ISSN: 0973-7510
E-ISSN: 2581-690X
The present study was carried out during February to May 2018 in Baghdad hospitals. A sample of urine has been collected from fifty patients with an infection in their urinary tract (UTIs) of both sexes and different ages. Bacteriological investigation of urine samples from UTIs patients is made to isolate and diagnose Proteus mirabilis bacterium. In addition, the study detects the phenotypic and genetic characteristic of Proteus mirabilis a-hemolysin activity. Moreover, for the study to prove its hypothesis, a molecular detection has been carried out utilizing specific primer to hpmB gene which encodes a-Hemolysin as a factor of virulence of Proteus mirabilis through the use of PCR. The results show that 7(%100) of isolates are positive for hpmB at 422 bp. Two isolates of P. mirabilis are sequenced as hpmB genes. The ratio of identity of the hpmB genes with the CP017085.1 and CP020052.1 stains at NCBI global databases copmrised 100%, 99% respectively.
P. mirabilis,virulence factors gene hpmB, Sequencing.
Proteus mirabilis refers to a Gram-negative bacterium1. Its harmful virulence factors can be recognized by P. mirabilis that is able to access and settle the host urinary tract. These include toxins like hemolysin and its function of pore formation, biofilm formation and regulation of pathogenesis2. Proteus mirabilis has numerous virulence factors that may inflict UTIs. These factors have an important role in causing an infection in varying spots of the urinary tract3. Alpha (a) hemolysin hpmA is created by P. mirabilis that leads to an injury in kidney tissues. This a-hemolysin belongs to the cell, independent of calcium, pores-former which is encoded by two genes, hpmA and hpmB, that control the hpmB (63kDa) proteins3. hpmA a-hemolysin accountable for destructive and triggering the tissue when its N-terminal peptide is slashed. So, the result can activate hpmA and hpmB that is responsible for hpmA activation and transport4. The present study aims to characterize the hpmB among P. mirabilis isolated from UTIs patients.
Patients with urinary tract infection are from Baghdad Teaching Hospital during the period from September to October 2018. Midstream urine samples are collected and controlled in sterilized wide-container from one hundred urinary tract infection patients. The extraction of the Proteus mirabilis DNA from bacterial cells is carries out by using Genomic DNA Mini kit which supplemented by the manufacturing company (Promega, US). DNA electrophoresis in agarose gel is performed according to Ouda, 20145.
Thermic cycles program for amplifying the DNA
Specific primers are used for detecting the Proteus mirabilis virulence gene encodes of b-hemolysin sequence according to Cestari et al., 20136. These primers are provided by Promega Company (USA) and prepared according to the information of the supplying company, which is listed below:
DNA Sequencing of hpmB gene
The process of sequencing through PCR-sequences included two. This is performed according to Macrogen company/Korea sending. The nucleotide replacement is decided by comber. The data that obtained are from gene bank which is available at NCBI https://www.ncbi. nlm.nih.gov.
The technique used to investigate the genes responsibility for the virulence factor in P. mirablis is Single Polymerase Chain Reaction Technique. This has been conducted through using segments of the DNA with restricted number of nucleotides (oligonucleotide). Their action is to be primers specialized for virulence genes in P. mirablis., hpmB is also included and it is responsible for producing hemolycin P. mirabilis b-hemolysin that is different from other Proteus spp. It is organized by two genes, (hpmA and hpmB) that encodes the hpmA and hpmB proteins respectively7,8.
The present study shows that hpmB gene is present in all 7 isolates at rate (100%) from urine samples of RA patients as shown in fig (1). The results contest the result which is verified by Al-Jumaily and Zgaer. In their study, it is declared that the frequency of this gene in P. mirablis isolates is %100; that is isolated from patients suffering from urinary tract infections9. While Cestari et al., 2013 find the ratio of this gene in bacterial isolates comprising 96.24 % existing amplification for the hpmA and hpmB genes by PCR.6. The a-hemolycin toxin acts as a destroyer to the leukocyte membrane through creating small holes in the leukocyte membrane and epithelial cell. So, its presence is a vital factor in supplying the bacteria with iron and because of having the cytotoxic effects, this could lead to the destruction of the host kidney tissue10. Isolates with hpmA gene in them is in compatibility with the characterization presented by Uphoff and Welch (1990) who state the necessity of cleaving the N-terminal peptide of the hpmA by hpmB for the purpose of activating and transporting the hemolytic hpmA protein out of the cell8. This standard indicates that hpmA is a factor in the pathogenesis of P. mirabilis samples isolated from human urine10. Among the other results are those reported by Swihart and Welch,1990 refering that all P. mirabilis strains as having hpmA but HlyA is not detected in P. mirabilis isolates and is found only in 2 of the 24 P. vulgaris strains examined. Since P. mirabilis composes most (97%) of the Proteus urinary tract isolates, this suggests that hpmA is the predominant Proteus hemolysin and might play a role in extra intestinal infections caused by Proteus spp11. These positive isolates with hpmB gene are also checked to confirm their ability to produce hemolysin on blood agar and it has been found that all the isolates (100%) have the ability to produce hemolysin. These results agree with the results of Sosa et al. (2006) and AL-Jumaa et al. (2011), who demonstrate that all isolates (100%) of Proteus bacterium which are isolated from different clinical sources exhibit hemolysis on blood agar plates, but Mishara et al. (2001) find that (85.14%) of Proteus isolates produce a-hemolysis while other isolates produce b –hemolysis on blood agar plate12,13,14. The results of the study demonstrate that the detection of hpmB gene by PCR is sensitive enough to be used for discovering these virulence factors produced by P. mirabilis. The PCR technique is shown to be precise, fast, cheap and more accurate therefore this suggests that hpmB could be used as a diagnostic tool for P. mirablis bacterium.
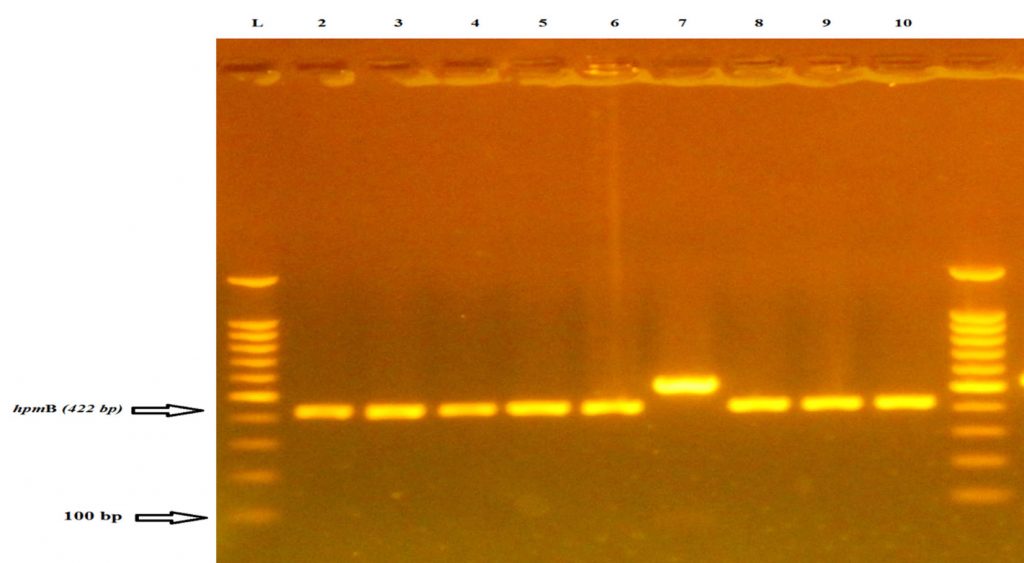
Fig. 1. Agarose gel electrophoresis (2% agarose, 75 V for 1:45 hour) of hpmB and PCR products (422bp) codify for a-hemolysin of P. mirabilis isolates. Lane 1DNA ladder), 100-1100bp molecular marker, lanes 2 ,3,4,5,6,7,8,9,10 isolates were positive results
DNA sequencing analysis
Analyzing DNA sequence of hpmB gene
Three isolates were sequenced by Macrogen/Korea. The nucleotide switch is firmed by comber. The data are obtained from gene bank available at NCBI (https://www.ncbi.nlm.nih.gov). The results of gene sequence analysis hpmB show that there are two polymorphism in 2 isolates of the gene hpmB as shown in fig (2) and table (1). In the isolation of P. mirabilis (MF993448) Thymine nucleotide substitutions to Adenine is found at locus 2389107 and Cytosine has substitutions to Thymine at locus 2389062 . Finally, the results show nonsense polymorphism as predict . Also, the results show silent variation and this type of variation doesn’t change the sequence of amino acid in the protein and doesn’t alter protein function as shown in table (1)15. Samples (MF993446) show 100 % identity for hpmB in comparison to the same genes of the CP017085.1. While samples, (MF993448) show 99 % identity for hpmB in comparison to the same genes of the : CP020052.1 strain.
Table (1):
Primer sequence of hpmB geneand PCR condition
| Genes | Sequence (5′ to 3′) | PCR condition | Size (bp) | References |
|---|---|---|---|---|
|
hpmB |
F:CAGTGGATTAAGCGCAAATG | 95 ⁰ C 5min 1x
95 ⁰ C 30sec 62⁰ C 30sec 30x 72 ⁰ C 20 sec 72 ⁰ C 5min 1x |
422
|
(Cestari etal.,2013) |
| R:CCTTCAATACGTTCAACAAACC |
Table (2):
Identity of hpmB gene sequence in
| Source | Identities | Expect | Score | Sequence ID | Range of nucleotide | |
|---|---|---|---|---|---|---|
| Proteus mirabilis
HPMB |
100% | 0.0 | 610 | CP017085.1 | 3196590 to 3196919 | |
| Proteus mirabilis
HPMB |
99% | 0.0 | 645 | CP020052.1 | 2389008 to 2389362 | |
Table (3):
polymorphism of hpmB gene sequence
| effect | Amino acid change | Nucleotide change | Location | Type of substitution | sample |
|---|---|---|---|---|---|
| non | 1 | ||||
| Silent | Threonine> Threonine | ACT>ACA | 2389107 | Transversion | 2 |
| Silent | Phenylalanine> Phenylalanine | TTC>TTT | 2389062 | Transition | |
Table (4):
Alignment of hpmB gene sequence Proteus mirabilis strain T18
| Sequence ID: CP017085.1Length: 4131426 Range 1: 3196590 to 3196919 | ||||
|---|---|---|---|---|
| Score | Expect | Identities | Gaps | Strand |
| 610 bits(330) | 8e-171 | 330/330(100%) | 0/330(0%) | Plus/Plus |
| Query 1 GAAATTAATCAATTAATAGAACAAAATCGCTATCAGCAACTGCAAGAAAAAGCGGTAAAT 60
Sbjct 3196590 GAAATTAATCAATTAATAGAACAAAATCGCTATCAGCAACTGCAAGAAAAAGCGGTAAAT 3196649 Query 61 ATTTCACCTACCCCAACTTTAATTACTGAGTCAGAACACTGTTTGCCTATAAAAGGCGTT 120 Sbjct 3196650 ATTTCACCTACCCCAACTTTAATTACTGAGTCAGAACACTGTTTGCCTATAAAAGGCGTT 3196709 Query 121 TATATTCAAGGTATTACTTTACTTACTGAGAAGGATCTCAATTCATTATCTCCGTTACCT 180 Sbjct 3196710 TATATTCAAGGTATTACTTTACTTACTGAGAAGGATCTCAATTCATTATCTCCGTTACCT 3196769 Query 181 GATCAATGTATTAAGAGTGCTGATATTAATCGCCTCGTGAAAGAACTCACTCAGCGTTAT 240 Sbjct 3196770 GATCAATGTATTAAGAGTGCTGATATTAATCGCCTCGTGAAAGAACTCACTCAGCGTTAT 3196829 Query 241 CTTCAACATGGTTATATTACCGCGCGTATCCAATTTTTACGTCCTAACCAACATGGCGAA 300 Sbjct 3196830 CTTCAACATGGTTATATTACCGCGCGTATCCAATTTTTACGTCCTAACCAACATGGCGAA 3196889 Query 301 TTAGGTCTGTATGCTATTGAAGGGTTTGTT 330 Sbjct 3196890 TTAGGTCTGTATGCTATTGAAGGGTTTGTT 3196919 |
||||
Table (5):
Alignment of hpmB gene sequence Proteus mirabilis strain AR_0059
| Sequence ID: CP020052.1Length: 4191021 Range 1: 2389008 to 2389362 | ||||
|---|---|---|---|---|
| Score | Expect | Identities | Gaps | Strand |
| 645 bits(349) | 0.0 | 353/355(99%) | 0/355(0%) | Plus/Minus |
| Query 1 GAAATTAATCAATTAATAGAACAAAATCGCTATCAGCAACTGCAAGAAAAAGCGGTAAAT 60
Sbjct 3196590 GAAATTAATCAATTAATAGAACAAAATCGCTATCAGCAACTGCAAGAAAAAGCGGTAAAT 3196649 Query 61 ATTTCACCTACCCCAACTTTAATTACTGAGTCAGAACACTGTTTGCCTATAAAAGGCGTT 120 Sbjct 3196650 ATTTCACCTACCCCAACTTTAATTACTGAGTCAGAACACTGTTTGCCTATAAAAGGCGTT 3196709 Query 121 TATATTCAAGGTATTACTTTACTTACTGAGAAGGATCTCAATTCATTATCTCCGTTACCT 180 Sbjct 3196710 TATATTCAAGGTATTACTTTACTTACTGAGAAGGATCTCAATTCATTATCTCCGTTACCT 3196769 Query 181 GATCAATGTATTAAGAGTGCTGATATTAATCGCCTCGTGAAAGAACTCACTCAGCGTTAT 240 Sbjct 3196770 GATCAATGTATTAAGAGTGCTGATATTAATCGCCTCGTGAAAGAACTCACTCAGCGTTAT 3196829 Query 241 CTTCAACATGGTTATATTACCGCGCGTATCCAATTTTTACGTCCTAACCAACATGGCGAA 300 Sbjct 3196830 CTTCAACATGGTTATATTACCGCGCGTATCCAATTTTTACGTCCTAACCAACATGGCGAA 3196889 Query 301 TTAGGTCTGTATGCTATTGAAGGGTTTGTT 330 Sbjct 3196890 TTAGGTCTGTATGCTATTGAAGGGTTTGTT 3196919 |
||||
Table (6):
Alignment of hemolysin activation protein [Proteus mirabilis]
| Sequence ID: WP_074561482. 1Length: 561 Identical proteins to WP_074561482.1 | ||||||
|---|---|---|---|---|---|---|
| Score | Expect | Method | Identities | Positives | Gaps | Frame |
| 228 bits(582) | 2e-70 | Compositional matrix adjust. | 110/110
(100%) |
110/110
(100%) |
0/110
(0%) |
+1 |
| Query 1 EINQLIEQNRYQQLQEKAVNISPTPTLITESEHCLPIKGVYIQGITLLTEKDLNSLSPLP 180
Sbjct 43 EINQLIEQNRYQQLQEKAVNISPTPTLITESEHCLPIKGVYIQGITLLTEKDLNSLSPLP 102 Query 181 DQCIKSADINRLVKELTQRYLQHGYITARIQFLRPNQHGELGLYAIEGFV 330 Sbjct 103 DQCIKSADINRLVKELTQRYLQHGYITARIQFLRPNQHGELGLYAIEGFV 152 |
||||||
Table (7):
Alignment of hemolysin activation protein [Proteus mirabilis]
| Sequence ID: WP_046334659.1Length: 561 Identical proteins to WP_046334659.1 | ||||||
|---|---|---|---|---|---|---|
| Score | Expect | Method | Identities | Positives | Gaps | Frame |
| 244 bits(623) | 2e-76 | Compositional matrix adjust. | 118/118(100%) | 118/118(100%) | 0/118(0%) | +2 |
| Query 2 RALQDSQREINQLIEQNRYQQLQEKAVNISPTPTLITESEHCLPIKGVYIQGITLLTEKD 181
Sbjct 35 RALQDSQREINQLIEQNRYQQLQEKAVNISPTPTLITESEHCLPIKGVYIQGITLLTEKD 94 Query 182 LNSLSPLPDQCIKSADINRLVKELTQRYLQHGYITARIQFLRPNQHGELGLYAIEGFV 355 Sbjct 95 LNSLSPLPDQCIKSADINRLVKELTQRYLQHGYITARIQFLRPNQHGELGLYAIEGFV 152 |
||||||
It is noticed, through the process of sequencing of the a-hemolysin at P. mirabilis, that it consists of two genes hpmA and hpmB7. P. mirabilis hemolysin hpmA investigation and description is necessary to clarify its significance as a virulence factor. Furthermore, it might have a possible association with other elements that are produced by P. mirabilis. These altogether could contribute to cytotoxicity in the UTIs of humans7. Mordi and Momoh.(2009) find that a change in the amino acid or replacement with other amino acid may lead to a change in the nature of protein or output and thus lead to the emergence of strains resistant or sensitive to antibiotics[16]. The hpmA and hpmB genes are sequenced from local isolated samples presented 98 % identity for hpmA and hpmB compared to the same genes of the HI4320 (wild strain) (NCBI GenBank Number NC_010554.1). When the samples of the study are compared with each other 100 % identity is found among these genes17. The results of the study display that the amino acid remained the same and that indicates variation from the silent type. In contrast, there is a study conducted on hpmB gene like Strauss et al., (1997) find that mutations enhance the function of hpmB (increase in hemolytic activity). This increasing in hemolytic activity could be a result of hpmB activating and secreting more hpmA18. Genotyping works to establish the relationship between bacteria strain on the basis of their genetic content uses. Many genotyping methods become important in the field of genealogy, classification of bacteria, identification of sources, method of infection and the differentiation of strain of high virulent bacteria to prevent their spread and elimination19. Therefore, the study finds a great importance in the genetic sequence of P. mirabilis virulence factors. The study finds that variant isolates possess polymorphism in hpmB genes.
A phylogenic tree based on the hpmB gene
Molecular phylogenetic is a branch of phylogeny that analyzes hereditary molecular differences mainly in DNA sequences to gain information on an organisms’ evolutionary relationships20. The identified genetic profile of any bacteria by a specific genotyping method can be as unique as fingerprint20.
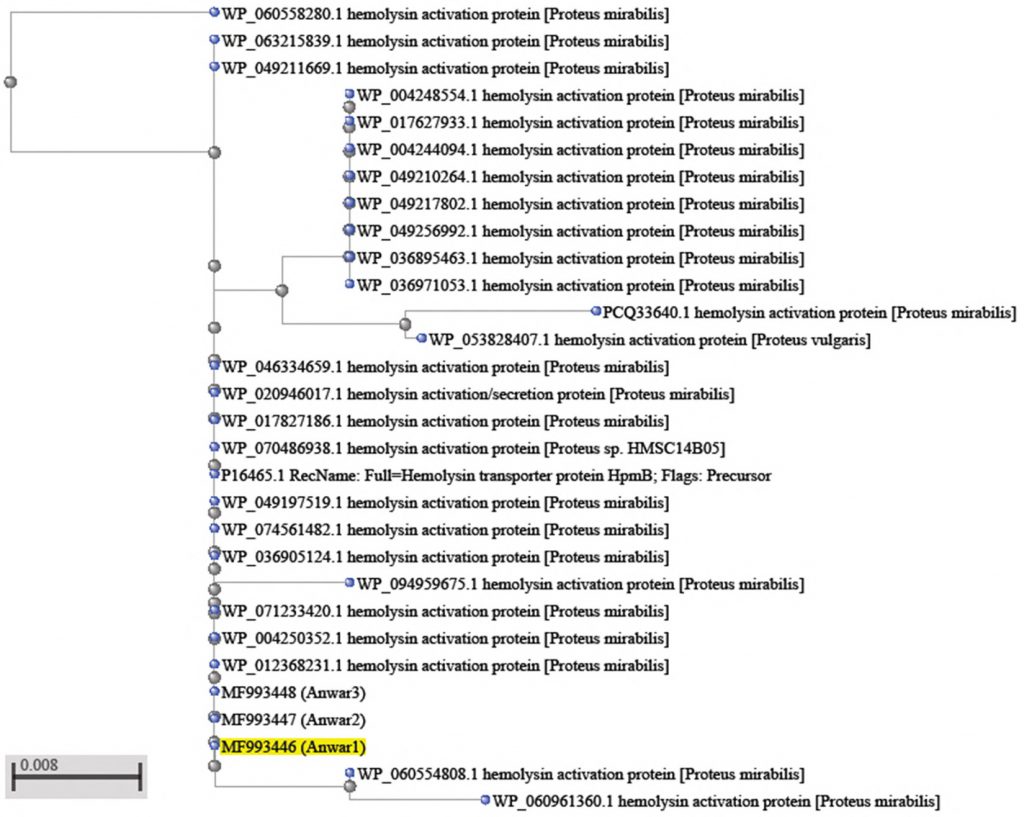
However, phylogeny estimated from a single gene should be treated with caution21,22. The phylogenetic tree derived from hpmB gene sequences of clinical strains of 2 samples Proteus mirabilis with other sequences is available at NCBI showed in (Fig.2). As to be seen in the this figure, P. mirabilis (MF993443 ) lies in the same branch of the phylogenetic tree with P.mirabilis (WP_088207120.1).
Sequences of 16SrRNA with the size of 1.5 Kb is considered and widely used in bacterial taxonomy because it contains high conservation region which has variable region in different species. Furthermore, the most important was that 16SrRNA gene which could be sequenced easily22. On the other hand, the sensitivity of this approach is questioned particularly among human bacterial closely related to Enterobacteriaceae, which includes many common pathogens because of the high degree of conservation in species22. Therefore, the use of other genes rather than16SrRNA gene and the distinction between bacteria at the species level is regarded as a very important issue23. Results indicate that since there is an increase in clinical significance of P. mirabilis, the choice of effectual molecular methods is of great epidemiological reputation. Bacterial genotyping opened new chances on epidemiological studies by the documentation of clinical and ecological isolates, the assessment of this association, the watching of clone propagation and the classification of bacterial populations within more or less constrained environments24. By joining molecular phylogeny with traditional approach such as morphological, physiological and biochemical characteristics, bacteria identification could be achieved in a more accurate way25,26.
The prevalence of UTIs was higher in females comparing of males, and the age group (35-49) years in adult diabetic patients. Escherichia coli was identified as the most common causative agent of UTIs in diabetes in Misan province. Although, the high rate of multi – drug resistance among bacterial isolates, but imipenem is a very effective antibiotic.
None
The authors declare no conflict of interest.
- Shakibaie, M., Forootanfar, H., Golkari, Y., Mohammadi-Khorsand, T., and Shakibaie, M. R. Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. J Trace Elem Med Biol. 2015; 29:235-241.
- Mohammed, G. J., Kadhim, M. J., and Hameed, I. H. Proteus species: Characterization and herbal antibacterial: A review. International J Pharmacog and Phytochem Res, 2016; 8(11):1844-1854.
- Stankowska, D., Kwinkowski, M., and Kaca, W. Quantification of Proteus mirabilis virulence factors and modulation by acylated homoserine lactones. J Microbiol Immunol Infect, 2008; 41(3):243-253.
- Howery, K. E., Clemmer, K. M., and Rather, P. N. The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr Genetics, 2016; 62(4):775-789.
- Ouda, S. M. Some nanoparticles effects on Proteus sp. and Klebsiella sp. isolated from water. Am J Infect Dis Microbiol, 2014; 2(1):4-10.
- Cestari ,S.E., Ludovico,M.S., Martins,F.H., Rocha,S.P.D., Elias,W.P. and Pelayo. J.S. Molecular Detection of HpmA and HlyA Hemolysin of Uropathogenic Proteus mirabilis. Curr Microbiol., 2013; 67:703–707.
- Rozalski, A., Torzewska, A., Moryl, M., Kwil, I., Maszewska, A., Ostrowska, K., .and Staחzek, P. Proteus sp.–an opportunistic bacterial pathogen–classification, swarming growth, clinical significance and virulence factors. Folia Biologica et Oecologica, 2012; 8(1):1-17.
- Uphoff, T. and Welch, R. Nucleotide sequencing of the Proteus mirabilis calcium independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with Serratia marcescens hemolysin genes (shlA and shlB). J Bacteriol., 1990; 172:1206–1216.
- Jumaily, E. F., and Zgaer, S. H. Characterization and molecular detection of purified Proteus mirabilis pmbs 41 alpha-hemolysin. Basrah J Veter Res, 2016; 15(3).
- Liaw, S. J., Lai, H. C., Ho, S. W., Luh, K. T., and Wang, W. B. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J Med Microbiol, 2003; 52(1):19-28
- Swihart, K. G., and Welch, R. A. The HpmA hemolysin is more common than HlyA among Proteus isolates. Infect and Immun, 1990; 58(6):1853-1860.
- Sosa, V.; Schlapp, G.and Zunino, P. Proteus mirabilis isolates of differentorigins do not show correlation with virulence attributes and can colonize the urinary tract of mice. Microbiol, 2006; 152(7):2149-2157.
- AL-Jumaa, M. H.; Bnyan, I. A. and Al-Khafaji, J. K. Bacteriological and Molecular Study of Some Isolates of Proteus mirabilis and Proteus vulgaris in Hilla Province. A thesis for the Degree of Master of Science in Microbiology. College of Medicine, University of Babylon, 2011.
- Mishara, M.; Thakar, Y.S. and Pathak, A.A. Haemagglutination, haemolysin production and serum resistance of Proteus and related species isolated from clinical sources, Indian J Med Microbiol. 2001; 19(2):5-11
- Abdul-Lateef, L. A. Sequencing of Proteus Toxic Agglutinin (Pta) Gene in Proteus Mirablis and Cytotoxic Effect of Pta on Human Colon Cancer Cell and Human Kidney Cell. J Global Pharma Technol, 2017; 9(9).
- Mordi, R. M., and Momoh, M. I. Incidence of Proteus species in wound infections and their sensitivity pattern in the University of Benin Teaching Hospital. African J Biotechnol, 2009; 8(5).
- Fraser GM, Claret L, Furness R, Gupta S, Hughes C. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiol, 2002; 148:2191–2201.
- Strauss, E. J., Ghori, N., and Falkow, S. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect and Immun, 1997; 65(9):3924-3932.
- Plomin, R., DeFries, J. C., Knopik, V. S., and Neiderheiser, J. Behavioral genetics. Palgrave Macmillan, 2013.
- Li, W., Raoult, D., and Fournier, P. E. Bacterial strain typing in the genomic era. FEMS Microbiol Revs, 2009; 33(5):892-916.
- Hadfield, J. D., and Nakagawa, S. General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi trait models for continuous and categorical characters. J Evolution Biol, 2010; 23(3), 494-508.
- Case, R. J., Boucher, Y., Dahllof, I., Holmstrצm, C., Doolittle, W. F., and Kjelleberg, S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environmen Microbiol, 2007; 73(1):278-288.
- Ghebremedhin, B., Layer, F., Konig, W., and Konig, B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J Clinic Microbiol, 2008; 46(3):1019-1025.
- Michelim, L., Muller, G., Zacaria, J., Delamare, A. P. L., Costa, S. O. P. D., and Echeverrigaray, S. Comparison of PCR-based molecular markers for the characterization of Proteus mirabilis clinical isolates. Brazilian J Infect Dis, 2008; 12(5), 423-429.
- Ma, R., Wu, X., Wang, R., Wang, C., and Zhu, J. Identification and phylogenetic analysis of a bacterium isolated from the cloaca of Chinese alligator. African J Biotechnol, 2008; 7(13).
- Brenner, D. J., Staley, J. T., and Krieg, N. R. Classification of Procaryotic Organisms and the Concept of Bacterial Speciation. Bergey’s Manual of Systematics of Archaea and Bacteria, 2015; 1-9.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.