ISSN: 0973-7510
E-ISSN: 2581-690X
The ability of Methicillin-resistant Staphylococcus aureus (MRSA) to form biofilms is one of the triggering factors for the emergence of MRSA resistance to antibiotics. Streptomyces W-5B has shown potency as an antibacterial producer against MRSA. However, the production of microbial bioactive compounds is strongly affected by the source of nutrients in the fermentation medium. Therefore, the objective of this study was to determine the optimal sources of carbon and nitrogen for the production of bioactive compounds with antibiofilm activities. The research method included cultivating Streptomyces W-5B, extract production, and variation of carbon (glucose, sucrose, starch) and nitrogen (casein, peptone, urea) sources for fermentation medium. Antibiofilm activities were measured based on inhibition of biofilm formation and biofilm degradation tests using the microtiter plate method with a crystal violet stain. The results showed that the highest inhibition of biofilm formation was 68.206 ± 1.750% after 12 days of incubation in a fermentation medium containing sucrose and urea. Meanwhile, the highest biofilm degradation was 73.023 ± 1.972% after nine days of incubation on a fermentation medium containing starch and urea. These findings indicated that Streptomyces W-5B has the potency to produce antibiofilm extract against MRSA.
Antibiofilm, carbon source, MRSA, nitrogen source, Streptomyces
Methicillin-resistant Staphylococcus aureus (MRSA) is an S. aureus bacterium resistant to β-lactam antibiotics such as penicillin and its derivatives, namely methicillin, oxacillin, dicloxacillin, nafcillin, and cephalosporins.1 Penicillin-resistant strain of S. aureus produces a plasmid encoding a lactamase (penicillinase) capable of hydrolyzing the β-lactam ring of penicillin, causing antibiotic resistance. Currently, MRSA is one of the leading causes of infection in humans. Research conducted by Basanisi et al.2 showed that of the 12.9% of samples infected with S. aureus, 8.3% were MRSA. The World Health Organization (WHO) already classifies MRSA as one of the priority pathogens that threaten human health due to antibiotic resistance.3 Resistance to MRSA increases due to the ability of MRSA to form biofilms.4
Biofilm is a matrix layer of extracellular polymeric substances (EPS) produced by bacteria. The EPS matrix generally consists of oligosaccharides, DNA, and proteins.5 Bacteria with the ability to form biofilms can withstand stress and harsh host environments. Biofilms provide protection against antibiotics, thereby increasing resistance compared to bacteria in a planktonic state.4 The ability of MRSA to produce biofilms will inhibit the penetration of antibiotics, thus increasing resistance properties. Therefore, the exploration of bioactive compounds with antibiofilm properties is essential as an alternative therapy for MRSA infections. Antibiofilm compounds play a role in inhibiting the formation of biofilms and the degradation of biofilms that have been formed.
Bioactive compounds from actinobacteria, particularly the genus Streptomyces, have been reported as antibiofilm against MRSA. Streptorubin B extracted from Streptomyces sp. MC11024 belongs to a group of antibiotics capable of inhibiting and damaging the MRSA N315 biofilm.6 Ethyl acetate extract of Streptomyces sp. SBT343 is known to significantly inhibit the biofilm of several Staphylococcal species, including MRSA USA300.7 Bhakyashree & Kannabiran8 also reported that ethyl acetate extract of Streptomyces gancidius has antibacterial activity against MRSA strains ATCC 43300 and ATCC 700699.
Furthermore, Singh & Dubey9, reported that Streptomyces californicus strain ADR1 had an antibiofilm activity that could inhibit and degrade biofilms of MRSA strains 562 and ATCC 43300. Asnani et al.10 succeeded in exploring actinobacteria from the mangrove area of Segara Anakan Cilacap, which has the potential as a source of antibacterial compounds, one of which is Streptomyces W-5B. Biosynthetic Gene Cluster (BGC) analysis of Streptomyces W-5B suggested eight BGCs that play a role in the biosynthesis of secondary metabolites.11 The potency of Streptomyces W-5B encourages further research to optimize the production of antibiofilm compounds against MRSA.
Extracts produced by a microorganism will have optimum bioactivities if nutrient sources for microbial growth are selected accordingly. Carbon and nitrogen sources are the main components in a culture medium grown by microorganisms for cell growth processes and production of secondary metabolites, particularly in Streptomyces.12 Carbon is the main component of all organic molecules in cells, whereas Nitrogen plays an essential role in preparing nucleic acids, amino acids, and enzymes.13 Hence, optimization of the fermentation process can be done by varying the composition of the media through simultaneous experiments to determine the interaction between variables. Therefore, this research aimed to optimize the production of antibiofilm extract from Streptomyces W-5B by varying carbon and nitrogen sources.
The research was carried out from December 2020 to May 2021 at the University of Jenderal Soedirman, Purwokerto, Indonesia. Streptomyces W-5B was isolated from mangrove area in Segara Anakan, Cilacap, Indonesia. The species identification based on the 16S rRNA gene of Streptomyces W-5B showed 99,92% similarity with Streptomyces cellulosae strain NBRC 13027.11 The isolate MRSA 2983 was collected from a clinical specimen from the pus of a female patient in Prof. Dr. Margono Soekarjo Hospital, Banyumas regency, Indonesia.14
Cultivation of Streptomyces W-5B
Cultivation of Streptomyces W-5B followed the procedure described by Asnani et al.10 Streptomyces W-5B was cultivated by quadrant streak on Starch Casein Nitrate (SCN) agar medium consisting of starch, casein, KNO3, K2HPO4, MgSO4.7H2O, NaCl, FeSO4.7H2O, and Bacto agar supplemented with nystatin. The culture was incubated for seven days at room temperature prior to being used.
A total of 10 plugs (6 mm in diameter) of Streptomyces W-5B culture were inoculated into 100 ml of liquid SCN medium [starch, casein, KNO3, K2HPO4, MgSO4.7H2O, NaCl, and FeSO4.7H2O] supplemented with nystatin. The mixture was incubated using an orbital shaker at 90 rpm for eight days at room temperature. After incubation, the culture was used as an inoculum for the fermentation process.
Determination of the Optimal Incubation Time
The optimal incubation time was determined by varying the incubation time in the fermentation process. As much as 10% inoculum was inoculated in a liquid SCN medium. The culture was then incubated using an orbital shaker at 90 rpm with various incubation times 0, 3, 6, 9, 12, and 15 days. After each incubation, the cultures were centrifuged at 4000 rpm for 10 minutes at 4°C, then filtered to obtain Streptomyces W-5B extracts. The extracts obtained from various incubation times were tested against MRSA 2983 to evaluate extract potency to inhibit biofilm formation and degrade biofilm. The precipitate obtained was dried at 80°C for 1 hour for the dry weight. The production curve was made using the dry weight of the precipitate (g) and the percentage of inhibition of biofilm formation or biofilm degradation against the incubation time (days). The incubation time with the highest % inhibition and % degradation against MRSA 2983 was then used as the incubation time for the production of extracts with various carbon and nitrogen sources.
Determination of Carbon and Nitrogen Sources
The fermentation experiment used a complete randomized design with two factorials to evaluate the optimal carbon and nitrogen sources. The first factor was three different carbon sources (K1 = glucose, K2 = sucrose, K3 = starch), and the second factor was three different nitrogen sources (N1 = casein, N2 = peptone, N3 = urea). All treatments were replicated three times, so there were 3 x 3 x 3 = 27 experimental units.
Up to 10% inoculum was inoculated in a liquid SCN medium with a combination of carbon and nitrogen sources in a fermentation medium. Cultures were incubated using an orbital shaker at 90 rpm at room temperature at the optimal incubation time. Each extract obtained was tested for the activity to inhibit biofilm formation and to degrade biofilm.
Inhibition of the biofilm formation test
The ability of the extract to inhibit biofilm formation was analyzed using a semi-quantitative microtiter plate assay following the procedure described by Dinda et al.14 The MRSA 2983 inoculated into Brain Heart Infusion (BHI) broth supplemented with 1% glucose (BHI-Glu). The culture was incubated at 37°C for 24 hours, adjusted to 0.5 McFarland standard, and diluted with BHI-Glu medium in a 1:100 ratio to become an MRSA subculture.
The inhibition of biofilm formation was carried out by adding 10µL of MRSA subculture and 100 µL of extract into 100 µL of BHI-Glu medium in each well in a 96-well microplate. The mixture was incubated at 37°C for 24 hours. After incubation, planktonic cells were carefully removed, and the microplate was rinsed with 300 µL of PBS twice and stained with 50 µL of 1% (w/v) crystal violet solution. The absorbance value was measured at 620 nm, and the percentage of inhibition of biofilm formation was calculated by the following formula:
% Inhibition=(OD control-OD test)/(OD control) x 100%
The minimum biofilm inhibition concentration of 50% (MBIC-50) was defined as extract concentration at which a 50% decrease in absorbance readings was detected in comparison to the absorbance of control biofilms formed in the absence of extract. MBIC-50 was determined by calculating the cut-off point at 50% inhibition by the following formula.15:
MBIC-50=(OD control-OD Blank) x 50/100
Biofilm Degradation Test
The ability of the extract to degrade biofilm was also analysed with a semi-quantitative microtiter plate biofilm assay.14 Up to 10 µL of MRSA subculture was added into 100 µL of BHI-Glu medium in each well on a 96-well microplate, and the mixture was incubated at 37°C for 24 hours. After incubation, the planktonic cells were carefully removed, and each well was added 100 µL of extract. The mixture was further incubated at 37°C for 24 h to expose the extract with the biofilm formed. After incubation, the microplate was washed similarly with the procedure mentioned above, and the absorbance value was measured at 620 nm. The percentage of biofilm degradation was calculated by the following formula:
% Degradation=(OD control-OD test)/(OD control) x 100%
Likewise, the minimum biofilm reduction concentration of 50% (MBRC-50) was defined as extract concentration at which a 50% decrease in absorbance readings were detected in comparison to the absorbance of control biofilms formed in the absence of extract. MBRC-50 was determined by calculating the cut-off point at 50% reduction by the following formula.15:
MBRC-50=(OD control-OD Blank) x 50/100
Data Analysis
All data were analyzed using analysis of variance (ANOVA) at a confidence level of 95%. The results of the ANOVA were significantly different, followed by Duncan’s test.
Streptomyces W-5B is a filamentous bacterium with brown pigmentation to medium, yellow substrate mycelium, and white aerial mycelium (Fig. 1). The colony is circular and has a powdery surface. Streptomyces W-5B has been reported to have antibacterial activity against MRSA 2983.11
The Optimum Incubation Time
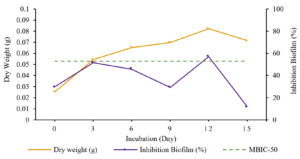
The production curve depicts the relationship between incubation time and Streptomyces W-5B extract production. The absorbance value represented the inhibitory activity, and biofilm degradation read at 620 nm. Then, the absorbance value was used to calculate the percentage of inhibition and degradation of the biofilm. The specific incubation time was determined to produce an extract with the highest activity, while the bacterial precipitate measured as dry weight was used to analyze the bacterial growth. The research results showed that the optimum incubation time to produce Streptomyces W-5B extract with the highest inhibition activity was 12 days, with the percentage of biofilm inhibition being 55,456 ± 3,306% (Fig. 2). This result followed the microbial growth, which showed optimum growth on 12-days based on the dry weight (0.0823 g). The percentage of biofilm inhibition at the incubation time of 12 days also had a value greater than the cut-off point of MBIC-50. These results indicated that an incubation time of 12 days would produce an extract that could inhibit biofilm formation by more than 50%.
The one-way ANOVA results showed a significant effect (Sig. <0.05) between the incubation time and the percentage of inhibition of biofilm formation. Further tests using Duncan’s test showed that treatment with an incubation time of 12 days was significantly different from other treatments (0, 6, 9, 15 days) with the highest percentage of biofilm inhibition (55,456 ± 3,306%).
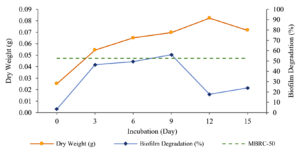
The percentage of biofilm degradation by Streptomyces W-5B extract increased from day 0 to the optimal incubation time on day 9 with a percentage value of biofilm degradation of 56.146 ± 8.784% (Fig. 3). The biofilm degradation that crossed the MBRC-50 boundary line was the extract produced on the 9th day, which indicated that extract produced with an incubation time of 9 days could degrade 50% of the biofilm.
The one-way ANOVA results showed a significant effect (Sig. <0.05) between the incubation time and the percentage of biofilm degradation. Further tests using Duncan’s test concluded that variations of incubation time show significantly different effects. However, the incubation time treatment on the 9th day showed no significant effect on the 3rd and 6th days. The optimal incubation time was on the ninth day, with the highest percentage of biofilm degradation being 56.146 ± 8.784%.
Based on these results, the fermentation process with various carbon and nitrogen sources used 12 days of incubation to produce an extract with inhibitory activity. In comparison, nine days of incubation was used to produce an extract with biofilm degradation activity.
The Optimum Carbon and Nitrogen Sources
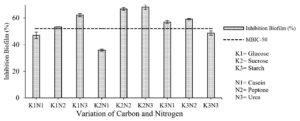
Variations of carbon and nitrogen sources determined the best combination of carbon and nitrogen sources in fermentation medium to produce an extract with the highest inhibition biofilm formation and biofilm degradation activities. Based on the research results (Fig. 4), the highest percentage of inhibition of biofilm formation (68.206 ± 1.750%) was produced from the K2N3 fermentation medium (sucrose-urea). The percentage of inhibition in the K2N3 medium was higher than the cut-off point of MBIC-50, which means that the extract produced in the K2N3 medium could inhibit biofilm formation by more than 50%. Other extracts with a higher value than the cut-off point of MBIC-50 were K1N2 (glucose-peptone), K1N3 (glucose-urea), K2N2 (sucrose-peptone), K3N1 (starch-casein), and K3N2 (starch-peptone).
Fig. 4. Correlation of combined carbon and nitrogen sources in fermentation medium with the percentage of inhibition of biofilm formation.
The results of the two-way ANOVA showed a significant effect (Sig. <0.05) between the interaction of carbon and nitrogen sources on the percentage of inhibition of biofilm formation. Further tests using Duncan’s test showed that the treatment with sucrose had a significantly different effect on other treatments.
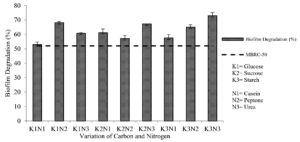
The results from the biofilm degradation test indicated that the highest percentage of biofilm degradation (73.023 ± 1.972%) was obtained from the extract produced by fermentation medium K3N3 (starch-urea). The MBRC-50 value determines the minimum concentration capable of degrading 50% of the biofilm. 7 Interestingly, all extracts had a higher value than MBRC-50, which means that they can degrade more than 50% of the biofilm formed. Still, the best carbon-nitrogen combination was K3N3, with the highest biofilm degradation activity (Fig. 5).
Fig. 5. Correlation of combined carbon and nitrogen sources in fermentation medium with the percentage of biofilm degradation
The two-way ANOVA results showed a significant effect (Sig. <0.05) between the interaction of carbon and nitrogen sources on the percentage of biofilm degradation. Further tests using Duncan’s test concluded that the percentage of biofilm degradation was significantly different with a combination of other carbon and nitrogen sources. The best combination for fermentation medium was K3N3 (starch-urea) which produced the highest (73.023 ± 1.972%) biofilm degradation activity.
Streptomyces W-5B has been identified as Streptomyces cellulosae strain NBRC 13027. It has been reported to have antibacterial activity against MRSA 2983 with an inhibitory index of 0.47 mm and contained biosynthetic gene clusters (BGC) that have antibacterial potential.11 According to Memariani et al.,16 biofilm inhibition can occur at a lower concentration than the concentration to kill planktonic bacteria so that antibacterial compounds can also be used to inhibit biofilm formation. Thus, this research further analyzed the potency of Streptomyces W-5B to produce antibiofilm extract against MRSA.
Streptomyces W-5B was cultivated on SCN medium, which is a suitable medium for the growth of actinobacteria. The screening and isolation of 36 actinobacterial isolates using SCN agar were successfully carried out by Rajaram et al.17 Recently, SCN agar was also used to isolate antibiotic-producing bacteria (APB) from river sediments of Bangladesh. The 16S rDNA sequencing analysis revealed APB as Streptomyces genus.18 Actinobacteria grown on SCN agar medium showed fast growth and were able to produce secondary metabolites. Vellingiri et al.19 reported that SCN medium has been effective for Streptomyces hygroscopicus AVS7 to produce bioactive metabolites.
The high production of bioactive compounds was correlated with the microbial growth phase during incubation. Ryandini et al.20 reported that incubation time affects the production of bioactive compounds from actinobacteria. However, different bacteria might have different optimal incubation times to produce extracts with antibiofilm activity. Bakkiyaraj & Pandian,21 have reported that Streptomyces akiyoshiensis CAA-3 produced an antibiofilm extract against S. aureus ATCC 11632 and MRSA ATCC 33591 with an incubation time of six days. Lee et al.22 reported that Streptomyces parvulus produced an antibiofilm compound against MRSA ATCC 33591 with an incubation time of seven days.
In general, the production of bioactive compounds occurs in the stationary phase or at the end of the exponential phase until the end of the microbial incubation period.23 Similarly, in this research, the extract with inhibition of biofilm formation activity occurred at the beginning of the stationary phase. In contrast, the extract with biofilm degradation activity occurred exponentially. This result suggested biofilm degradation might correlate with the hydrolytic enzyme produced during the exponential phase. Indeed, Millenbaugh et al.24 stated that biofilm degradation activity is related to the enzymatic activity produced by bacteria to reduce biofilms.
Antibiofilm compounds inhibit biofilm formation by preventing the adhesion of planktonic bacteria to the surface or host tissues to form biofilms. The mechanism of inhibition occurs at the beginning of the reversible attachment of planktonic bacteria to the surface before the biofilm matrix maturation occurs. Inhibition takes place because biofilm-producing bacteria are not yet protected by a mature biofilm matrix; hence they are still very susceptible to antibiotic compounds.25
Carbon and nitrogen are macronutrients that affect the production of microbial bioactive compounds.13 In this research, the combination of sucrose and urea produces extract with the highest activity to inhibit biofilm formation. Sucrose is a better carbon source than other carbon sources for producing secondary metabolites that function as antibacterial.26 The presence of sucrose in the fermentation medium can stimulate the activity of the phenylalanine ammonia lyase (PAL) enzyme involved in producing secondary metabolites. This PAL enzyme plays a vital role in microbial defense mechanisms against environmental stresses.27 The addition of sucrose can also suppress the salt content in the fermentation medium. The lack of salt will activate genes that produce secondary metabolites such as compounds from the polyketide group.28 Urea acts as a simple nitrogen source that is easily used by actinobacteria so that actinobacteria can reach the stationary phase more quickly to produce secondary metabolite compounds that play a role in inhibiting the formation of MRSA biofilms.
Based on the research results, the combination of starch and urea produced the extract with the highest biofilm degradation activity. The mechanism of biofilm degradation is carried out by weakening hydrogen bonds, which are considered necessary for the stability of the EPS matrix.29 Starch, a source of carbon in the form of polysaccharides, can help bacteria produce organic molecules in cells, including anti-adhesion polysaccharides or antibiofilm polysaccharides. This antibiofilm polysaccharide can act as a biofilm degrader by damaging cell-to-cell interactions in the biofilm matrix.30 Suzuki et al.6 reported that Streptomyces grown in media containing starch could produce compounds with degradation activity against S. aureus biofilms. Nandhagopal et al.31 have used starch as a carbon source to produce biofilm-degrading compounds against MRSA.
Urea as a nitrogen source will help bacteria synthesize essential components such as amino acids and proteins. These components play a significant role in producing bioactive metabolites such as enzymes and other metabolites that degrade biofilms.32 Urea has been used in bacterial growth medium to produce an extracellular enzyme complex that can degrade S. aureus biofilms up to 80%.33 The enzymes effectively degrade biofilms because they lyse cells and tissues to damage the biofilm matrix.
Our results conclude that Streptomyces W-5B can produce MRSA antibiofilm compounds. The antibiofilm activity consists of antibacterial, inhibitor of biofilm formation, and biofilm degrading. Actinobacteria are known as antibiotic-producing bacteria. Many commercial antibiotics include important antimicrobials such as β-lactams, rifamycin, aminoglycosides, macrolides, tetracyclines, erythromycin, vancomycin, and streptomycin are derived from the secondary metabolism of actinobacteria. In the class Actinobacteria, the genus Streptomyces is the primary producer of various secondary metabolites and many antibiotics. One of the determining factors for producing primary and secondary metabolites is the fermentation media formulation. The fermentation medium can also act as an induction to obtain the desired metabolite optimally. In the future, it is necessary to identify and characterize secondary metabolites that specifically function as MRSA antibiofilm.
ACKNOWLEDGMENTS
The authors would like to express their gratitude to Ms. Sinta Prima Ardinas for assisting with biofilm measurements in the Research Laboratory, Faculty of Medicine, UNSOED, Purwokerto, Indonesia. The authors also appreciate Associate Prof. Amin Fatoni and Associate Prof. Zusfahair for valuable input in reviewing the research results.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AA supervised the entire work. DUA led the antibiofilm work. AP and WAB performed all experiments. AA wrote and revised the manuscript. DUA reviewed the manuscript. AA and DUA approved the manuscript for publication.
FUNDING
This study was funded by the Ministry of National Education, Culture, Research and Technology of the Republic of Indonesia for the Fundamental Research Grant with the grant No. /1411/UN23.18/PT.01.01/2021.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated and analyzed during this research are included in the manuscript.
- Lee AS, de Lencastre H, Garau J, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Prim. 2018;4:18033.
Crossref - Basanisi MG, La Bella G, Nobili G, Franconieri I, La Salandra G. Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 2017;62:141-146.
Crossref - WHO. World Health Organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. 2017
- Craft KM, Nguyen JM, Berg LJ, Townsend SD. Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm. 2019;10(8):1231-1241.
Crossref - Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563-575.
Crossref - Suzuki N, Ohtaguro N, Yoshida Y, et al. A compound inhibits biofilm formation of Staphylococcus aureus from Streptomyces. Biol Pharm Bull. 2015;38(6):889-892.
Crossref - Balasubramanian S, Skaf J, Holzgrabe U, et al. A new bioactive compound from the marine sponge-derived Streptomyces sp. SBT348 inhibits Staphylococcal growth and biofilm formation. Front Microbiol. 2018;9:1473.
Crossref - Bhakyashree K, Kannabiran K. Actinomycetes mediated targeting of drug resistant MRSA pathogens. J King Saud Univ – Sci. 2020;32(1):260-264.
Crossref - Singh R, Dubey A. Isolation and characterization of a new endophytic actinobacterium Streptomyces californicus strain ADR1 as a promising source of antibacterial, anti-biofilm and antioxidant metabolites. Microorganisms. 2020;8(6):929.
Crossref - Asnani A, Luviriani E, Oedjijono O. Activity of actinomycetes isolated from mangrove Segara Anakan Cilacap toward methicillin-resistant Staphylococcus aureus (MRSA). J Kim Sains dan Apl. 2020;23(1):1-7.
Crossref - Asnani A, Amaliyah R, Yuniaty A. Screening anti-MRSA activities of indigenous microbes and prediction of the Biosynthetic Gene Clusters. Journal of Physics: Conference Series. 2020:1665.
Crossref - Romero-Rodriguez A, Maldonado-Carmona N, Ruiz-Villafan B, Koirala N, Rocha D, Sanchez S. Interplay between carbon, nitrogen and phosphate utilization in the control of secondary metabolite production in Streptomyces. Antonie Van Leeuwenhoek. 2018;111(5):761-781.
Crossref - Al-Ghazali LH, Omran R. Optimization of medium composition for antibacterial metabolite production from Streptomyces sp. Asian J Pharm Clin Res. 2017;10(9):381-385.
Crossref - Dinda AP, Asnani A, Anjarwati DU. The Activities of Streptomyces W-5A as antibacterial and antibiofilm towards methicillin-resistant Staphylococcus aureus 2983. In: Proceedings of the 1st Jenderal Soedirman International Medical Conference in Conjunction with the 5th Annual Scientific Meeting (Temilnas) Consortium of Biomedical Science Indonesia. SCITEPRESS – Science and Technology Publications. 2021:109-115.
Crossref - Pierce CG, Uppuluri P, Tummala S, Lopez-Ribot JL. A 96 well microtiter plate-based method for monitoring formation and antifungal susceptibility testing of Candida albicans biofilms. J Vis Exp. 2010;(44):2287.
Crossref - Memariani H, Memariani M, Ghasemian A. An overview on anti-biofilm properties of quercetin against bacterial pathogens. World J Microbiol Biotechnol. 2019;35(9):143.
Crossref - Rajaram SK, Ahmad P, Keerthana SSS, Jeya Cressida P, Moorthy IG, Suresh RSS. Extraction and purification of an antimicrobial bioactive element from lichen associated Streptomyces olivaceus LEP7 against wound inhabiting microbial pathogens. J King Saud Univ – Sci. 2020;32(3):2009-2015.
Crossref - Arefa N, Sarker AK, Rahman MA. Resistance-guided isolation and characterization of antibiotic-producing bacteria from river sediments. BMC Microbiol. 2021;21(1):116.
Crossref - Vellingiri MM, Arockiam Jeyasundar PGS, Venkatesan BP, Liu W, Balasubramanian B. Statistical optimization of parameters for enhanced bioactive metabolites produced by Streptomyces hygroscopicus AVS7. Arab J Sci Eng. 2021;46(6):5345-5360.
Crossref - Ryandini D, Radjasa OK, Oedjijono. Isolate actinomycetes SA32 origin of Segara Anakan mangrove rhizosphere and its capability in inhibiting multi-drugs resistant bacteria growth. J Microb Biochem Technol. 2018;10(1):1-7.
Crossref - Bakkiyaraj D, Pandian STK. In vitro and in vivo antibiofilm activity of a coral associated actinomycete against drug resistant Staphylococcus aureus biofilms. Biofouling. 2010;26(6):711-717.
Crossref - Lee J-H, Kim Y-G, Lee K, et al. Streptomyces-derived actinomycin D inhibits biofilm formation by Staphylococcus aureus and its hemolytic activity. Biofouling. 2016;32(1):45-56.
Crossref - Singh N, Rai V. Optimization of cultural parameters for antifungal and antibacterial metabolite from microbial isolate; Streptomyces rimosus MTCC 10792 from soil of Chhattisgarh. Int J Pharm Pharm Sci. 2012;4(4):94-101.
- Millenbaugh N, Watters C, Burton T, Kirui D. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect Drug Resist. 2016;9:71-78.
Crossref - Estela L, Rivera C, Ramos P. Biofilms: A Survival and resistance mechanism of microorganisms. In: Antibiotic Resistant Bacteria – A Continuous Challenge in the New Millennium. IntechOpen. 2012;576:159-178.
Crossref - Iniyan AM, Joseph FRS, Kannan RR, Vincent SGP. Anti-MRSA potential of phenolic compound isolated from a marine derived actinomycete Micromonospora sp. ICN36. Indian J Geo-Marine Sci. 2016;45(10):1279-1287.
- Formela-Luboińska M, Chadzinikolau T, Drzewiecka K, et al. The role of sugars in the regulation of the level of endogenous signaling molecules during defense response of Yellow Lupine to Fusarium oxysporum. Int J Mol Sci. 2020;21(11):4133.
Crossref - Yi JS, Kim M, Kim S-J, Kim B-G. Effects of sucrose, phosphate, and calcium carbonate on the production of pikromycin from Streptomyces venezuelae. J Microbiol Biotechnol. 2015;25(4):496-502.
Crossref - Brindle ER, Miller DA, Stewart PS. Hydrodynamic deformation and removal of Staphylococcus epidermidis biofilms treated with urea, chlorhexidine, iron chloride, or Dispersin B. Biotechnol Bioeng. 2011;108(12):2968-2977.
Crossref - Rendueles O, Kaplan JB, Ghigo J-M. Antibiofilm polysaccharides. Environ Microbiol. 2013;15(2):334-346.
Crossref - Nandhagopal S, Iniyan AM, Kannan RR, Vincent SGP. In vivo evaluation of anti-MRSA compound from Streptomyces collinus ICN1 in zebrafish embryos. Indian J Geo-Marine Sci. 2017;46(6):1155-1161.
- Sharma G, Karnwal A. Biological strategies against biofilms. In: Singh J, Vyas A, Wang S, Prasad R, eds. Microbial Biotechnology: Basic Research and Applications. Environmental and Microbial Biotechnology. Springer, Singapore. 2020:205-232.
Crossref - Nagraj AK, Gokhale D. Bacterial biofilm degradation using extracellular enzymes produced by Penicillium janthinellum EU2D-21 under submerged fermentation. Adv Microbiol. 2018;08(09):687-698.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.