, while for smear-negative cases are 67% and 99%, respectively.128 RT-PCR, however, has certain limitations, as it needs costly, specialized equipment, skilled personnel, and an amplified target size range to identify multiple mutations effectively.127
The GeneXpert MTB/RIF assay is based on NAAT, particularly RT-PCR, with results available within two hours. It is a completely automated technology that requires no human intervention other than inserting the sample into the cartridge.129 GeneXpert MTB/RIF test was first suggested in 2010 for adult sputum specimen-based diagnosis of PTB.GeneXpert MTB/RIF test can identify MTB-specific DNA sequences in sputum samples.130 More than 80% of smear-negative and 99% of smear-positive TB patients could be diagnosed by a single GeneXpert MTB/RIF test performed directly using the sputum sample.131,132 GeneXpert MTB/RIF assay can be utilized as an additional test after microscopic MTB examination, replacing AFB smear microscopy and identifying MTB in both AFB smear-positive and smear-negative culture-positive cases. It can also detect MTB in pleural fluid, a lymph node biopsy or fine-needle aspiration, gastric juice, cerebrospinal fluid and tissue samples. Still, GeneXpert MTB/RIF assay can be used as the initial test to diagnose PTB-suspected people.133
The GeneXpert MTB/RIF test can be utilized as an initial or a further test after a negative AFB smear microscopy result.134,135 Notably, despite the high cost, the median time to therapy for AFB smear-negative TB has decreased from 56 days (range 39 – 81 days) to 5 days (range 2 – 8 days) owing to the GeneXpert MTB/RIF test implementation.136,137 However, the quick sputum-based test would be more cost-effective than the traditional sputum smear microscopy.138,139 Although the proficiency in lowering the early missing to follow-up patients and shortening therapy period commencement, the introduction of GeneXpert MTB/RIF would not be able to enhance controlling drug-resistant TB.140
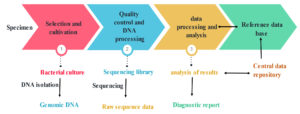
Whole-genome sequencing (WGS) as a diagnostic approach for TB
The investigation of genetic markers of organisms for diagnosis, therapy and follow-up infection prognosis has become possible through advances in microbial genomics.141 As MTB lineages spread among people, whole-genome sequencing (WGS) is developing into an accessible and inexpensive technique for detecting microevolution within those lineages.142 Figure 4 shows the outlines of the WGS process from the specimen collection to the diagnostic report. There are two DNA sequencers: first-generation and second-generation sequencers (widely known as the next-generation sequencer [NGS]). The first-generation sequencer has a low cost (about $65 per bacterial genome) and remarkable throughput but relative slowness. The second generation can sequence many genomes in less than a day but has a lower throughput and higher cost (the IlluminaR MiSeq costs around $150 per genome).143 Table 2 describes the principles of first-generation and next-generation DNA sequencing techniques.
Figure 4. Whole-genome sequencing workflow of MTB from specimen processing until the diagnostic report
Molecular-based determination of drug susceptibility of MTB
TB has become resistant to clinically used drugs to cure the disease. MTB strains likely gain drug resistance during patient treatment because of delayed diagnosis, unsuitable medicine, or poor adherence to the treatment regimen. Thus, the rapid detection of TB helps timely access to the proper treatment, decreases transmission rates and improves treatment outcomes.100 The progress in molecular biology techniques and knowledge of drug resistance on the molecular levels have provided advanced approaches for rapidly detecting drug resistance in MTB.144,145
Single nucleotide polymorphisms (SNPs) within the MTB chromosome are the primary means by which MTB strains develop drug resistance owing to insertions or deletions events.146,147 Mutations in one or more genes have been identified for anti-TB drugs. Moreover, each mutation is associated with varying levels of drug resistance. For example, rpoB gene alterations, predominantly in an 81 bp hotspot area, are responsible for 97 % of resistance to RIF. Mutations in the katG and inhA genes or inhA promoter region cause MTB to acquire resistance to INH.148 RIF resistance in MTB can be considered a surrogate marker for MDR strains; consequently, the molecular-based method should detect the mutations leading to RIF resistance, which indicates MDR TB.149,150
In 2020, the WHO recommended using molecular assays for rapidly diagnosing MDR TB.151 Many molecular-based techniques have been developed to simultaneously identify MTB and its drug resistance to RIF and INH. These assays detect resistance-related mutations in genes encoding resistance of MTB isolates. In addition, they have the advantage of yielding results in one to two days and may be employed directly for smear-positive sputum and other clinical specimens.152 Therefore, several highly sensitive and specific nucleic acid-based assays that identify mutations linked to resistance to antitubercular drugs have been designed and developed; thus, that allows for low-cost, reliable, straightforward, and quick results despite the high-cost laboratory infrastructures (Table 3).153
Solid-phase hybridization or line probe assays
Line probe assays are a group of modern DNA strip tests that utilize PCR and reverse hybridization assays. These tests include several steps: extracting DNA from both mycobacterial isolate or clinical specimens, then PCR-based nucleic acid amplification and hybridizing PCR products with oligonucleotide probes immobilized on a strip. Completely, hybridization is indicated by developing a coloured reaction on the strip as lines where the probes are located (thus, the term “line-probe”).100,154
The commercially available solid phase reverse hybridization assays include the Line Probe Assay (LiPA) (INNO-LiPA Rif TB Assay, Innogenetics, Zwijndrecht, Belgium) for identifying MTB and screening for RIF resistance.INNO-LIPA has ten probes on its strip, one identifying MTBc, five highlighting specific regions and four detecting mutations of the rpoB gene. It showed high sensitivity when used on mycobacterial culture but less in direct specimens. Only the rpoB flashpoint area of mutations (codon 509 to codon 534; Asp516Val, His526Tyr, His526Asp, and Ser531Leu) is examined.155 A meta-analysis suggested that the LiPA assay is highly sensitive and specific for detecting RIF-resistant MTB. In this analysis, 12 of 14 published studies showed a sensitivity> 95% with a specificity of 100% when the assay was employed on isolates but was less sensitive when applied to clinical specimens. However, four studies on clinical samples revealed 100% specificity, but the sensitivity ranged from 80% to 100%.156 However, another study that evaluated LiPA for determining RIF resistance in 420 sputum samples revealed 99.6% agreement between culture-based identification and LiPA. The study addressed that with satisfactory DNA extraction, LiPA allows rapid detection of resistance to RIF when employed directly on sputum.157
AID TB Resistance LPA (Aid Diagnostika GmbH) is used to identify resistance to first- and second-line anti-TB drugs in clinical specimens and culture via three modules. Module 1 detects rpoB, katG, and inhA promoter genes; module 2 examines rpsL and rrs to identify aminoglycoside resistance (STR, AMK, CAP), and module 3 examines gyrA and embB to find FQ and EMB resistance.158 The three modules comprise both wild-type and mutant probes. The AID TB Resistance LPA showed remarkable specificity and sensitivity for detecting resistance to RIF, INH, STR, FQs, and second-line injections. (ranged from 90% to 100%), but with reduced sensitivity to detect resistance to EMB of 72.9 %.159
WHO suggested GenoType MTBDRplus assay (Hain LifeScience GmbH, Germany) and Nipro NTMCMDRTB (Nipro Co., Osaka, Japan)for the early screening for drug resistance in sputum smear-positive samples.160 The GenoType MTBDRplus assay identifies rpoB mutations in addition to INH resistance by examining the inhA gene (which encodes low-level resistance) and the katG gene (which encodes high-level resistance). It has probes for wild-type areas of susceptible strains and their associated mutations. WHO approved GenoType MTBDRplus assay in 2008 and may be utilized on clinical samples directly or cultures with high sensitivity and specificity for RIFresistance (88.2% and 89.5%, respectively) and INHresistance (91.7 and 97.2%, respectively). It delivers results in 48 to 72 hours.161,162
Several studies performed on MTBDRplus VER2.0 assay have shown high sensitivity (83.3% to 96.4%) and specificity (98.6% to 100%) for detecting MDR isolates in smear-positive specimens.100,162 GenoType MTBDRsl VER2.0 can detect resistance to second-line anti-TB drugs such as CP, AK, and KM (rss gene), fluoroquinolones (gyrA and gyrB genes), and KAN (eis promoter gene).66 It has variable sensitivity and specificity when screening for resistance to second-line injectable drugs. Still, it has excellent sensitivity and specificity (91% to 100%) for identifying resistance to fluoroquinolone.163 Although; there are variations in the overall specificity and sensitivity of GenoType MTBDRsl VER2.0 for identifying XDR MTB isolates. Additionally, results for smear-negative sputum samples were difficult to interpret.104 In addition, Nipro NTMCMDR-TB detects MDR-TB by targeting the genes rpoB, katG, and inhA and can differentiate four important Mycobacterium species (MTB, M. avium, M. intracellulare, and M. kansasii) that cause diseases in humans.164 Solid-phase hybridization assays are reasonably simple, rapid and straightforward. Yet, fundamental expertise in molecular-based methods is necessary. In addition, the test sensitivity varies with the amount of DNA in the sample, and the presence of inhibitors might lead to false-negative results. Furthermore, LPA methods target only the primary mutation; thus, the specificity and sensitivity may vary if mutations happen in target regions.160
Multiple allele-specific PCR (MAS-PCR)
MAS-PCR can concurrently detect common mutations in the RIF resistance gene, thus reducing the cost and practical procedures. MAS-PCR aims to detect RIF resistance-conferring mutations in codons 435, 445, and 450 of the rpoB gene using pure DNA extracted from mycobacterial culture.165,166 These point mutations have been detected according to the wild-type sequences of strain H37Rv. The 3 prime ends of allele-specific primers pair with the nucleotide base of the respective codon. If the investigated strain carries the targeted position as the wild-type, the allele-specific region can be amplified by PCR to produce a visible DNA fragment. In contrast, if the target DNA sequence contains a mutation, it will block the PCR amplification, giving no DNA band.165 In a study evaluating the effectiveness, MAS-PCR showed a sensitivity of 88.3% and 100% specificity compared to the DST by proportion method.166 In another study, the three RIF resistance-related mutations were identified by MAS-PCR with 97.9% sensitivity and 100% specificity compared to the standard DST. MAS-PCR could be a suitable method for routinely detecting RIF-resistant MTB, providing a fast, cost-effective and straightforward method.167
RT-PCR-based techniques for detecting drug resistance
The GeneXpert MTB/RIF (Cepheid, Sunnyvale, California, USA) is recommended by WHO as a rapid molecular test for identifying MTB complex and screening for RIF resistance in MTB isolates.160 The Xpert MTB/RIF test employs semi-quantitative nested RT-PCR technology to detect the rpoB gene 81 bp hotspot region to identify mutations linked to RIF resistance.168,169 The WHO recommended this approach as the primitive diagnostic step in high-risk regions of MDR TB.170 In addition, Gonחalves et al. study findings confirmed that RT-PCR detects RIF resistance with a sensitivity of 99% and specificity of 100% in less than four hours.171 However, some studies reported false-positive results by GeneXpert MTB/RIF owing to silent mutations and false-negative results because it was incapable of identifying RIF resistance-related mutations beyond the hotspot region.168,172
The next-generation Xpert MTB/RIF Ultra system (Cepheid, Sunnyvale, California, USA) includes two more targets for MTB complex identification (IS1081 and IS6110) with a tenfold improvement in analytical sensitivity.173 Xpert MTB/RIF Ultra undertakes Xpert MTB/RIF limitations by omitting silent rpoB mutations Q513Q and F514F.174 The next-generation Ultra system was recommended as a first diagnostic test for all patients, whatever the age, with TB symptoms and for testing extrapulmonary specimens, including tissue and lymph node samples and cerebrospinal fluid.160
The Genedrive MTB/RIF ID Kit (Epistem, UK) can detect MTB and RIF resistance in MTB from raw sputum samples. The system employs a straightforward paper-based DNA extraction process, asymmetric RT-PCR, and a patented hybridization probe technology (Highlighter Probes).175 The system has an overall sensitivity of 72.3 % for rpoB mutation detection, as it can detect the mutations in the 81-bp hotspot area of rpoB at codons 516, 526, and 531. This system has the advantage of a short time round and low cost; thus, it is accessible to low-income communities.93
The Abbott RealTime MTB RIF/INH Resistance can detect resistance to MTB and RIF by targeting rpoB and INH resistance genes by targeting katG and inhA upper promoter region for INH. The sensitivity of detecting each RIF and INH resistance is 100% and 94.3%., respectively, and the specificity is 100% and 94.3%, respectively. It was configured as a companion assay for the Abbott Real-time MTB assay.176 Both assays are used on the high-throughput automated Abbott m2000 system and can handle three to 96 samples in one run, with controls and specimens.177
Anyplex II MTB/MDR/XDR (Seegene, South Korea) is a multiplex RT-PCR that detects MTBc and resistance to RIF, INH, FQs, and injectable drugs. Anyplex is programmed to identify 18 mutations in rpoB, seven mutations in the katG and inhA promoters, three mutations in gyrA, and three mutations in the rrs and eis promoter regions, which cause resistance to RIF, INH, FQs, and aminoglycosides, respectively. This test depends on two techniques that aid in identifying particular mutations in target genes: dual-priming oligonucleotides and tagging oligonucleotide cleavage and extension. Specificity was between 94% and 100%, while sensitivity ranged between 50% and 100% to detect MTB and drug resistance.72
FluoroType MTBDR (Hain Lifescience, Nehren, Germany) is a new assay with a different technology. This test incorporates the linear-after-the-exponential PCR (LATE-PCR) with probes using the Lights-on/lights-off technique.178 One tube can detect the rpoB, katG, and inhA regulatory region mutations in respiratory and non-respiratory clinical samples. The absence of a wild-type band may indicate other mutations within the amplified region of the target genes. The test is performed in a FluoroCycler96 instrument (Hain Lifescience), allowing testing of up to 96 samples within three to four hours from DNA extraction. Melting curves are the data of the FluoroType MTBDR, and the shapes indicate either wild types or the existence of definite mutations.179 Compared to the Genotype MTBDRplus, the FluoroType MTBDR provides several advantages: fewer hands-on hours, quicker outcomes, automated interpretation with the capability of instantly importing findings into a lab information system, and a lower possibility of DNA contamination. FluoroType MTBDR showed excellent sensitivity and specificity for determining RIF and INH resistance when using culture isolates. A study reported the sensitivity and specificity of FluoroType MTBDR to be 91.7% and 97% for INH, respectively, and 98.9% and 95.6% for RIF, respectively. Another study revealed sensitivity and specificity as 98.9% and 100% for RIF, respectively, and 98.8% and 100% for INH, respectively.180 A recent study detected sensitivity and specificity for RIF resistance in smear-positive specimens of 100% and 97.8%, respectively, and in smear-negative samples of 100% and 96.9%, respectively. As for INH, both detection sensitivity and specificity were 100% in smear-positive specimens and 93.8% and 97.4% in smear-negative samples, respectively.181
BD MAX MDR-TB assay (BD MAX) (Becton and Dickinson, 2018) is an automated, qualitative diagnostic test for the direct detection of MTBc DNA and mutations in rpoB and katG genes and the inhA promoter area in sputum from patients with clinical suspicion of TB disease.182 The assay employs RT-PCR of particular DNA targets and fluorogenic target-specific hybridization probes. The assay is fully automated and requires a steady electricity supply and laboratory worker training. The pre-validation study of the assay demonstrated high sensitivity and specificity similar to the Abbott RealTime MTB RIF/INH Resistance assay and the FluoroType MTBDR for detecting MTBc DNA and mutations associated with resistance to RIF and INH.183 A recent multicenter study performed on 1053 participants with presumptive TB reported that the sensitivity and specificity of BD MAX MDR-TB for RIF resistance compared with phenotypic DST were 90% and 95%, respectively. Sensitivity and specificity for detecting INH resistance were 82% and 100%, respectively.173 It has a faster turnaround time than the GeneXpert MTB/RIF, i.e., four hours for testing 24 samples, compared to the GeneXpert MTB/RIF TAT, which takes around 2 hours for testing one sample. However, like other molecular assays, it does not recognize new mutations, and thus the sample needs sequencing.184
Droplet digital PCR-based technique (DDPCR)
Pholwat et al. developed a digital PCR-based method to detect and quantify various resistance subpopulations in a mycobacterial community containing even one XDR-MTB among thousands of susceptible MTB, i.e., heteroresistance.185,186 DDPCR combines microfluidic technology and PCR, enabling precise quantification of target DNA with high sensitivity and specificity.162 DDPCR can split the sample into thousands of drops and run PCR for individual sub-reactions with little or no off-target sequence in each sub-reaction.187 The fluorescence signal is detected in each droplet following the PCR procedures. Poisson statistical analysis is used for positive droplets to quantify the target sequence accurately. The PCR reaction setup and amplification are identical to quantitative PCR, except some contain the DNA target while others do not. This reproducible method detects bacilli at a count as low as 1000 CFU/ml. With these benefits, DDPCR enables the early detection of emerging mutations that evolve in treatment and may necessitate a medication adjustment.185
DNA sequencing-based identification of drug resistance
DNA sequencing is considered the gold standard molecular approach because it visualizes the nucleotide sequence of the target DNA position in a short time (10 to 12 hours). As in INH resistance, many genes may be involved in a single drug resistance, or the mutations may be distributed across a large gene segment. On the other hand, PCR-based sequencing (PCR amplification followed by gene sequencing) is a suitable approach for particular applications such as investigating rpoB hotspots, where mutations linked with RIF resistance are localized in a concise portion of the gene. However, unfortunately, sequencing is not as simply applicable for routine diagnosis of TB and identification of drug resistance-related mutations.55
Developing Next Generation Sequencing (NGS) has enabled bacterial species identification, showing all (known and novel) mutations, including synonymous and non-synonymous mutations, insertions and deletions in a sample, determination of antimicrobial resistance profile and so optimal therapy, and prediction of organism evolution.188,189 The AmpliSeq for Illumina TB Research Panel (Illumina) amplifies and sequences full-length of eight genes (rpoB, katG, and inhA, pncA, gyrA, eis, embB, and rpsL), not of entire genomes. Continuous advances in whole genome sequencing (WGS) techniques and bioinformatics analysis software have extended the chances of follow-up drug resistance in TB. Thus, it is possible to accurately detect resistance within one to three days after sample collection to most antituberculosis drugs used for treatment based on the quality and quantity of extracted mycobacterial DNA and the sequencing platform. Unlike conventional molecular assays that detect specific targets, WGS offers detailed and precise sequencing information for whole genomes.190
Several researchers reported that WGS showed good sensitivity and specificity in determining resistance to the first-line drugs RIF and INH; nevertheless, there was a significant difference in WGS accuracy for the other first- and second-line drugs.191-193 A recent study revealed that WGS has a 20% advantage over traditional genotypic techniques in finding drug resistance outside the suspected drug resistance-related region.194 Direct sequencing from sputum samples will enable rapid and complete resistance profiling in a therapeutically relevant time frame. However, direct sputum DNA extraction presents significant obstacles due to the restricted number of bacilli in sputum, particularly in smear-negative TB cases, resulting in a poor yield of genomic DNA accessible for sequencing. Furthermore, the results varied due to the high frequency of human genome contamination.153 A study by Colman et al. evaluated the performance of direct WGS and automated data analysis for drug resistance prediction in sputum tuberculosis samples (the Next Gen- RDST assay). The assay demonstrated high sensitivity and specificity (above 95 %) for detecting resistance to RIF, INH, and KM; still, low sensitivity in detecting resistance to amikacin, capreomycin, and fluoroquinolones compared to the phenotypic DST.194
Compared to the Xpert® MTB/RIF assay, the WGS is more accurate in identifying various gene mutations. Moreover, WGS disallowed false positive results in detecting the polymorphism in the rifampicin-resistance determining region (RRDR) of rpoB.195 However, because the workflow to extract DNA requires MTB culturing, which takes several weeks before an appropriate amount of DNA can be recovered, WGS has not been employed as a usual TB diagnostic method.143,153 Although one mL of early-positive MGIT cultures can be used to reliably and reasonably extract DNA for WGS to identify mycobacterial species and know its drug resistance.196 A technique that enables WGS without prior specimen culturing has recently been developed. The method uses biotinylated RNA baits unique to MTB DNA to extract entire MTB genomes from uncultured sputum samples.197 WGS data are collected several weeks before DST results are available.198 DNA sequencing might be utilized to confirm RIF resistance identified by Xpert MTB/RIF.199
The regular WGS for diagnosis is only available in high-income nations due to the high costs, which can reach hundreds of dollars.160,190,200 The use of WGS on a broad scale, particularly in middle- and low-income nations, remains problematic for the following reasons: the lack of standardization in WGS analysis directly from sputum, including specific procedures to remove non-mycobacterial genetic material and enrich the genome of M. tuberculosis, software and database must be established of this technology in this area of medicine; technical personnel and bioinformatics facilities are required, data acquisition and analysis are needed to determine whether new mutations confer anti-TB drug, in addition to the high cost of this technology.189,201,202
MDR TB is an infectious bacterial disease caused by MTB, one of the deadliest contagious illnesses worldwide. The inappropriate therapy contributes to the development of drug resistance in MTB, leading to a reduction in available treatments and an increase in the mortality rate. Thus, for better treatment and management, an early, quick, and accurate diagnosis of TB and determination of the antimicrobial susceptibility of the infecting strain are essential. However, clinical diagnosis is usually insufficient as MTB infection is commonly asymptomatic. In addition, the conventional culture procedures for bacterial isolation, identification and drug susceptibility testing are time-consuming and require specialized laboratories and well-trained personnel due to MTB sluggish growth. Molecular-based technologies have sped up turnaround times after sample collection, which could enhance the control of drug-resistant TB. Although molecular-based diagnostic methods have high sensitivity and specificity, they require high-cost laboratory infrastructures and skilled personnel to perform tests. However, despite their several drawbacks, traditional laboratory techniques for TB diagnosis are still widely utilized because they are feasible, trustworthy, and cost-effective. In addition, phenotypic susceptibility testing is still required to determine its drug susceptibility. Therefore, nucleic acid amplification tests (NAATs), culture-based procedures and acid-fast smear microscopy are currently employed for the definitive laboratory diagnosis of TB.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers. Cells. 2020;9(10):2160.
Crossref - World Health Organization. Global tuberculosis report 2021: supplementary material. https://apps.who.int/iris/bitstream/handle/10665/360605/9789240046856-eng.pdf. Accessed: October 4, 2021.
- Sanford CA, Jong EC, Pottinger P. The travel and tropical medicine manual E-Book. Elsevier Health Sciences. 2016.
- Pai M, Behr MA, Dowdy D, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076.
Crossref - Pelaez Coyotl EA, Barrios Palacios J, Mucino G, et al. Antimicrobial Peptide against Mycobacterium Tuberculosis That Activates Autophagy Is an Effective Treatment for Tuberculosis. Pharmaceutics. 2020;12(11):1071.
Crossref - Natarajan K, Kundu M, Sharma P, Basu J. Innate immune responses to M. tuberculosis infection. Tuberculosis (Edinb). 2011;91(5):427-431.
Crossref - Xpert MTB/RIF Implementation Manual: Technical and Operational ‘How-To’; Practical Considerations. Geneva: World Health Organization. 2014.
- Chuchottaworn C, Thanachartwet V, Sangsayunh P, et al. Risk Factors for Multidrug-Resistant Tuberculosis among Patients with Pulmonary Tuberculosis at the Central Chest Institute of Thailand. PLoS One. 2015;10(10):e0139986.
Crossref - Pradipta IS, Forsman LD, Bruchfeld J, Hak E, Alffenaar JW. Risk factors of multidrug-resistant tuberculosis: A global systematic review and meta-analysis. J Infect. 2018;77(6):469-478.
Crossref - Parwati I, Alisjahbana B, Apriani L, et al. Mycobacterium tuberculosis Beijing genotype is an independent risk factor for tuberculosis treatment failure in Indonesia. J Infect Dis. 2010;201(4):553-557.
Crossref - Lan NT, Lien HT, Tung le B, Borgdorff MW, Kremer K, van Soolingen D. Mycobacterium tuberculosis Beijing genotype and risk for treatment failure and relapse, Vietnam. Emerg Infect Dis. 2003;9(12):1633-1635.
Crossref - Drobniewski F, Balabanova Y, Nikolayevsky V, et al. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA. 2005;293(22):2726-2731.
Crossref - Dormans J, Burger M, Aguilar D, et al. Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin Exp Immunol. 2004;137(3):460-468.
Crossref - Centres for Disease Control and Prevention (CDC). Epidemiology of Tuberculosis. Reichman and Hershfield’s Tuberculosis: A Comprehensive, International Approach, Third Edition. https://www.cdc.gov/mmwr/volumes/68/wr/mm6811a3.htm. 2019; 8(11):263-266.Accessed: March 22.
- Drewe JA. Who infects whom? Social networks and tuberculosis transmission in wild meerkats. Proc Biol Sci. 2010;277(1681):633-642.
Crossref - Paleckyte A, Dissanayake O, Mpagama S, Lipman MC, McHugh TD. Reducing the risk of tuberculosis transmission for HCWs in high incidence settings. Antimicrob Resist Infect Control. 2021;10(1):106.
Crossref - Mounchili A, Perera R, Lee RS, Njoo H, Brooks J. Chapter 1: Epidemiology of tuberculosis in Canada. Can J Respir Crit Care, Sleep Med. 6:8-21.
Crossref - Machado D, Couto I, Viveiros M. Advances in the molecular diagnosis of tuberculosis: From probes to genomes. Infect Genet Evol. 2019;72:93-112.
Crossref - Shaikh A, Sriraman K, Vaswani S, Oswal V, Mistry N. Detection of Mycobacterium tuberculosis RNA in bioaerosols from pulmonary tuberculosis patients. Int J Infect Dis. 2019;86:5-11.
Crossref - World Health Organization. Latent Tuberculosis Infection (LTBI). http://www.who.int/tb/chal-lenges/ltbi/en/. Accessed: February 2, 2017.
- Organization Mundial de la Salud (OMS). Tuberculosis. http://www.who.int/topics/tuberculosis/es/. Accessed: October 15, 2016.
- Maiga M, Abaza A, Bishai WR. Current tuberculosis diagnostic tools & role of urease breath test. Indian J Med Res. 2012;135(5):731-736.
- Starke JR. Tuberculosis skin testing: new schools of thought. Pediatrics. 1996;98(1):123-125.
Crossref - Piccazzo R, Paparo F, Garlaschi G. Diagnostic accuracy of chest radiography for the diagnosis of tuberculosis (TB) and its role in the detection of latent TB infection: a systematic review. J Rheumatol Suppl. 2014;91:32-40.
Crossref - Drobniewski FA, Watterson SA, Wilson SM, Harris GS. A clinical, microbiological and economic analysis of a national service for the rapid molecular diagnosis of tuberculosis and rifampicin resistance in Mycobacterium tuberculosis. J Med Microbiol. 2000;49(3):271-278.
Crossref - Nijiati M, Ma J, Hu C, et al. Artificial Intelligence Assisting the Early Detection of Active Pulmonary Tuberculosis From Chest X-Rays: A Population-Based Study. Front Mol Biosci. 2022;9:874475.
Crossref - Organization, World Health. Chest radiography in tuberculosis detection: summary of current WHO recommendations and guidance on programmatic approaches. https://iris.who.int/handle/10665/252424. Accessed: December 14, 2016.
- Vaezipour N, Fritschi N, Brasier N, et al. Towards Accurate Point-of-Care Tests for Tuberculosis in Children. Pathogens. 2022;11(3):327.
Crossref - Rajpurkar P, O’Connell C, Schechter A, et al. CheXaid: deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. NPJ Digit Med. 2020;3:115.
Crossref - WHO consolidated guidelines on tuberculosis: Module 2: screening – systematic screening for tuberculosis disease. Geneva: World Health Organization. https://www.who.int/publications/i/item/9789240022676. Accessed: March 22, 2021.
- Gill CM, Dolan L, Piggott LM, McLaughlin AM. New Developments in Tuberculosis Diagnosis and Treatment. Breathe. 2022;18(1):210149.
Crossref - Baptista-Rosas RC, Hinojosa A, Riquelme M. Ecological niche modeling of Coccidioides spp. in western North American deserts. Ann N Y Acad Sci. 2007;1111:35-46.
Crossref - McHugh KE, Sturgis CD, Procop GW, Rhoads DD. The cytopathology of Actinomyces, Nocardia, and their mimickers. Diagn Cytopathol. 2017;45(12):1105-1115.
Crossref - Bento CM, Gomes MS, Silva T. Looking beyond Typical Treatments for Atypical Mycobacteria. Antibiotics (Basel). 2020;9(1):18.
Crossref - Moore DA, Evans CA, Gilman RH, et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355(15):1539-1550.
Crossref - Steingart KR, Henry M, Ng V, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(9):570-581.
Crossref - Hendry C, Dionne K, Hedgepeth A, Carroll K, Parrish N. Evaluation of a rapid fluorescent staining method for detection of mycobacteria in clinical specimens. J Clin Microbiol. 2009;47(4):1206-1208.
Crossref - Martinez-Martinez YB, Martinez-Rodriguez HG, Said-Fernandez SL. Conventional and Molecular Diagnosis of Drug-Sensitive and Drug-Resistant Pulmonary Tuberculosis. Mycobacterium-Research and Development. Intech Open. 2018.
Crossref - Gilpin C, Kim SJ, Lumb R, Rieder HL, Van Deun and Working Group on Sputum Smear Microscopy. Critical appraisal of current recommendations and practices for tuberculosis sputum smear microscopy [Workshop Report]. Int J Tuberc Lung Dis. 2007;11(9):946-952.
- Anthony RM, Kolk AH, Kuijper S, Klatser PR. Light emitting diodes for auramine O fluorescence microscopic screening of Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2006;10(9):1060-1062.
- Viveiros M, Machado D, Couto I, Amaral L. Improving on the LJ slope. Automated liquid culture. Tuberculosis: laboratory diagnosis and treatment strategies, Advances in Molecular and Cellular Microbiology Series; McHugh, TD, Ed. 2013;34-45.
Crossref - Procop GW. Laboratory Diagnosis and Susceptibility Testing for Mycobacterium tuberculosis. Microbiol Spectr. 2016;4(6):10-11.
- Kudoh S, Kudoh T. A simple technique for culturing tubercle bacilli. Bull World Health Organ. 1974;51(1):71-82.
- Morris TC, Hoggart CJ, Chegou NN, et al. Evaluation of Host Serum Protein Biomarkers of Tuberculosis in sub.Saharan Africa. Front Immunol. 2021;12:639174.
Crossref - MacLean E, Broger T, Yerlikaya S, et al. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol. 2019;4(5):748-758.
Crossref - Shete PB, Ravindran R, Chang E, et al. Evaluation of antibody responses to panels of M. tuberculosis antigens as a screening tool for active tuberculosis in Uganda. PLoS One. 2017;12(8):e0180122.
Crossref - Grace PS, Dolatshahi S, Lu LL, et al. Antibody Subclass and Glycosylation Shift Following Effective TB Treatment. Front Immunol. 2021;12:679973.
Crossref - Minion J, Leung E, Talbot E, Dheda K, Pai M, Menzies D. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur Respir J. 2011;38(6):1398.1405.
Crossref - Musvosvi M, Duffy D, Filander E, et al. T-cell biomarkers for diagnosis of tuberculosis: candidate evaluation by a simple whole blood assay for clinical translation. Eur Respir J. 2018;51(3):1800153.
Crossref - Silveira-Mattos PS, Barreto-Duarte B, Vasconcelos B, et al. Differential Expression of Activation Markers by Mycobacterium tuberculosis-specific CD4+ T Cell Distinguishes Extrapulmonary From Pulmonary Tuberculosis and Latent Infection. Clin Infect Dis. 2020;71(8):1905-1911.
Crossref - Acharya MP, Pradeep SP, Murthy VS, et al. CD38+CD27-TNF-a + on Mtb.specific CD4+ T Cells Is a Robust Biomarker for Tuberculosis Diagnosis. Clin Infect Dis. 2021;73(5):793.801.
Crossref - Comella-Del-Barrio P, Bimba JS, Adelakun R, et al. Fujifilm SILVAMP TB.LAM for the Diagnosis of Tuberculosis in Nigerian Adults. J Clin Med. 2021;10(11):2514.
Crossref - World Health Organization. Tuberculosis prevalence surveys: a handbook. World Health Organization; 2011. https://apps.who.int/iris/bitstream/handle/10665/44481/?sequence=1
- Devasia RA, Blackman A, May C, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis: an assessment of MGIT 960, MODS and nitrate reductase assay and fluoroquinolone cross.resistance. J Antimicrob Chemother. 2009;63(6):1173-1178.
Crossref - Lange C, Abubakar I, Alffenaar JW, et al. Management of patients with multidrug.resistant/extensively drug.resistant tuberculosis in Europe: a TBNET consensus statement. Eur Respir J. 2014;44(1):23-63.
Crossref - Nguyen TNA, Anton-Le Berre V, Banuls AL, Nguyen TVA. Molecular Diagnosis of Drug. Resistant Tuberculosis; A Literature Review. Front Microbiol. 2019;10:794.
Crossref - Zaman K. Tuberculosis: a global health problem. J Health Popul Nutr. 2010;28(2):111-113.
Crossref - World Health Organization. Treatment of Tuberculosis. Guidelines for National Programmes. 4th ed. Geneva: WHO Library Cataloguing-in-Publication Data. http://apps.who.int/iris/bitstream/10665/44165/1/9789241547833_eng.pdf. 2009
- Farmer P, Kim JY. Community based approaches to the control of multidrug resistant tuberculosis: introducing “DOTS-plus”. BMJ. 1998;317(7159):671-674.
Crossref - Lee SH, Choi HB, Yu SY, Chang UJ, Kim CK, Kim HJ. Detection of first-line anti-tuberculosis drug resistance mutations by allele.specific primer extension on a microsphere-based platform. Ann Lab Med. 2015;35(5):487.493.
Crossref - Coll F, McNerney R, Preston MD, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7(1):51.
Crossref - World Health Organization. Rapid סommunication: key changes to treatment of multidrug. and rifampicin-resistant tuberculosis (MDR/RR.TB). https://apps.who.int/iris/handle/10665/275383. Accessed: 10 October 2018.
- Gupta A, Anupurba S. Detection of drug resistance in Mycobacterium tuberculosis: Methods, principles and applications. Indian J Tuberc. 2015;62(1):13-22.
Crossref - Richter E, Rusch-Gerdes S, Hillemann D. Drug.susceptibility testing in TB: current status and future prospects. Expert Rev Respir Med. 2009;3(5):497-510.
Crossref - Engstrom A, Morcillo N, Imperiale B, Hoffner SE, Jureen P. Detection of first- and second-line drug resistance in Mycobacterium tuberculosis clinical isolates by pyrosequencing. J Clin Microbiol. 2012;50(6):2026-2033.
Crossref - Amini S, Hoffner S, Allahyar Torkaman MR, et al. Direct drug susceptibility testing of Mycobacterium tuberculosis using the proportional method: A multicenter study. J Glob Antimicrob Resist. 2019;17:242-244.
Crossref - World Health Organization; International Union Against Tuberculosis and Lung Disease; Royal Netherlands Tuberculosis Association. Revised international definitions in tuberculosis control. Int J Tuberc Lung Dis. 2001;5(3):213.215.
- Heifets L, Cangelosi G. Drug resistance assays for Mycobacterium tuberculosis. Antimicrobial Drug Resistance: Clinical and Epidemiological Aspects. 2009:1161-1170.
Crossref - Canetti G, Fox W, Khomenko A, et al. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ. 1969;41(1):21-43.
- Musa HR, Ambroggi M, Souto A, Angeby KA. Drug susceptibility testing of Mycobacterium tuberculosis by a nitrate reductase assay applied directly on microscopy.positive sputum samples. J Clin Microbiol. 2005;43(7):3159-3161.
Crossref - Gupta A, Sen MR, Mohapatra TM, Anupurba S. Evaluation of the performance of nitrate reductase assay for rapid drug-susceptibility testing of Mycobacterium tuberculosis in north India. J Health Popul Nutr. 2011;29(1):20-25.
Crossref - Martin A, Panaiotov S, Portaels F, Hoffner S, Palomino JC, Angeby K. The nitrate reductase assay for the rapid detection of isoniazid and rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta.analysis. J Antimicrob Chemother. 2008;62(1):56-64.
Crossref - Joloba ML, Bajaksouzian S, Jacobs MR. Evaluation of E test for susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38(10):3834-3836.
Crossref - Hausdorfer J, Sompek E, Allerberger F, Dierich MP, Rusch-Gerdes S. E-test for susceptibility testing of Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 1998;2(9):751.755.
- Liu Z, Guo S, Ji M, Sun K, Li Z, Fan X. Progresses of mycobacteriophage.based Mycobacterium tuberculosis detection. Biocell. 2020;44(4):683-694.
Crossref - Albert H, Trollip A, Seaman T, Mole RJ. Simple, phage.based (FASTPplaque) technology to determine rifampicin resistance of Mycobacterium tuberculosis directly from sputum. Int J Tuberc Lung Dis. 2004;8(9):1114-1119.
- Singh JP. A study of efficacy of phage amplification technique in diagnosis of pulmonary and extra-pulmonary tuberculosis. 2019;6(1):2349-3933.
Crossref - Baylan O, Kisa O, Albay A, Doganci L. Evaluation of a new automated, rapid, colorimetric culture system using solid medium for laboratory diagnosis of tuberculosis and determination of anti-tuberculosis drug susceptibility. Int J Tuberc Lung Dis. 2004;8(6):772-777.
- Palomino JC, Martin A, Von Groll A, Portaels F. Rapid culture.based methods for drug-resistance detection in Mycobacterium tuberculosis. J Microbiol Methods. 2008;75(2):161-166.
Crossref - Abate G, Aseffa A, Selassie A, et al. Direct colorimetric assay for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2004;42(2):871-873.
Crossref - Yajko DM, Madej JJ, Lancaster MV, et al. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J Clin Microbiol. 1995;33(9):2324-2327.
Crossref - Helal ZH, Menofy GE, Ibrahim, ZA et al. Comparative evaluation of Anyplex II MTB/MDR/XDR and resazurin microtiter assay for detection of drug resistant Mycobacterium tuberculosis. J Microbiol Biotechnol Food Sci. 2019;8(5):1150-1155.
Crossref - Noordhoek GT, Kolk AH, Bjune G, et al. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. J Clin Microbiol. 1994;32(2):277-284.
- Gazi MA, Islam MR, Kibria MG, Mahmud Z. General and advanced diagnostic tools to detect Mycobacterium tuberculosis and their drug susceptibility: a review. Eur J Clin Microbiol Infect Dis. 2015;34(5):851-861.
Crossref - Kadam M, Govekar A, Shenai S, et al. Can cord formation in BACTEC MGIT 960 medium be used as a presumptive method for identification of M. tuberculosis complex? Indian J Tuberc. 2010;57(2):75-79.
- Rasslan O, Hafez SF, Hashem M, et al. Microscopic observation drug susceptibility assay in the diagnosis of multidrug-resistant tuberculosis. Int J Tuberc Lung Dis. 2012;16(7):941-946.
Crossref - Bemer P, Bodmer T, Munzinger J, Perrin M, Vincent V, Drugeon H. Multicenter evaluation of the MB/BACT system for susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2004;42(3):1030-1034.
Crossref - Rusch-Gerdes S, Domehl C, Nardi G, Gismondo MR, Welscher HM, Pfyffer GE. Multicenter evaluation of the mycobacteria growth indicator tube for testing susceptibility of Mycobacterium tuberculosis to first.line drugs. J Clin Microbiol. 1999;37(1):45-48.
Crossref - Reisner BS, Gatson AM, Woods GL. Evaluation of mycobacteria growth indicator tubes for susceptibility testing of Mycobacterium tuberculosis to isoniazid and rifampin. Diagn Microbiol Infect Dis. 1995;22(4):325-329.
Crossref - Pfyffer GE, Welscher HM, Kissling P, et al. Comparison of the Mycobacteria Growth Indicator Tube (MGIT) with radiometric and solid culture for recovery of acid.fast bacilli. J Clin Microbiol. 1997;35(2):364-368.
Crossref - Palomino JC, Traore H, Fissette K, Portaels F. Evaluation of Mycobacteria Growth Indicator Tube (MGIT) for drug susceptibility testing of Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 1999;3(4):344-348.
- Johansen IS, Thomsen VO, Marjamaki M, Sosnovskaja A, Lundgren B. Rapid, automated, nonradiometric susceptibility testing of Mycobacterium tuberculosis complex to four first-line antituberculous drugs used in standard short-course chemotherapy. Diagn Microbiol Infect Dis. 2004;50(2):103-107.
Crossref - El-Sayed Zaki M, Goda T. Rapid phenotypic assay of antimycobacterial susceptibility pattern by direct mycobacteria growth indicator tube and phage amplified biological assay compared to BACTEC 460 TB. Tuberculosis (Edinb). 2007;87(2):102-108.
Crossref - Mugusi F, Villamor E, Urassa W, Saathoff E, Bosch RJ, Fawzi WW. HIV co.infection, CD4 cell counts and clinical correlates of bacillary density in pulmonary tuberculosis. Int J Tuberc Lung Dis. 2006;10(6):663-669.
- Steingart KR, Flores LL, Dendukuri N, et al. Commercial serological tests for the diagnosis of active pulmonary and extrapulmonary tuberculosis: an updated systematic review and meta-analysis. PLoS Med. 2011;8(8):e1001062.
Crossref - Krupinski EA, Williams MB, Andriole K, et al. Digital radiography image quality: image processing and display. J Am Coll Radiol. 2007;4(6):389-400.
Crossref - Yeager H Jr, Lacy J, Smith LR, LeMaistre CA. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95(6):998-1004.
Crossref - Cruciani M, Scarparo C, Malena M, Bosco O, Serpelloni G, Mengoli C. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J Clin Microbiol. 2004;42(5):2321-2325.
Crossref - Mgode GF, Weetjens BJ, Nawrath T, et al. Diagnosis of tuberculosis by trained African giant pouched rats and confounding impact of pathogens and microflora of the respiratory tract. J Clin Microbiol. 2012;50(2):274-280.
Crossref - Alcaide F, Coll P. Advances in rapid diagnosis of tuberculosis disease and anti-tuberculous drug resistance. Enferm Infecc Microbiol Clin. 2011;29(Suppl 1):34-40.
Crossref - Imaeda T, Kirchheimer WF, Barksdale L. DNA isolated from Mycobacterium leprae: genome size, base ratio, and homology with other related bacteria as determined by optical DNA-DNA reassociation. J Bacteriol. 1982;150(1):414-417.
Crossref - Collins DM, De Lisle GW. DNA restriction endonuclease analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG. J Gen Microbiol. 1984;130(4):1019-1021.
Crossref - Kim JY, Ferraro MJ, Branda JA. False-negative results obtained with the Gen-Probe Amplified Mycobacterium tuberculosis direct test caused by unrecognized inhibition of the amplification reaction. J Clin Microbiol. 2009;47(9):2995-2997.
Crossref - Lumb R, Lanser JA, Lim IS. Rapid identification of mycobacteria by the Gen.Probe Accuprobe system. Pathology. 1993;25(3):313-315.
Crossref - Eddabra R, Ait Benhassou H. Rapid molecular assays for detection of tuberculosis. Pneumonia (Nathan). 2018;10:4.
Crossref - Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol. 2002;118(5):796-801.
Crossref - Adekambi T, Drancourt M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. 2004;54(Pt 6):2095-2105.
Crossref - Qin L, Zheng R, Fan C, et al. Identification and evaluation of a new nucleic acid amplification test target for specific detection of Mycobacterium tuberculosis. Clin Chem Lab Med. 2010;48(10):1501-1505.
Crossref - Niemz A, Boyle DS. Nucleic acid testing for tuberculosis at the point.of.care in high-burden countries. Expert Rev Mol Diagn. 2012;12(7):687-701.
Crossref - Tiwari S, Nataraj G, Kanade S, Mehta P. Diagnosis of pediatric pulmonary tuberculosis with special reference to polymerase chain reaction based nucleic acid amplification test. Int J Mycobacteriol. 2015;4(1):48-53.
Crossref - Sia IG, Wieland ML. Current concepts in the management of tuberculosis. Mayo Clin Proc. 2011;86(4):348-361.
Crossref - Policy Statement: Automated Real.Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF System. Geneva: World Health Organization. 2011.
- Bergmann JS, Woods GL. Clinical evaluation of the Roche AMPLICOR PCR Mycobacterium tuberculosis test for detection of M. tuberculosis in respiratory specimens. J Clin Microbiol. 1996;34(5):1083-1085.
Crossref - Kunduracıoglu A, Karasu I, Bicmen C, Ozsoz A, Erbaycu AE. Comparison of the performances of MTD Gene-Probe® test, BACTEC 960™ system and Löwenstein-Jensen culture methods in the diagnosis of smear-negative tuberculosis cases. Mikrobiyol Bul. 2013;47(3):417-431.
Crossref - Woods GL. Molecular methods in the detection and identification of mycobacterial infections. Arch Pathol Lab Med. 1999;123(11):1002-1006.
Crossref - Neonakis IK, Spandidos DA, Petinaki E. Use of loop.mediated isothermal amplification of DNA for the rapid detection of Mycobacterium tuberculosis in clinical specimens. Eur J Clin Microbiol Infect Dis. 2011;30(8):937-942.
Crossref - Ou X, Song Y, Zhao B, et al. A multicenter study of cross-priming amplification for tuberculosis diagnosis at peripheral level in China. Tuberculosis (Edinb). 2014;94(4):428-433.
Crossref - Castan P, de Pablo A, Fernandez-Romero N, et al. Point of care system for detection of Mycobacterium tuberculosis and rifampin resistance in sputum samples. J Clin Microbiol. 2014;52(2):502-507.
Crossref - Cheng VC, Yew WW, Yuen KY. Molecular diagnostics in tuberculosis. Eur J Clin Microbiol Infect Dis. 2005;24(11):711.720.
Crossref - Chakaya J, Khan M, Ntoumi F, et al. Global Tuberculosis Report 2020-Reflections on the Global TB burden, treatment and prevention efforts. Int J Infect Dis. 2021;113(Suppl 1):S7.S12.
Crossref - Laraque F, Griggs A, Slopen M, Munsiff SS. Performance of nucleic acid amplification tests for diagnosis of tuberculosis in a large urban setting. Clin Infect Dis. 2009;49(1):46.54.
Crossref - Centers for Disease Control and Prevention (CDC). Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009;58(1):7.10.
- Centers for Disease Control and Prevention (CDC). Update: Nucleic acid amplification tests for tuberculosis. MMWR Morb Mortal Wkly Rep. 2000;49(26):593.594.
- Nurwidya F, Handayani D, Burhan E, Yunus F. Molecular Diagnosis of Tuberculosis. Chonnam Med J. 2018;54(1):1-9.
Crossref - Dylewski J. Nucleic Acid amplification testing for the diagnosis of tuberculosis: not for all. Clin Infect Dis. 2009;49(9):1456-1457.
Crossref - Ling DI, Flores LL, Riley LW, Pai M. Commercial nucleic.acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta.analysis and meta-regression. PLoS One. 2008;3(2):e1536.
Crossref - Kralik P, Ricchi M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front Microbiol. 2017;8:108.
Crossref - Sharma SK, Kohli M, Yadav RN, et al. Evaluating the Diagnostic Accuracy of Xpert MTB/RIF Assay in Pulmonary Tuberculosis. PLoS One. 2015;10(10):e0141011.
Crossref - Centers for Disease Control and Prevention (CDC). Availability of an assay for detecting Mycobacterium tuberculosis, including rifampin.resistant strains, and considerations for its use . United States, 2013 [published correction appears in MMWR Morb Mortal Wkly Rep. 2013;62(41):821.827.
- Friedrich SO, Rachow A, Saathoff E, et al. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 2013;1(6):462.470.
Crossref - Zeka AN, Tasbakan S, Cavusoglu C. Evaluation of the GeneXpert MTB/RIF assay for rapid diagnosis of tuberculosis and detection of rifampin resistance in pulmonary and extrapulmonary specimens. J Clin Microbiol. 2011;49(12):4138-4141.
Crossref - Automated Real.Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children. Geneva: World Health Organization. 2013.
- World Health Organization. Companion handbook to the WHO guidelines for the programmatic management of drug.resistant tuberculosis. World Health Organization. 2014. https://apps.who.int/iris/bitstream/handle/10665/130918/?sequence=1
- Kohli M, Schiller I, Dendukuri N, et al. Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance. Cochrane Database Syst Rev. 2018;8(8):CD012768.
Crossref - Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;2014(1):CD009593.
Crossref - Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377(9776):1495-1505.
Crossref - Steingart KR, Sohn H, Schiller I, et al. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2013;(1):CD009593.
Crossref - Pantoja A, Kik SV, Denkinger CM. Costs of novel tuberculosis diagnostics-will countries be able to afford it? J Infect Dis. 2015;211(Suppl 2):S67-S77.
Crossref - Bodmer T, Strohle A. Diagnosing pulmonary tuberculosis with the Xpert MTB/RIF test. J Vis Exp. 2012;(62):e3547.
Crossref - Churchyard GJ, Stevens WS, Mametja LD, et al. Xpert MTB/RIF versus sputum microscopy as the initial diagnostic test for tuberculosis: a cluster-randomised trial embedded in South African roll.out of Xpert MTB/RIF. Lancet Glob Health. 2015;3(8):e450-e457.
Crossref - Kwong JC, McCallum N, Sintchenko V, Howden BP. Whole genome sequencing in clinical and public health microbiology. Pathology. 2015;47(3):199-210.
Crossref - Walker TM, Ip CL, Harrell RH, et al. Whole.genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13(2):137-146.
Crossref - Koser CU, Ellington MJ, Peacock SJ. Whole.genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30(9):401-407.
Crossref - Palomino JC, Martin A. Drug Resistance Mechanisms in Mycobacterium tuberculosis. Antibiotics (Basel). 2014;3(3):317-340.
Crossref - Tiwari RP, Hattikudur NS, Bharmal RN, Kartikeyan S, Deshmukh NM, Bisen PS. Modern approaches to a rapid diagnosis of tuberculosis: promises and challenges ahead. Tuberculosis. 2007;87(3):193-201.
Crossref - Stucki D, Gagneux S. Single nucleotide polymorphisms in Mycobacterium tuberculosis and the need for a curated database. Tuberculosis (Edinb). 2013;93(1):30-39.
Crossref - McGrath M, Gey van Pittius NC, van Helden PD, Warren RM, Warner DF. Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2014;69(2):292-302.
Crossref - Laurenzo D, Mousa SA. Mechanisms of drug resistance in Mycobacterium tuberculosis and current status of rapid molecular diagnostic testing. Acta Trop. 2011;119(1):5-10.
Crossref - Zaczek A, Brzostek A, Augustynowicz-Kopec E, Zwolska Z, Dziadek J. Genetic evaluation of relationship between mutations in rpoB and resistance of Mycobacterium tuberculosis to rifampin. BMC Microbiol. 2009;9:10.
Crossref - Wilson ML. Recent advances in the laboratory detection of Mycobacterium tuberculosis complex and drug resistance. Clin Infect Dis. 2011;52(11):1350-1355.
Crossref - World Health Organization. Geneva. WHO/CDS/TB/2019.14. Licence: CC BY.NCSA 3.0 IGO, 2019. Available at: https://apps.who.int/iris/bitstream/handle/10665/329388/WHO.CDS.TB.2019.14.eng.pdf?ua=1. Accessed: 7 January 2020.
- Parsa S, Soleimanpour S, Derakhshan M, Babaei Nik L, Mir R, Izadi N. A Review of Phenotypic and Genotypic Methods for Detection of Drug Resistance in Mycobacterium tuberculosis. Iran J Med Microbiol. 2020;14(2):108-124.
Crossref - Bjorn-Mortensen K, Zallet J, Lillebaek T, et al. Direct DNA Extraction from Mycobacterium tuberculosis Frozen Stocks as a Reculture.Independent Approach to Whole. Genome Sequencing. J Clin Microbiol. 2015;53(8):2716-2719.
Crossref - Makinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H. Comparison of two commercially available DNA line probe assays for detection of multidrug.resistant Mycobacterium tuberculosis. J Clin Microbiol. 2006;44(2):350-352.
Crossref - Viveiros M, Leandro C, Rodrigues L, et al. Direct application of the INNO-LiPA Rif. TB line-probe assay for rapid identification of Mycobacterium tuberculosis complex strains and detection of rifampin resistance in 360 smear.positive respiratory specimens from an area of high incidence of multidrug-resistant tuberculosis. J Clin Microbiol. 2005;43(9):4880-4884.
Crossref - Morgan M, Kalantri S, Flores L, Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta.analysis. BMC Infect Dis. 2005;5:62.
Crossref - Traore H, van Deun A, Shamputa IC, Rigouts L, Portaels F. Direct detection of Mycobacterium tuberculosis complex DNA and rifampin resistance in clinical specimens from tuberculosis patients by line probe assay. J Clin Microbiol. 2006;44(12):4384-4388.
Crossref - Ritter C, Lucke K, Sirgel FA, et al. Evaluation of the AID TB resistance line probe assay for rapid detection of genetic alterations associated with drug resistance in Mycobacterium tuberculosis strains. J Clin Microbiol. 2014;52(3):940-946.
Crossref - Molina-Moya B, Lacoma A, Prat C, et al. AID TB resistance line probe assay for rapid detection of resistant Mycobacterium tuberculosis in clinical samples. J Infect. 2015;70(4):400-408.
Crossref - Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15(12):1767-1776.
Crossref - Bedewi Omer Z, Mekonnen Y, Worku A, et al. Evaluation of the GenoType MTBDRplus assay for detection of rifampicin. and isoniazid.resistant Mycobacterium tuberculosis isolates in central Ethiopia. Int J Mycobacteriol. 2016;5(4):475-481.
Crossref - Meaza A, Kebede A, Yaregal Z, et al. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR.TB in smear positive and negative sputum samples. BMC Infect Dis. 2017;17(1):280.
Crossref - Gardee Y, Dreyer AW, Koornhof HJ, et al. Evaluation of the GenoType MTBDRsl Version 2.0 Assay for Second.Line Drug Resistance Detection of Mycobacterium tuberculosis Isolates in South Africa. J Clin Microbiol. 2017;55(3):791-800.
Crossref - Nathavitharana RR, Hillemann D, Schumacher SG, et al. Multicenter Noninferiority Evaluation of Hain GenoType MTBDRplus Version 2 and Nipro NTM+MDRTB Line Probe Assays for Detection of Rifampin and Isoniazid Resistance. J Clin Microbiol. 2016;54(6):1624-1630.
Crossref - Yang Z, Durmaz R, Yang D, et al. Simultaneous detection of isoniazid, rifampin, and ethambutol resistance of Mycobacterium tuberculosis by a single multiplex allele.specific polymerase chain reaction (PCR) assay. Diagn Microbiol Infect Dis. 2005;53(3):201-208.
Crossref - Ullah I, Ahmad W, Shah AA, et al. Detection of rifampicin resistance of Mycobacterium tuberculosis using multiplex allele specific polymerase chain reaction (MAS.PCR) in Pakistan. Infect Genet Evol. 2019;71:42-46.
Crossref - Wang X, Jiao J, Xu W, Chai X, Li Z, Wang Q. A simple, rapid and economic method for detecting multidrug.resistant tuberculosis. Braz J Infect Dis. 2013;17(6):667-671.
Crossref - Bunsow E, Ruiz-Serrano MJ, Lopez Roa P, Kestler M, Viedma DG, Bouza E. Evaluation of GeneXpert MTB/RIF for the detection of Mycobacterium tuberculosis and resistance to rifampin in clinical specimens. J Infect. 2014;68(4):338-343.
Crossref - Ochang EA, Udoh UA, Emanghe UE, et al. Evaluation of rifampicin resistance and 81-bp rifampicin resistant determinant region of rpoB gene mutations of Mycobacterium tuberculosis detected with XpertMTB/Rif in Cross River State, Nigeria. Int J Mycobacteriol. 2016;5 Suppl 1:S145-S146.
Crossref - Van Rie A, Page.Shipp L, Scott L, Sanne I, Stevens W. Xpert(®) MTB/RIF for point-of-care diagnosis of TB in high.HIV burden, resource-limited countries: hype or hope? Expert Rev Mol Diagn. 2010;10(7):937-946.
Crossref - Goncalves MG, Fukasawa LO, Oliveira RS, et al. Fast test for assessing the susceptibility of Mycobacterium tuberculosis to isoniazid and rifampin by real.time PCR. Mem Inst Oswaldo Cruz. 2012;107(7):903-908.
Crossref - Sanchez-Padilla E, Merker M, Beckert P, et al. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med. 2015;372(12):1181-1182.
Crossref - Perez-Risco D, Rodriguez-Temporal D, Valledor-Sanchez I, Alcaide F. Evaluation of the Xpert MTB/RIF Ultra Assay for Direct Detection of Mycobacterium tuberculosis Complex in Smear-Negative Extrapulmonary Samples. J Clin Microbiol. 2018;56(9):e00659.18.
Crossref - Chakravorty S, Simmons AM, Rowneki M, et al. The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing. mBio. 2017;8(4):e00812-17.
Crossref - Castan P, de Pablo A, Fernandez-Romero N, et al. Point-of-care system for detection of Mycobacterium tuberculosis and rifampin resistance in sputum samples. J Clin Microbiol. 2014;52(2):502-507.
Crossref - Kostera J, Leckie G, Tang N, et al. Analytical and clinical performance characteristics of the Abbott RealTime MTB RIF/INH Resistance, an assay for the detection of rifampicin and isoniazid resistant Mycobacterium tuberculosis in pulmonary specimens. Tuberculosis (Edinb). 2016;101:137.143.
Crossref - Tang N, Frank A, Pahalawatta V, et al. Analytical and clinical performance of Abbott RealTime MTB, an assay for detection of Mycobacterium tuberculosis in pulmonary specimens. Tuberculosis (Edinb). 2015;95(5):613-619.
Crossref - Rice JE, Reis AH Jr, Rice LM, Carver-Brown RK, Wangh LJ. Fluorescent signatures for variable DNA sequences. Nucleic Acids Res. 2012;40(21):e164.
Crossref - Hillemann D, Haasis C, Andres S, Behn T, Kranzer K. Validation of the FluoroType MTBDR Assay for Detection of Rifampin and Isoniazid Resistance in Mycobacterium tuberculosis Complex Isolates. J Clin Microbiol. 2018;56(6):e00072.18.
Crossref - de Vos M, Derendinger B, Dolby T, et al. Diagnostic Accuracy and Utility of FluoroType MTBDR, a New Molecular Assay for Multidrug-Resistant Tuberculosis. J Clin Microbiol. 2018;56(9):e00531-18.
Crossref - Dippenaar A, Derendinger B, Dolby T, et al. Diagnostic accuracy of the FluoroType MTB and MTBDR VER 2.0 assays for the centralized high-throughput detection of Mycobacterium tuberculosis complex DNA and isoniazid and rifampicin resistance. Clin Microbiol Infect. 2021;27(9):1351-e1.1351.e4.
Crossref - BD MAX™ MDR.TB Assay Package InsertBD Life Sciences, Sparks, MD, (2019) Available at: https://www.bd.com/resource.aspx?IDX=35759. Accessed 21 May 2019].
- Zimmermann S, Dalpke A, Murray P. Pre-validation of the BD Max MDR.TB assay for the rapid detection of MTBc DNA and mutations associated with rifampin and isoniazid resistance. In 28th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID). 2018:21-24.
- Shah M, Paradis S, Betz J, et al. Multicenter Study of the Accuracy of the BD MAX Multidrug-resistant Tuberculosis Assay for Detection of Mycobacterium tuberculosis Complex and Mutations Associated With Resistance to Rifampin and Isoniazid. Clin Infect Dis. 2020;71(5):1161-1167.
Crossref - Pholwat S, Stroup S, Foongladda S, Houpt E. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One. 2013;8(2):e57238.
Crossref - Morley AA. Digital PCR: A brief history. Biomol Detect Quantif. 2014;1(1):1-2.
Crossref - Phenix-Lan Q, Martin S, Eric B. dPCR: A technology review. Sensors. 2018;18(4):1271.
Crossref - CRyPTIC Consortium and the 100,000 Genomes Project, Allix-Bיguec C, Arandjelovic I, et al. Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing. N Engl J Med. 2018;379(15):1403-1415.
Crossref - Zignol M, Cabibbe AM, Dean AS, et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance. Lancet Infect Dis. 2018;18(6):675-683.
Crossref - Rizzo JM, Buck MJ. Key principles and clinical applications of “next-generation” DNA sequencing. Cancer Prev Res (Phila). 2012;5(7):887-900.
Crossref - Satta G, Atzeni A, McHugh TD. Mycobacterium tuberculosis and whole genome sequencing: a practical guide and online tools available for the clinical microbiologist. Clin Microbiol Infect. 2017;23(2):69-72.
Crossref - Walker TM, Merker M, Kohl TA, Crook DW, Niemann S, Peto TE. Whole genome sequencing for M/XDR tuberculosis surveillance and for resistance testing. Clin Microbiol Infect. 2017;23(3):161-166.
Crossref - Lam C, Martinez E, Crighton T, et al. Value of routine whole genome sequencing for Mycobacterium tuberculosis drug resistance detection. Int J Infect Dis. 2021;113(Suppl 1):S48-S54.
Crossref - Colman RE, Anderson J, Lemmer D, et al. Rapid Drug Susceptibility Testing of Drug-Resistant Mycobacterium tuberculosis Isolates Directly from Clinical Samples by Use of Amplicon Sequencing: a Proof-of-Concept Study. J Clin Microbiol. 2016;54(8):2058-2067.
Crossref - Witney AA, Cosgrove CA, Arnold A, Hinds J, Stoker NG, Butcher PD. Clinical use of whole genome sequencing for Mycobacterium tuberculosis. BMC Med. 2016;14:46.
Crossref - Votintseva AA, Pankhurst LJ, Anson LW, et al. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53(4):1137-1143.
Crossref - Brown AC, Bryant JM, Einer-Jensen K, et al. Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples. J Clin Microbiol. 2015;53(7):2230-2237.
Crossref - Murphy SG, Smith C, Lapierre P, et al. Direct detection of drug-resistant Mycobacterium tuberculosis using targeted next generation sequencing. Front Public Health. 2023;11:1206056.
Crossref - McAlister AJ, Driscoll J, Metchock B. DNA sequencing for confirmation of rifampin resistance detected by Cepheid Xpert MTB/RIF assay. J Clin Microbiol. 2015;53(5):1752-1753.
Crossref - Takiff HE, Feo O. Clinical value of whole-genome sequencing of Mycobacterium tuberculosis. Lancet Infect Dis. 2015;15(9):1077-1090.
Crossref - Phelan J, O’Sullivan DM, Machado D, et al. The variability and reproducibility of whole genome sequencing technology for detecting resistance to anti-tuberculous drugs. Genome Med. 2016;8(1):132.
Crossref - Dookie N, Khan A, Padayatchi N, Naidoo K. Application of Next Generation Sequencing for Diagnosis and Clinical Management of Drug-Resistant Tuberculosis: Updates on Recent Developments in the Field. Front Microbiol. 2022;13:775030.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.