ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is an opportunistic pathogen which is commonly associated with healthcare associated infection. They possess multiple pathogenic factors which play a role in causing invasive infections such as surgical site infection, pneumonia, and blood stream infection. There were two hospital outbreaks caused by sensitive strains P. aeruginosa between 2016 and 2017 involving 17 patients. The outbreak investigation by Pulsed Field Gel Electrophoresis (PFGE) revealed seven clonally related P. aeruginosa strains (A-G). This study aims to determine the virulence factors acquired by the P. aeruginosa isolates and describe the clinical outcome of the patients. Seventeen P. aeruginosa isolates from the stocked collection were retrieved for six virulence genes, namely ToxA, ExoS, LasI, LasB, OprI, and OprL by PCR. Ten out of 17 of the P. aeruginosa isolates were able to revive. The ExoS, LasI, LasB, OprI, and OprL genes, respectively, were detected in all isolates, while ToxA gene was detected in six isolates which belonged to clone A (one isolate) and clone C (five isolates). The isolate from clone A caused pneumonia and isolates from clone C caused surgical site infections which led to disseminated infections and death. The presence of multiple virulence genes in these P. aeruginosa isolates may have contributed to the invasiveness, and the outcome of the infection. More studies with a larger number of patients will give a better insight regarding the actual role of these genes in different clinical manifestations caused by sensitive strain P. aeruginosa.
Pseudomonas aeruginosa, Virulence Genes, Outbreak, Pulsed-field Gel Electrophoresis
Pseudomonas aeruginosa is a major human pathogen among the Pseudomonas species. It is an aerobic gram-negative, rod-shaped bacterium that is motile and does not ferment lactose.1 P. aeruginosa is ubiquitous in the environment, including soil, water, and plants.1 This is due to the simple growth requirement and nutritional versatility. P. aeruginosa are able to use many organic compounds as the source of carbon and nitrogen, and utilizes minimal traces of nutrients.2 In hospital settings, P. aeruginosa is commonly isolated from reservoirs such as sinks, respiratory equipment as well as surgical equipment.3 It is an opportunistic organism which causes infection in patients with underlying comorbidities such as burns, malignancies, human immunodeficiency virus (HIV) infection, surgical site infections (SSI), patients with catheter in situ, post-solid organ transplant as well as in cystic fibrosis patients.4,5 A systematic literature review and meta-analysis by Ling et al. on the burden of HAI in South-East Asia showed that the prevalence of overall HAI is 9.0%, and the estimated incidence of SSI was 8.6% with P. aeruginosa being one of the most common microorganisms identified for overall HAIs, among Klebsiella spp. and Acinetobacter baumanii. P. aeruginosa accounts for 11% of all nosocomial infections causing surgical and wound infections, urinary tract infection, pneumonia and also bacteraemia.6-8
P. aeruginosa is not a common bacterium that colonizes healthy human hosts and it is suggested that long term colonization can occur when there is disruption of the microbiome by antimicrobial agents, medications, and the host having pre-existing diseases or being immunocompromised.2,3
P. aeruginosa contains numerous virulence factors which are commonly encountered in other bacteria. They form biofilm, exotoxin, pili, flagella and through a quorum sensing system making them resistant to multiple antibiotics which have contributed to the infections in vulnerable hospitalized patients.7 Many studies have shown that different virulence factors of P. aeruginosa contribute to different types of infections such as acute invasive or persistent infections.5,8 They are basically grouped into three main categories, namely bacterial surface structures (OprI, and OprL), secreted factors (ToxA, ExoS, LasB) and bacterial cell-to-cell interaction (LasI).9
Exotoxin A, encoded by ToxA, is a polypeptide that catalyses ADP-ribosylation of Elongation Factor 2 (EF2) causing inhibition of protein synthesis and cell death.10 It is secreted by type II secretion system (T2SS).11 It is one of the main virulence factors for P. aeruginosa which contribute to the toxicity trait causing delayed wound contraction and healing.12 This toxin causes dermatonecrosis in a burn wound, damages the cornea in ocular infection, and damages the tissue in pulmonary infection.2
LasB encodes elastase, an enzyme which degrades elastin causing damage to elastin-containing tissue.2 The enzyme also assists in bacterial attachment and immune system disruption by splitting collagen, IgG, IgA, and complement and also destruction of fibronectin to uncover ligands for bacterial adhesion.13 This further results in dissemination of infection and tissue destruction.2
P. aeruginosa virulence also depends on its cell-to-cell communication system or quorum sensing system (QS) which uses diffusible signalling molecules that accumulate with increasing cell density and allow it to respond to the host and environment by regulating gene expression accordingly.11,13,14 There are currently three known quorum sensing systems which includes LasI/R, Pseudomonas quinolone signal (PQS) and RhII/R system. Quorum sensing inhibitors, an antiphagocytic drug, are the most studied alternative for therapeutic drugs to overcome increasing antibiotic resistance in P. aeruginosa.11
OprI and OprL gene encodes for peptidoglycan related outer membrane protein which mediate resistance to antibiotic by efflux mechanism and alteration of membrane permeability rendering infections due to P. aeruginosa becomes more difficult to be treated.12 The outer membrane protein also plays an important role in the organism’s interaction with the environment.15 P. aeruginosa forms an immune-resistant aggregate by forming biofilms where it produces an extracellular matrix of exopolysaccharides, protein, and deoxyribonucleic acid. In persistent respiratory infection caused by P. aeruginosa, which was often seen in cystic fibrosis patients, often, this organism overproduces polysaccharide alginate causing the mucoid phenotype of the bacteria.16 This biofilm formed by P. aeruginosa caused the organism to be highly resistant to antibiotic treatment.13 A study also has shown that P. aeruginosa isolated from acute infection and chronic infection show different phenotype characteristics. Isolates from acute infection express more virulence factors while isolates from chronic infection lack some of the inflammatory features such as flagella and pili and down-regulate other virulence mechanisms such as type 3 secretion system (T3SS).4 In our study, the objective is to determine the virulence factors acquired by P. aeruginosa isolated from the clinical samples during the outbreak and to describe the clinical outcome associated with these infections.
Setting
The Hospital UiTM or formerly known as Pusat Perubatan Universiti Teknologi MARA (PPUiTM), Sungai Buloh, was a training centre for medical and cardiothoracic discipline. The infection control Unit initiated the outbreak investigations following increase in the number of P. aeruginosa infection cases after coronary artery bypass grafting (CABG) procedures and some from the general ward medical patients.
Study Design and Data Collection
This is a retrospective study involving all 17 patients who were involved in the outbreaks. The first outbreak occurred between November 2016 to December 2016 and the second outbreak occurred between February 2017 to April 2017. The P. aeruginosa isolates were recovered from the patients’ specimens from eight sternotomy wound pus swab, one sternal tissue, one pleural fluid, one mediastinal fluid, two blood culture, one bone specimen, one pericardial fluid, one sputum, and one tracheal aspirate specimen. The isolates were processed at the Medical Microbiology and Parasitology Laboratory, Department of Clinical and Diagnostic Laboratories, UiTM Medical Faculty, Sungai Buloh and Institute of Medical Molecular Biotechnology (IMMB), UiTM Medical Faculty, Sungai Buloh. Patients’ medical records were reviewed to collect the demographic data and patients’ clinical information. The data collected were patients’ age, underlying illness, history of presenting illness and clinical progress during admission, length of stay, antibiotic history, imaging findings and clinical outcome.
Inclusion criteria
- P. aeruginosa strains that were isolated from patients who were admitted to PPUiTM from November 2016 until April 2017.
- New infection with P. aeruginosa occurred during the admission.
Exclusion criteria
- Patients with polymicrobial infections occurring at the same time of the P. aeruginosa infection.
- Patients who had a known case of chronic infection with P. aeruginosa.
Ethical approval
The ethical approval was obtained from Universiti Teknologi MARA (UiTM) Institutional Ethics Board Committee for retrieval of isolates and patients’ medical records. Ethics No: REC/04/2020 (MR/75).
Bacterial strains, identification and anti-susceptibility test
All the P. aeruginosa isolates stored in inoculated cryobeads were retrieved and inoculated on blood agar. The blood agar plates were incubated at 37°C. The isolates which showed a pure culture of the morphology characterized as P. aeruginosa were reidentified by gram stain followed by automated identification instrument VITEK®2 COMPACT (bioMérieux, Durham, USA) using GN card. Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method and interpreted according to Clinical and Laboratory Standard Institute (CLSI) guidelines. The P. aeruginosa colony were inoculated into broth suspension equivalent to 0.5 McFarland and cultured on Mueller-Hinton Agar with piperacillin-tazobactam disk 110 µg, ceftazidime disk 30 µg, imipenem disk 10 µg, gentamicin disk 10 µg, and ciprofloxacin disk 5 µg. After 24 hours incubation at 37°C, the diameter of the zone of inhibition for each disc was measured and interpreted according to the CLSI breakpoint for P. aeruginosa.
Typing method
PFGE was performed as part of the outbreak investigation to assess the relatedness of the P. aeruginosa isolates. This typing method was outsourced to the National Public Reference Laboratory (Makmal Kesihatan Awam-MKAK) in Sungai Buloh.
Preparation of bacterial DNA
All isolates were inoculated aerobically on tryptase soy broth and bacterial DNA extraction was performed using the Deoxyribonucleic E.Z.N.A® DNA Mini Kit (Omega Bio-tec, Georgia, USA) according to the manufacturer’s protocols. The extracted DNA was preserved at -20°C until further use for PCR test.
Amplification of virulence genes
PCR amplification of the P. aeruginosa virulence genes ToxA, ExoS, LasI, LasB, OprI, and OprL were performed in 25 µL reaction mixture containing master mix, primer and the extracted DNA following the PCR protocol (ThermoScientific, USA). The primer sequences were retrieved from previous study (Table 1).15,17-19 The PCR product was visualized using a gel documentation system (GelDoc XR, BioRad Inc, Hercules, CA, USA). P. aeruginosa ATCC 27853 was used as positive control for all the virulence genes studied.
Table (1):
Primer sequences used in the amplification of ToxA, ExoS, LasB, LasI, OprL and OprI virulence genes of P. aeruginosa
| Virulence gene | Primers | Size, bp | Ref. | |
|---|---|---|---|---|
| ToxA | FWD [5’ – CTG CGC GGG TCT ATG TGCC – 3’] REV [5’ – GAT GCT GGA CGG GTC GAG – 3’] |
270 | [15] | |
| ExoS | FWD [5’ – ATC CTC AGG CGT ACA TCC – 3’] REV [5’ – ACG ACG GCT ATC TCT CCAC – 3’] |
328 | [17] | |
| LasB | FWD [5’ – TTC TAC CCG AAG GAC TGA TAC – 3′] REV [5’ – AAC ACC CAT GAT CGC AAC – 3’] |
153 | [18] | |
| LasI | FWD [5’ – CGT GCT CAA GTG TTC AAGG – 3′] REV [5’ – TAC AGT CGG AAA AGC CCAG – 3′] |
295 | [18] | |
| OprL | FWD [5’ – ATG GAA ATG CTG AAA TTC GGC – 3′] REV [5’ – CTT CTT CAG CTC GAC GCG ACG – 3′] |
504 | [19] | |
| OprI | FWD [5’ – ATG AAC AAC GTT CTG AAA TTC TCT GCT – 3’] REV [5’ – CTT GCG GCT GGC TTT TTC CAG – 3’] |
249 | [19] | |
Note: bp, base pair.
Patients’ demographic
Of the 17 patients, 15 were male (88%), and two were female (12%). The patient’s age ranged from 24 to 72 years old with a mean of 57.8 years, and the majority (47.1%) of the patients were aged between 60 to 69. The existing co-morbidities in the patients as follows: hypertension (58.8%), diabetes mellitus (41.2%), dyslipidemia (29.4%), ischaemic heart disease (17.6%), and chronic kidney disease (17.6%). 15 patients (88.0%) underwent the CABG procedure during the hospital stay while two patients (12.0%) did not undergo the CABG procedure. One patient was admitted due to an alleged stab wound over the chest wall, and another patient was admitted for fast atrial fibrillation and later developed HAP. Most of the P. aeruginosa were isolated from specimens taken from sternal wound swabs (47.1%), followed by sterile body fluids (17.6%), blood cultures (11.7%), sputum (5.9%), tracheal aspirate (5.9%), tissue (5.9%) and bone (5.9%). In terms of the type of infections; they constituted soft tissue infections (29.4%), both superficial wound infection and osteomyelitis (23.5%), bacteraemia (11.8%), and HAP and VAP (11.8%). 15 patients (88.2%) had prolonged hospital stays of more than two weeks and eight of them (47.0%) were admitted for more than four weeks. Out of these 17 patients, two patients (12.0%) succumbed to death due to HAI (Table 2).
Table (2):
The demographic parameters, clinical characteristics and outcome of the total study population
Parameter/Clinal characteristic |
Number (n=17) |
Percentage (%) |
|---|---|---|
Gender: Male Female |
15 2 |
88.0 12.0 |
Age: <50 50-59 60-69 >70 |
3 5 8 1 |
17.6 29.4 47.1 5.9 |
Comorbidities: Hypertension Diabetes mellitus Dyslipidaemia Ischaemic heart disease Chronic kidney disease |
10 7 5 3 3 |
58.8 41.2 29.4 17.6 17.6 |
Type of specimens: Swab Sterile body fluid Blood Sputum Tracheal aspirate Tissue Bone |
8 3 2 1 1 1 1 |
47.1 17.6 11.7 5.9 5.9 5.9 5.9 |
Type of infections: Soft tissue infection Superficial wound infection Osteomyelitis Bacteraemia HAP/VAP |
5 4 4 2 2 |
29.4 23.5 23.5 11.8 11.8 |
Antimicrobial treatment received: Amoxicillin-clavulanate Ampicillin-sulbactam Piperacillin- tazobactam Ceftazidime Cefepime Ciprofloxacin Imipenem Colistin |
1 4 11 7 4 7 1 1 |
2.8 11.1 30.6 19.4 11.1 19.4 2.8 2.8 |
Length of hospital stay: < 2 weeks 2 – 4 weeks > 4 weeks |
2 7 8 |
11.8 41.2 47.0 |
Clinical outcome: Alive Death |
15 2 |
88 12 |
Note: HAP, hospital acquired pneumonia; VAP, ventilator associated pneumonia.
Seventeen clinical samples, two environmental, and two instruments sampling were taken from the sink pipe, the sink drain hole, a swab from the general operation theatre suction catheter, and a swab taken from the harvest cone which is an instrument used during bypass grafting were subjected to PFGE. The suction catheter was used for suctioning of fluids or blood to aid during the surgical procedure and the harvest cone was used during CABG surgery to harvest the veins (commonly from saphenous vein) that will be used as a graft for the blocked artery in the heart. The results of the PFGE from the two outbreaks found that the P. aeruginosa belonged to seven different clones (A, B, C, D, E, F, G). There was one isolate from clone A, one isolate from clone B, 14 isolates from clone C, one isolate from clone D, two isolates from clone E, one isolate from clone F, and one isolate from clone G. P. aeruginosa strains belonging to clone C has more than 99% similarity and they were isolated from 12 different patients and two medical instruments. P. aeruginosa isolated from clinical samples taken from Patients 1 until 12 belonged to clone C. The type of infection for Patients 1, 3, 6, 9 were surgical site infections without disseminated infections and with disseminated infections for Patients No. 2, 4, 5, 7, 8, 10, 11, and 12. Patients 2 and 4 had the longest hospital stay which was eight weeks and they both had disseminated P. aeruginosa infections. Both Patients No. 6 and 10 succumbed to the infection with Patient 6 not having any disseminated infection. P. aeruginosa isolated from Patient 13 which was from clone F also had a long hospital stay which was eight weeks duration. Patients 16 and 17 in which the P. aeruginosa was isolated belonged to clones A and B, both patients did not have surgical site infection but had pneumonia instead. They had the shortest hospital stay which was less than two weeks duration (Table 3).
Table (3):
Patients’ demographic data, clinical characteristics and outcomes listed according to cases
Patient |
Isolate |
Clone |
Age |
Gender |
Co-morbidities |
Specimen |
Type of infection |
Length of hospital stay (weeks) |
Number of Antibiotics treatments |
Antibiotic duration (weeks) |
Clinical outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
5 |
C |
72 |
M |
HTN |
Pus swab |
Sternal wound infection |
2 |
1 |
1 |
Survive |
2 |
6 |
C |
59 |
M |
HTN Dyslipidaemia |
Sternal tissue |
Sternal wound infection with osteomyelitis and mediastinitis |
8 |
5 |
9 |
Survive |
3 |
N/A |
C |
60 |
M |
HTN CKD Dyslipidaemia |
Pus swab |
Sternal wound infection |
2 |
1 |
1 |
Survive |
4 |
N/A |
C |
53 |
M |
HTN IHD |
Pleural fluid |
Sternal wound infection with pleural effusion |
8 |
4 |
8 |
Survive |
5 |
N/A |
C |
56 |
M |
HTN Dyslipidaemia Psoriasis |
Mediastinal fluid |
Sternal wound infection with mediastinitis |
5 |
2 |
5 |
Survive |
6 |
7 |
C |
67 |
M |
DM CKD |
Pus swab |
Sternal wound infection |
4 |
5 |
4 |
Died |
7 |
N/A |
C |
67 |
M |
NKMI |
Pus swab |
Sternal osteomyelitis |
6 |
1 |
6 |
Survive |
8 |
N/A |
C |
60 |
M |
HTN DM IHD |
Blood |
Sternal wound infection with osteomyelitis, mediastinitis and sepsis |
6 |
2 |
6 |
Survive |
9 |
2 |
C |
51 |
M |
HTN DM Dyslipidaemia |
Pus swab |
Sternal wound infection |
1 |
1 |
1 |
Survive |
10 |
N/A |
C |
66 |
F |
HTN DM |
Bone |
Sternal wound infection with osteomyelitis |
3 |
4 |
3 |
Died |
11 |
N/A |
C |
47 |
M |
NKMI |
Pus swab |
Sternal wound infection with mediastinitis and left pleural effusion |
6 |
2 |
10 |
Survive |
12 |
3 |
C |
59 |
M |
NKMI |
Blood |
Sternal wound infection with Pseudomonas bacteraemia |
3 |
1 |
2 |
Survive |
13 |
10 |
F |
67 |
M |
HTN DM Dyslipidaemia |
Pericardial fluid |
Sternal wound infection with mediastinitis |
8 |
2 |
12 |
Survive |
14 |
9 |
G |
46 |
M |
NKMI |
Pus swab |
Sternal wound infection |
2 |
1 |
1 |
Survive |
15 |
4 |
D |
61 |
M |
HTN DM IHD |
Pus swab |
Sternal wound infection with pleural effusion |
5 |
2 |
3 |
Survive |
16 |
1 |
A |
67 |
F |
DM CKD |
Sputum |
HAP |
2 |
1 |
1 |
Survive |
17 |
8 |
B |
24 |
M |
Schizophrenia |
Tracheal aspirate |
VAP |
1 |
1 |
1 |
Survive |
Note: N/A, not available; F, female; M, male; DM, diabetes mellitus; CKD, chronic kidney disease; HTN, hypertension; NKMI, no known medical illness; IHD, ischaemic heart disease; HAP, hospital acquired pneumonia; VAP, ventilator associated pneumonia.
P. aeruginosa virulence genes detection
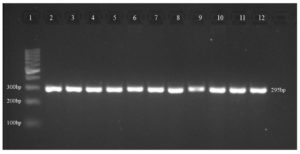
Ten isolates were subjected to PCR for the detection of the six virulence genes ToxA, ExoS, LasI, LasB, OprI, and OprL. ToxA gene was detected for isolates 1, 2, 3, 5, 6, and 7 which belong to clones A and C (Table 4). While all the other genes of ExoS, LasI, LasB, OprI and OprL were detected for all the ten isolates (Figure).
Table (4):
The virulence genes detected among the P. aeruginosa isolates
| Isolate | Patient | Clone | Type of specimen | Virulence Genes | |||||
|---|---|---|---|---|---|---|---|---|---|
| ToxA | ExoS | LasI | LasB | OprI | OprL | ||||
| 1 | 16 | A | Sputum | + | + | + | + | + | + |
| 2 | 9 | C | Sternal tissue | + | + | + | + | + | + |
| 3 | 12 | C | Pus swab | + | + | + | + | + | + |
| 4 | 15 | D | Pleural fluid | – | + | + | + | + | + |
| 5 | 1 | C | Pus swab | + | + | + | + | + | + |
| 6 | 2 | C | Pus swab | + | + | + | + | + | + |
| 7 | 6 | C | Blood | + | + | + | + | + | + |
| 8 | 17 | B | Tracheal aspirate | – | + | + | + | + | + |
| 9 | 14 | G | Pus swab | – | + | + | + | + | + |
| 10 | 13 | F | Pus swab | – | + | + | + | + | + |
Note: +, virulence gene detected; -, virulence gene not detected.
Figure. Representative of virulence genes (LasI) in P. aeruginosa isolates
Lane 1, DNA Ladder (100 bp); lane 2, positive control (P. aeruginosa ATCC 27853); lane 3, isolate 1; lane 4, isolate 2; lane 5, isolate 3; lane 6, isolate 4; lane 7, isolate 5; lane 8, isolate 6; lane 9, isolate 7; lane 10, isolate 8; lane 11, isolate 9; lane 12, isolate 10
The Virulence Genes and Clinical Outcome of Patients
From the ten P. aeruginosa isolates tested for the virulence genes, six of the isolates which belong to clones A and C were positive for ToxA gene while in other isolates from clone B, D, G and F, the ToxA genes were not detected. All ten isolates were positive for ExoS, LasI, LasB, OprI and OprL genes. The type of infections caused by clones A and C were similar to the other P. aeruginosa strains from other clones. They caused surgical site infections, where two patients had disseminated P. aeruginosa infection (Patients 6 and 12) while one patient had a lung infection (Patient 16) which was hospital-acquired. However, other P. aeruginosa isolates which did not harbour ToxA gene from clone F and D also had disseminated infections similar to Patients 13 and 15. In Patient 17, P. aeruginosa which belonged to clone B also had a lung infection (ventilator-associated pneumonia). Two patients in which the P. aeruginosa was isolated belonged to clone C (ToxA detected) and clone F (ToxA not detected) had similar length of hospital stay which was eight weeks duration. The number of classes of antibiotics used for treatment was highest in Patient 2 and Patient 6 amounting to five different types of antibiotics, and both patients had infections associated with P. aeruginosa which possessed ToxA gene. Patient 13 (ToxA not detected) had the longest duration of antibiotic treatment of 12 weeks. All patients except one (Patient 6) survived the infection (Table 5).
Table (5):
The P. aeruginosa isolated from 17 patients with its virulence genes and their clinical parameters and clinical outcomes
Patient |
Isolate |
Clone |
Tox A |
Virulence genes Exo S Las I Las B Opr I Opr L |
Type of infections |
Extent of involvement/Involvement of / distant / adjacent /other organs |
Length of Hospital stay (weeks) |
Number of different classes of antibiotics treatment |
Antibiotic duration (weeks) |
Clinical Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
1 |
5 |
C |
+ |
+ |
Surgical site infection |
Confined to sternal surgical wound site |
2 |
1 |
1 |
Survive |
2 |
6 |
C |
+ |
+ |
Surgical site infection |
Sternal wound infection involving soft tissue and fistula formation at the surgical site |
8 |
5 |
9 |
Survive |
3 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Confined to sternal surgical wound site |
2 |
1 |
1 |
Survive |
4 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with pleural effusion |
8 |
4 |
8 |
Survive |
5 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with mediastinitis |
5 |
2 |
5 |
Survive |
6 |
7 |
C |
+ |
+ |
Surgical site infection |
Sternal wound infection with sepsis and systemic involvement |
4 |
5 |
4 |
Died |
7 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with sternal osteomyelitis |
6 |
1 |
6 |
Survive |
8 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with sternal osteomyelitis, mediastinitis, and sepsis |
6 |
2 |
6 |
Survive |
9 |
2 |
C |
+ |
+ |
Surgical site infection |
Confined to sternal surgical wound site |
1 |
1 |
1 |
Survive |
10 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with sternal osteomyelitis |
3 |
4 |
3 |
Died |
11 |
N/A |
C |
N/A |
N/A |
Surgical site infection |
Sternal wound infection with mediastinitis and left pleural effusion |
6 |
2 |
10 |
Survive |
12 |
3 |
C |
+ |
+ |
Surgical site infection |
Sternal wound infection with bacteraemia |
3 |
1 |
2 |
Survive |
13 |
10 |
F |
– |
+ |
Surgical site infection |
Sternal wound infection with mediastinitis |
8 |
2 |
12 |
Survive |
14 |
9 |
G |
– |
+ |
Surgical site infection |
Confined to sternal surgical wound site |
2 |
1 |
1 |
Survive |
15 |
4 |
D |
– |
+ |
Surgical site infection |
Sternal wound infection with pleural effusion |
5 |
2 |
3 |
Survive |
16 |
1 |
A |
+ |
+ |
Hospital acquired
pneumonia |
Respiratory system involvement |
2 |
1 |
1 |
Survive |
17 |
8 |
B |
– |
+ |
Ventilator associated pneumonia |
Respiratory system involvement |
1 |
1 |
1 |
Survive |
Note: +, virulence gene detected; -, virulence gene not detected; N/A, not available.
Antimicrobial Susceptibility Pattern of the P. aeruginosa Isolates
The result showed that all the isolates were susceptible towards all the antibiotics tested (Table 6).
Table (6):
Antimicrobial susceptibility test results for P. aeruginosa isolates
| Isolate |
Patient |
Clone |
Zone Diameter (mm) | ||||
|---|---|---|---|---|---|---|---|
| Piperacillin- tazobactam, 110 µg | Ceftazidime, 30 µg | Imipenem, 10 µg | Gentamicin, 10 µg | Ciprofloxa-cin, 5 µg | |||
| 1 | 16 | A | 28(S) | 26(S) | 23(S) | 22(S) | 35(S) |
| 2 | 9 | C | 27(S) | 26(S) | 23(S) | 23(S) | 34(S) |
| 3 | 12 | C | 27(S) | 25(S) | 24(S) | 22(S) | 34(S) |
| 4 | 15 | D | 28(S) | 26(S) | 23(S) | 22(S) | 35(S) |
| 5 | 1 | C | 24(S) | 24(S) | 28(S) | 22(S) | 30(S) |
| 6 | 2 | C | 27(S) | 25(S) | 23(S) | 23(S) | 34(S) |
| 7 | 6 | C | 28(S) | 26(S) | 24(S) | 23(S) | 35(S) |
| 8 | 17 | B | 28(S) | 25(S) | 23(S) | 33(S) | 35(S) |
| 9 | 14 | G | 28(S) | 25(S) | 24(S) | 22(S) | 30(S) |
| 10 | 13 | F | 29(S) | 26(S) | 28(S) | 25(S) | 33(S) |
Note: (S), Susceptible. Breakpoint for susceptibility to piperacillin-tazobactam >21mm, ceftazidime > 18mm, imipenem > 19mm, gentamicin > 15mm, ciprofloxacin > 25mm.
In this study, we were able to detect and analyse the virulence genes of ten out of 17 P. aeruginosa that were isolated from patients’ clinical samples during both outbreaks from 2016 to 2017. The P. aeruginosa isolates that were revived had at least one representative from each clone (A, B, D, F and G) and five representatives from clone C. In addition, there were seven other clinical isolates and two non-clinical isolates which were isolated from the medical instruments used during the CABG but failed to grow. We were not able to investigate the virulence genes from the endoscopic vein harvest cone which was believed to be the source of the P. aeruginosa in this outbreak. Those isolates from the medical instruments also belonged to clone C. By virtue of the P. aeruginosa isolated from the harvest cone belonged to clone C and we had observed the same pattern of virulence genes in all five isolates from clone C, therefore we postulated that the P. aeruginosa isolated from the harvest cone is highly likely to have the same virulence genes pattern.
Our focus in this study is to determine six significant virulence genes which were responsible for tissue injury, invasiveness, and disseminated infection in P. aeruginosa. Six isolates, one from clone A and five from clone C were positive for ToxA gene while the other four isolates, from clone B, D, F, G did not harbor this gene. All isolates included in the study showed presence of ExoS, LasI, LasB, OprI, and OprL genes. This result showed that the isolates of P. aeruginosa causing the outbreak in this study possessed almost all the virulence genes tested and there was minimum variation of the virulence gene pattern observed with different clones of the isolates.
In studies by Ertugrul et al. and Badr et al., ToxA gene was found to be more prevalent in diabetic foot infection and burn wound patients and most of them develop retardation of wound healing.10,17 In our study, ToxA genes were detected in six out of ten isolates. The P. aeruginosa isolates with and without ToxA gene showed the ability to cause invasive infection of the surgical site which extended to the mediastinal and pleura. Other studies also associated exotoxin A with antibiotic resistance but in our study all the P. aeruginosa isolates were sensitive strains.17 A recent study found that secretion of exotoxin A by P. aeruginosa was reduced with negative pressure wound therapy.20 Therefore, by detecting the presence of ToxA gene in P. aeruginosa isolate could be an added value in the management of wound infection caused by P. aeruginosa.
Many studies correlate the presence of different virulence genes with toxigenesis, invasiveness and antibiotic resistance3,12,15,16,21 Choy et al. found that ExoS genes were common in non-contact lens related keratitis probably due to the association of ExoS with invasiveness of the infection.22 Another type of dissemination infection by P. aeruginosa was also described by Faraji et al. where he found that the prevalence of ToxA, LasB, and ExoS genes were higher in cystic fibrosis patients compared to burn wound patients.23 However, we did not have a cystic fibrosis patient in our cohort.
As we have described so far, our isolates possess multiple virulence factors. The conservation of the virulence genes among these isolates can be attributed to the fact that these P. aeruginosa cause healthcare associated outbreak in our Medical Centre and they had caused invasive infections which resulted in death in two patients despite being multi-drug sensitive strain. Matthew et al. also investigated whether there was presence of genomic variation in P. aeruginosa that were associated with different types of infection. The study examined 18 strains of P. aeruginosa by whole genome sequencing, and surprisingly they found that there was remarkable conservation of the virulence genes encoding the well-known virulence factors (≈ 97%).24 Thus, the disease-causing ability of P. aeruginosa may rely on the highly conserved pathogenic mechanism, and the specific features of infection may be influenced by a small number of strain-specific virulence genes.24
The risk factors that contributed to this outbreak involve both the microbial agent as well as the patient’s susceptibility towards infection. A study by Thu et al. shows that apart from patient’s demographic characteristics and presence of underlying co-morbidities, patients who underwent surgery or invasive procedure were also at increased risk of HAI.25 In this study, most of the patients had multiple risk factors for HAI due to advanced age, having underlying co-morbidities, and being involved with CABG procedure. Majority were between 60 to 69 of age and mostly had hypertension and diabetes mellitus. Furthermore, out of the 17 patients, 15 patients underwent CABG procedure during their admission. Thus, the patient population in this study were at higher risk to develop hospital acquired infection and at the same time were exposed to an environment contaminated with P. aeruginosa.
When an infection sets in, the roles of the virulence factors in P. aeruginosa help in the persistence and immune evasion of the organism which results in deep seated infection and systemic infection causing multiple and prolonged course of antibiotics and prolonged hospital stay. The OprL and OprI genes mediate resistance to antibiotic by efflux mechanism and alteration of membrane permeability causing difficulty to cure the infection.12 ExoS toxin disrupts actin cytoskeleton causing invasive infection.14 LasB assist in the attachment and immune system disruption which contribute to persistent infection.13 ToxA causes delayed wound contraction and healing as well as being responsible for immune evasion and persistence.12,14 Thus, the presence and conservation of these virulence genes in all the P. aeruginosa isolates in this study explains the prolonged and invasive infection in some of the patients despite the isolate were tested susceptible for the antimicrobial drug used as what we have seen in Patient 6 in our study. These two waves of outbreak mainly involved patients who underwent CABG procedure at two different time frames from November to December 2016 and February to April 2017. The type of infection usually acquired in the healthcare setting are urinary tract infection, surgical site infection, respiratory infection, vascular catheter infection and septicaemia. 15 of the patients with P. aeruginosa isolates from clone C, F, G and D initially had SSI that later develop into various degree of soft tissue infection with some extended to the adjacent pleura, mediastinum, and bone. While the other two patients with P. aeruginosa isolates from clone A and B had respiratory infection where one patient had HAP and another one patient had VAP respectively. The impact of healthcare associated infection is not only a burden to the healthcare organization but also to the patients. Functional disabilities, emotional stress that leads to reduced quality of life, or even death is among the effects that can occur to the patients. While for the healthcare organisation, this can result in increased length of hospital stay, increased antimicrobial and drugs usage, need for isolation, and additional laboratories and other diagnostic studies.26 In this study, only two patients (Patient 9 and Patient 17) were able to be discharged in less than two weeks from admission, while seven patients (Patients 1, 3, 6, 10, 12, 14, and 16) had prolonged hospital stay of two to four weeks and another eight patients (Patients 2, 4, 5, 7, 8, 11, 13, and 15) were staying at the hospital for more than four weeks. This prolonged hospital stay led to additional consumption of resources such as isolation bed, medical equipment, and medication usage, and increased the workload of the healthcare worker. Most of these patients received multiple courses of antimicrobial drugs and some ended up needing broad spectrum antimicrobial treatments such as carbapenem and colistin. Furthermore, two of the patients (Patient 6 and Patient 10) succumbed to death due to the infection.
Without elimination of the source or stopping the transmission of the P. aeruginosa, the outbreak can continue to persist even for years.27 P. aeruginosa has a range of mechanisms of adaptation and survival in the environment. Some of the survival mechanisms of P. aeruginosa are the biofilm formation, quorum sensing (QS) system, and viable but not culturable (VBNC) state of the organism.28 The biofilm is responsible for the survival and ability to adhere to wet surfaces or liquids, it is rich in nutrients and it can protect the microorganism against disinfectant.28 This favours P. aeruginosa colonization on medical equipment particularly in the harvest cone tip which was found to be the most likely source of this outbreak. A study by Wang et al. in 2015 proves that P. aeruginosa colonization of the endoscopic lumen was persistent even after proper cleaning with water, detergent, and ortho-phthalaldehyde (OPA) was performed in accordance with the Centre for Disease Control (CDC) guideline.29
The presence of multiple virulence genes in the P. aeruginosa isolates may have contributed to the invasiveness, and the outcome of the infection in our study. However, more studies with a larger number of patients will give better insight and the association regarding the actual role of these genes in different clinical manifestations caused by sensitive strain P. aeruginosa.
ACKNOWLEDGMENTS
The authors would like to thank the Institute of Medical Molecular and Biotechnology and the Hospital UiTM Infection Control Team for their technical support and retrieval of patients’ data.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Board Committee, Universiti Teknologi MARA (UiTM), Malaysia, with ethics approval number REC/04/2020 (MR/75).
- Bennett JE, Dolin R, Blaser MJ. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 9th Edition; 2019.
- Leifson SRC, and Fulton M. Murray Med. Microbiol. vol 1. Pseudomonas and Related Bacteria. 2013:288-295. https://www.clinicalkey.com/#!/content/book/3-s2.0-B9780323673228000270
- Noh RM, Shaari SA, Nawi SFAM, Adnan A. An Outbreak of Pseudomonas aeruginosa Infection in Coronary Artery Bypass Graft Patients Related to Endoscopic Vein Harvesting Equipment. Open Forum Infect Dis. 2017;4(Suppl 1):S174.
- Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67(3):159-173.
Crossref - Hogardt M, Heesemann J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol. 2010;300(8):557-562.
Crossref - Ling ML, Apisarnthanarak A, Madriaga G. The Burden of Healthcare-Associated Infections in Southeast Asia: A Systematic Literature Review and Meta-analysis. Clin Infect Dis. 2015;60(11):1690-1699.
Crossref - Khan HA, Ahmad A, Mehboob R. Nosocomial infections and their control strategies. Asian Pac J Trop Biomed. 2015;5(7):509-514.
Crossref - Bleves S, Viarre V, Salacha R, Michel GPF, Filloux A, Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int J Med Microbiol. 2010;300(8):534-543.
Crossref - Liao C, Huang X, Wang Q, Yao D, Lu W. Virulence Factors of Pseudomonas Aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front Cell Infect Microbiol. 2022;12:926758.
Crossref - Badr RI, el. Nagdy M, el. Sabagh A, el. Din AB. Pseudomonas aeruginosa Exotoxin A as a Virulence Factor in Burn Wound Infections Research article. Egypt J Med Microbiol. 2008;17(1):8.
- Chatterjee M, Anju cp, Biswas L, Kumar VA, Mohan CG, Biswas R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol. 2015;306(1):48-58.
Crossref - Al-Dahmoshi HOM, Al-Khafaji NS, Jeyad AA, Shareef HK, Al-Jebori RF. Molecular Detection of Some Virulence Traits among Pseudomonas aeruginosa Isolates, Hilla-Iraq. Biomed Pharmacol J. 2018;11(2):835-842.
Crossref - Bradbury RS, Roddam LF, Merritt A, Reid DW, Champion AC. Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J Med Microbiol. 2010;59(Pt 8):881-890.
Crossref - Veesenmeyer JL, Hauser AR, Lisboa T, Rello J. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit Care Med. 2009;37(5):1777-1786.
Crossref - De Vos D, Lim Jr A, Pirnay JP, et al. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35(6):1295-1299.
Crossref - Anderson G, Akhand S, Pettit R, Gardner T. New treatments in development for Pseudomonas aeruginosa infections in the lungs of individuals with cystic fibrosis. Orphan Drugs: Research and Reviews. 2014;4:71-81.
Crossref - Ertugrul BM, Oryasin E, Lipsky BA, Willke A, Bozdogan B. Virulence genes fliC, toxA and phzS are common among Pseudomonas aeruginosa isolates from diabetic foot infections. Infect Dis. 2018;50(4):273-279.
Crossref - Finnan S, Morrissey JP, O’Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42(12):5783-5792.
Crossref - Zhu H, Bandara R, Conibear TCR, et al. Pseudomonas aeruginosa with LasI Quorum-Sensing Deficiency during Corneal Infection. Invest Ophthalmol Vis Sci. 2004;45(6):1897-1903.
Crossref - Wang GQ, Li TT, Li ZR, et al. Effect of Negative Pressure on Proliferation, Virulence Factor Secretion, Biofilm Formation, and Virulence-Regulated Gene Expression of Pseudomonas aeruginosa In Vitro. Biomed Res Int. 2016;7986234.
Crossref - Bahador N, Shoja S, Faridi F, et al. Molecular detection of virulence factors and biofilm formation in Pseudomonas aeruginosa obtained from different clinical specimens in Bandar Abbas. Iran J Microbiol. 2019;11(1):25-30.
Crossref - Choy MH, Stapleton F, Willcox MDP, Zhu H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J Med Microbiol. 2008;57(Pt 12):1539-1546.
Crossref - Faraji F, Mahzounieh M, Ebrahimi A, Fallah F, Teymournejad O, Lajevardi B. Molecular detection of virulence genes in Pseudomonas aeruginosa isolated from children with Cystic Fibrosis and burn wounds in Iran. Microbial Pathogenesis. 2016;99:1-4.
Crossref - Wolfgang MC, Kulasekara BR, Liang X, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100(14):8484-8489.
Crossref - Thu TA, Hung NV, Quang NN, et al. A point-prevalence study on healthcare-associated infections in Vietnam: public health implications. Infect Control Hosp Epidemiol. 2011;32(10):1039-1041.
Crossref - Dellinger EP. Prevention of Hospital-Acquired Infections. Surg Infect (Larchmt). 2016;17(4):422-426.
Crossref - Snyder LA, Loman NJ, Faraj LA, et al. Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Euro Surveill. 2013;18(42)
Crossref - Spagnolo AM, Sartini M, Cristina ML. Pseudomonas aeruginosa in the healthcare facility setting. Rev Med Microbiol. 2021;32(3):169-175.
Crossref - Wang W-Y, Chiueh T-S, Lee Y-T, Tsao S-M. Persistent colonization of clonal Pseudomonas aeruginosa in endoscopic lumen despite repeating appropriate cleaning and disinfection. J Microbiol Immunol Infect. 2015;48(2)S174.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.