ISSN: 0973-7510
E-ISSN: 2581-690X
This study investigated the expression profiles of two specific microRNAs (miRNAs), miR-1271-5p and miR-10a-3p, in patients with bacterial vaginosis (BV) and in healthy controls. One hundred participants (50 patients with BV and 50 controls) were recruited. Bacterial species were identified using multiplex PCR of the 16S rRNA gene from vaginal swabs, and miRNA expression was quantified from blood samples using RT-qPCR. Gardnerella vaginalis was the most prevalent species (66.0%) in patients with BV. The expression levels of miR-1271-5p and miR-10a-3p were significantly higher in patients with BV than in healthy controls (p < 0.001). Receiver operating characteristic curves revealed promising diagnostic values: an miR-1271-5p cutoff of <5.42-fold achieved 94.0% sensitivity and specificity (area under the curve (AUC), 0.951), whereas an miR-10a-3p cutoff of <3.57-fold showed 96.0% sensitivity and 92.0% specificity (AUC, 0.970). Interestingly, miR-1271-5p and miR-10a-3p levels were the highest in patients with Megasphaera colonization. Thus, miR-1271-5p and miR-10a-3p are potential biomarkers for the diagnosis of BV and offer high diagnostic value. These miRNAs may play a crucial role in understanding BV pathogenesis and serve as targets for future diagnostic strategies.
miRNAs, miR-1271-5p, miR-10a-3p, Expression, Bacterial Vaginosis
Bacterial vaginosis (BV) is a common polymicrobial vaginal infection affecting approximately 30% of women of reproductive age.1 It is characterized by a significant alteration in the vaginal microbiota, specifically a reduction in beneficial Lactobacillus species and an overgrowth of various anaerobic bacteria.2 This microbial imbalance can lead to symptoms such as vaginal discharge, odor, and pain. Although the exact etiology of BV is not fully understood, factors such as sexual behavior, douching, and antibiotic use have been implicated.1 A shift in the microbiome, marked by the proliferation of facultative and obligate anaerobic microorganisms, particularly Gardnerella spp., is a key indicator of BV.3
The vaginal microbiome plays a crucial role in the maintenance of vaginal health. Recent studies have highlighted the intricate relationship between microRNAs (miRNAs) and the vaginal microbiome, demonstrating that miRNA expression patterns are significantly affected by the composition of this microbial community.4 miRNAs are small, non-coding RNAs that regulate gene expression.5,6 Evidence suggests that miRNAs may be involved in the pathogenesis of BV, with studies reporting altered levels of some miRNAs in women with BV compared to healthy individuals.4-6 This dysregulation of miRNAs, along with immune mediators, contributes to both anaerobic bacterial overgrowth and BV-related inflammation.
The precise mechanisms by which miRNAs contribute to BV pathogenesis and their potential as diagnostic biomarkers remain to be fully elucidated.7,8 The diagnostic utility of specific miRNAs in BV infections, particularly in Iraqi women, warrants further investigation. This study aimed to investigate the expression profiles of specific miRNAs, miR-1271-5p and miR-10a-3p, in patients with BV compared to healthy controls, and to evaluate their potential as diagnostic biomarkers for the disease.
Vaginal swab and blood sample collection
One hundred participants were recruited between January and October 2023 from the Microbiology Laboratory of Al-Diwanyiah Hospital, Al-Qadisiyah Province, Iraq. The cohort included 50 women aged 30-50 years diagnosed with BV and 50 healthy controls aged 30-45 years. Patient selection criteria were based on a confirmed diagnosis of BV using the Nugent score, which involves Gram staining of vaginal smears followed by microscopic evaluation to quantify bacterial morphotypes. A score of 0-3 indicates normal flora dominated by Lactobacillus morphotypes, 4-6 indicates intermediate flora, and 7-10 confirms BV with predominant Gardnerella/Bacteroides morphotypes and curved gram-variable rods. Healthy controls were women without any symptoms of vaginal infection and negative for BV according to the Nugent score.
All participants provided written informed consent prior to sample collection. The study protocol was reviewed and approved by the Institutional Review Board of the Iraqi Ministry of Health, Human Development and Training Center (Approval No. 1/2024/49). Patients were screened for recent antibiotic use and sexual lifestyle; only those who had not used antibiotics for more than six months and reported a generally normal sexual lifestyle were included. Demographic information including age and sex was also collected. Vaginal swabs were collected for bacterial species identification using multiplex PCR targeting the 16S rRNA gene. Blood samples were obtained for the gene expression analysis of miR-1271-5p and miR-10a-3p. Blood samples were immediately stored in TRIzol reagent at -20 °C, while vaginal swab samples were stored in PBS at -84 °C for subsequent analysis.
Multiplex polymerase chain reaction (PCR)
Multiplex-PCR was performed to identify microbial vaginosis using 16S ribosomal nucleic acid genetic factors. Microbial genomic DNA was extracted from the vagina using the Presto Mini gDNA Bacteria kit (Geneaid, New Taipei City, Taiwan). Extracted DNA was validated through NanoDrop (Thermo Scientific NanoDrop Lite UV Visible Spectrophotometer, UK) and quantified in ng/µL. Master mix of multiplex-PCR was prepared using GoTaq Green PCR master kit (Promega, Madison, USA) according to the instructions provided. DNA template samples (5-50 ng), forward and reverse primers of six bacterial species (Megasphaera sp., Atopobium sp., Lactobacillus sp., Erysipelothrix sp., Cutibacterium sp., and Gardnerella vaginalis) (10 pmol each), GoTaq Green PCR Master Mix 25 µL, (Promega, Madison, USA), and nuclease-free water were used. The total content of the master mix was 50 µL. This procedure was used for the prospective detection and identification of these six species within a DNA sample. Gene amplification was performed using a T100 PCR Thermocycler (Bio-Rad, California, USA). Finally, the PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide.
Stem-loop RT-qPCR
Stem-loop reverse transcription quantitative PCR (RT-qPCR) was used to quantify the expression of miR-1271-5p and miR-10a-3p in serum samples. Total RNA was isolated from 250 µL of blood using the TRIzol reagent kit (AccuZol RNA Extraction Kit, Bioneer, South Korea) following the manufacturer’s protocol. The extracted RNA was treated with DNase I (DNase I enzyme kit, Promega, Madison, USA) to eliminate trace genomic DNA contamination according to the manufacturer’s instructions. Subsequently, cDNA was synthesized from RNA using the M-MLV Reverse Transcriptase kit according to the manufacturer’s guidelines. The RT-qPCR mix was prepared using the GoTaq qPCR Master Mix kit, which relies on SYBR Green dye for the real-time detection of gene amplification. Data analysis for the target genes and the housekeeping gene, coding for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was performed using the relative quantification method based on the ΔCT approach. The calculations were as follows: ִCT (test) = CT (target gene, test) – CT (housekeeping gene, test); ΔCT (Control) = CT (target gene, control) – CT (housekeeping gene, control); ΔΔCT = ΔCT (test) – ΔCT (control); and fold change (target/housekeeping gene) = 2 – CT (-ΔΔCT).
Statistical methods
All statistical analyses were conducted using IBM SPSS software, version 22. Data for continuous variables were analyzed as the mean ± standard deviation (x ± SD), while categorical variables were summarized using frequencies and percentages. A p-value threshold of less than 0.05 was established for statistical significance. To compare demographic characteristics between the BV group and healthy controls, the Chi-square test was applied for categorical variables (e.g., age distribution), and independent sample t-tests were employed for continuous variables (e.g., mean age).
Given the non-normal distribution of miRNA expression results the miR-1271-5p and miR-10a-3p expression levels between the BV and healthy control groups were compared using the Mann-Whitney U test.
Receiver operating characteristic (ROC) curve analysis was performed to assess the diagnostic potential of miR-1271-5p and miR-10a-3p. The area under the curve (AUC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated to determine the optimal cutoff values.9,10 To compare miR-1271-5p levels across different bacterial species within the BV group, a one-way analysis of variance (ANOVA) was used, followed by appropriate post-hoc tests if significant differences were found.
Demographic characteristics of patients with BV and healthy control subjects
The demographic characteristics of the 50 patients with BV and the 50 healthy controls included in this study are presented in Table 1. Analysis of these characteristics revealed no statistically significant differences in age or age distribution between the two groups (p > 0.05), indicating a comparable demographic profile and minimizing the potential age-related bias in subsequent analyses.
Table (1):
Demographic characteristics of patients with bacterial vaginosis and healthy control
| Characteristic | Patients (n = 50) Mean ± SD / n (%) | Healthy control (n = 50) Mean ± SD / n (%) | p-value |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 31.40 ± 8.81 | 29.66 ± 8.60 | 0.320† |
| Range | 17-65 | 17-66 | |
| Age distribution | |||
| <30, n (%) | 23 (46.0%) | 29 (58.0%) | 0.377¥ |
| 30-39, n (%) | 18 (36.0%) | 16 (32.0%) | |
| ≥40, n (%) | 9 (18.0%) | 5 (10.0%) | |
n: number of cases; SD: standard deviation; †: independent samples t-test; ¥: Chi-square test
Detection of bacterial species in patients with BV using 16S rRNA multiplex-PCR
Multiplex PCR analysis of the 16S rRNA genes was performed to identify the prevalence of various bacterial species in patients with BV. The results illustrated in Figures 1A and 1B indicate diverse bacterial profiles. G. vaginalis was the most frequently detected species, present in 33 (66.0%) patients with BV. Atopobium and Megasphaera were also highly prevalent and were detected in 20 (40.0%) and 19 (38.0%) patients, respectively. Erysipelothrix and Cutibacterium were identified in 17 (34.0%) patients, whereas Lactobacilli spp. was found in 10 (20.0%) patients.
Figure 1. (A) Etiology and frequency of bacterial species infections according to results of multiplex-PCR. (B) Multiplex-PCR amplification of the 16S rRNA genes on agarose gel electrophoresis for the detection of BV bacterium. M: Marker ladder 100-2000 bp. Samples with Cutibacterium, Megasphaera, Atopobium, Lactobacillus sp., Erysipelothrix, and Gardnerella vaginalis infections were showed positive at 320 bp, 413 bp, 473 bp, 520 bp, 609 bp and 720 bp specific PCR product size respectively
Expression of miR-1271-5p in patients with BV and in healthy control subjects
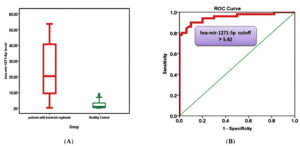
The expression levels of miR-1271-5p in the blood of patients with BV and healthy controls were quantified using stem-loop RT-qPCR. As shown in Figure 2A, the median miR-1271-5p level was significantly higher in patients with BV (20.33; IQR, 11.76) than in healthy controls (2.01; IQR, 0.98) (p < 0.001). This significant upregulation suggests a potential role of miR-1271-5p in BV pathogenesis or as a biomarker.
To evaluate the diagnostic utility of miR-1271-5p in BV, an ROC curve analysis was performed (Figure 2B). A cut-off value of <5.42-fold for miR-1271-5p demonstrated high diagnostic accuracy, with sensitivity, 94.0%; specificity, 94.0%; PPV, 94.0%; NPV, 94.0%; and AUC, 0.951 (95% CI, 0.909-0.994). These metrics indicate that miR-1271-5p is a promising diagnostic marker for BV.
Figure 2. (A) Median miR-1271-5p level of patients and healthy controls. (B) Receiver operator characteristic curve analysis of miR-1271-5p for the calculation of possible diagnostic cutoff value
Furthermore, the frequency distribution of miR-1271-5p levels across the different bacterial species identified in patients with BV was analyzed (Table 2). Mean miR-1271-5p levels varied among species, with the highest levels observed in patients colonized by Megasphaera (31.49 ± 7.84). However, one-way ANOVA revealed no statistically significant difference in miR-1271-5p levels among the different bacterial species (p = 0).
Table (2):
Frequency distribution of miR-1271-5p according to VB species of bacteria
Species of bacteria |
miR-1271-5p level Mean ± SE |
|---|---|
Gardnerella vaginitis |
21.72 ± 2.06 |
Lactobacilli sp. |
20.20 ± 1.30 |
Erysipelothrix |
26.25 ± 2.08 |
Cutibacterium |
29.42 ± 2.79 |
Atopobium |
22.03 ± 1.30 |
Megasphaera |
31.49 ± 2.84 |
p-value |
0.225† |
SE: standard Error; †: One way ANOVA test
Expression of miR-10a-3p in patients with BV and in healthy control subjects
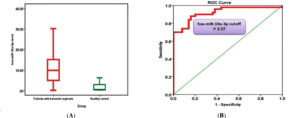
The expression of miR-10a-3p was evaluated. The miR-10a-3p median levels were significantly elevated in patients with BV (9.85; IQR, 6.71) compared to those in healthy controls (0.56; IQR, 0.32) (p < 0.001) (Figure 3A). This substantial increase further supported the potential involvement of miR-10a-3p in BV.
ROC curve analysis of miR-10a-3p (Figure 3B) demonstrated excellent diagnostic performance. A cutoff value of <3.57-fold for miR-10a-3p yielded sensitivity, 96.0%; specificity, 92.0%; PPV, 92.3%; NPV, 95.8%; and AUC, 0.970 (95% CI, 0.938-1.000). These results highlighted miR-10a-3p as another highly effective diagnostic biomarker of BV.
Figure 3. (A) Median miR-10a-3p level of patients and healthy controls. (B) Receiver operator characteristic curve analysis of miR-10a-3p for the calculation of possible diagnostic cutoff value
Furthermore, the levels of miR-10a-3p were compared across different bacterial species identified in patients with BV, as detailed in Table 3. Although mean levels varied, with the highest observed in patients with Megasphaera colonization (15.14 ± 2.49), the one-way ANOVA indicated no statistically significant difference in miR-10a-3p levels among the various bacterial species (p = 0.574).
Table (3):
Frequency distribution of miR-10a-3p according to bacterial species in BV patients
Species of bacteria |
miR-10a-3p level Mean ± SE |
|---|---|
Gardnerella vaginitis |
10.78 ± 1.54 |
Lactobacilli sp. |
11.33 ± 2.18 |
Erysipelothrix |
13.05 ± 3.41 |
Cutibacterium |
10.68 ± 1.84 |
Atopobium |
10.41 ± 1.71 |
Megasphaera |
15.14 ± 2.49 |
p-value |
0.574† |
SE: standard Error; †: One way ANOVA test
BV is a prevalent and complex vaginal disorder characterized by a significant shift in the vaginal microbiota, where protective Lactobacillus species are replaced by an overgrowth of diverse anaerobic bacteria.11 This dysbiosis is associated with various adverse reproductive health outcomes, including increased susceptibility to sexually transmitted infections and preterm birth.12 Understanding the intricate microbial communities involved in BV pathogenesis is crucial for effective diagnosis and treatment.
This study used multiplex PCR targeting the 16S rRNA gene for the identification of bacterial species within the vaginal microbiota of patients with BV. This molecular approach offers superior sensitivity and specificity compared to conventional methods.13 Our analysis revealed G. vaginalis as the most common species (66.0% of patients), consistent with its established role as a key initiator of BV biofilms.11,14 However, the polymicrobial nature of BV was further highlighted by the detection of Atopobium (40.0%) and Megasphaera (38.0%), species increasingly recognized for their contribution to the inflammatory environment associated with BV.15,16 The presence of Lactobacillus spp. (34.0%) in some patients with BV, underscores the dynamic and complex nature of the vaginal microbiome.17 The detection of Erysipelothrix (20.0%) also warrants further investigation into its potential role in vaginal dysbiosis.18 These findings collectively emphasize the need to move beyond a narrow focus on Gardnerella to fully understand the complex bacterial ecology in BV.
A central finding of this study was the significant upregulation of both miR-1271-5p and miR-10a-3p in patients with BV compared to that in healthy controls. miRNAs are small, non-coding RNA molecules that play critical roles in post-transcriptional gene regulation, affecting host immune responses, and modulating bacterial interactions within the vaginal mucosa.19 The substantial upregulation of miR-1271-5p (median 20.33 vs. 2.01) and miR-10a-3p (median 9.85 vs. 0.56) suggested their active involvement in BV pathogenesis. These miRNAs may contribute to the disease by promoting the growth of pathogenic bacteria and suppressing protective Lactobacillus species, potentially by downregulating genes essential for Lactobacillus adhesion or survival.4,20 Furthermore, the potential of miR-1271-5p to modulate inflammatory pathways could explain its contribution to the vaginal irritation symptoms common in BV.20 The precise mechanisms require further elucidation; however, these findings open new avenues for understanding host-pathogen interactions in BV.
This study highlights the diagnostic potential of miR-1271-5p and miR-10a-3p. Both miRNAs demonstrated high sensitivity and specificity, and the ROC curve analysis yielded an AUC of 0.951 for miR-1271-5p (sensitivity, specificity, PPV, and NPV, 94.0%) and a similarly impressive performance for miR-10a-3p (sensitivity, 92.0%; specificity, 90.0%; PPV, 90.2%; and NPV, 91.8%). These results indicated that both miRNAs surpassed the diagnostic accuracy of the traditional Nugent score (typically 80-90%), positioning them as robust, non-invasive biomarkers for BV.11,21 The combined use of miR-1271-5p and miR-10a-3p further enhanced the diagnostic accuracy, achieving sensitivity, 94.1%; specificity, 92.9%; and AUC, 0.957. This supports the growing trend of using miRNA panels for improved diagnostic precision in various conditions.22 The observed variations in miRNA levels, particularly the highest average levels of miR-10a-3p in patients with Megasphaera (15.14 ± 2.49), suggest a potential link between specific bacterial colonization and host miRNA expression. Although not statistically significant in this cohort (p = 0.574), this warrants further investigation in larger studies, as such associations could lead to personalized diagnostics by combining bacterial identification with miRNA profiling to better characterize BV subtypes and guide targeted therapies.
Despite these promising findings, this study has several limitations that must be acknowledged. The relatively small sample size (50 infected patients and 50 controls) and single-center design in Iraq may limit the generalization of these results to broader and more diverse populations. The absence of validation in a real-world clinical context further emphasizes the need for additional studies to evaluate the performance of these biomarkers in routine clinical practice. Future research should prioritize larger, multicenter studies with diverse cohorts and rigorous clinical validation.22,23 Additionally, the close association of miR-1271-5p with the estrogen receptor-α highlights the importance of considering confounding factors such as hormonal fluctuations and menstrual cycle stage, as these can affect miRNA expression independently of the bacterial presence.19 Investigating the target genes of these miRNAs could further elucidate their precise roles in BV pathogenesis and their interactions with various BV-associated bacteria, potentially revealing mechanisms by which miRNAs suppress host defense mechanisms against pathogens such as G. vaginalis.12 Addressing these limitations will be crucial for translating these promising research findings into clinically applicable diagnostic tools and improving patient management for BV.
This study demonstrated the significant involvement of miRNAs in the pathophysiology of BV. These findings indicate that hsa-miR-1271-5p and hsa-miR-10a-3p are significantly upregulated in patients with BV compared to healthy controls, suggesting their crucial role in the development of the disease. Both miRNA biomarkers show great promise as highly sensitive, specific, and noninvasive diagnostic tools. However, several challenges must be addressed before implementing BV diagnostic tests in routine laboratory practice, including cost, scalability, and infrastructure requirements. Our results underscore the diagnostic utility of immune mediators and miRNAs, and provide valuable insights into immunological dysregulation in BV infections. Future research should focus on addressing these limitations to provide more comprehensive and generalizable findings, ultimately paving the way for improved BV diagnosis and patient management.
ACKNOWLEDGMENTS
The authors would like to thank the Tunisian Ministry of Higher Education and Scientific Research, Tunisia [GRANT UR17/ES30] for supporting this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This research was funded by the Tunisian Ministry of Higher Education and Scientific Research, Tunisia [GRANT UR17/ES30].
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Review Board of Iraqi Ministry of Health, Human Development and Training Center (Approval No. 1/2024/49).
INFORMED CONSENT
Written informed consent was obtained from all patients and from healthy volunteers involved in this study.
- Chacra LA, Fenollar F, Diop K. Bacterial vaginosis: What do we currently know? Front Cell Infect Microbiol. 2022;11:672429.
Crossref - Onderdonk AB, Delaney ML, Fichorova RN. The Human Microbiome during Bacterial Vaginosis. Clin Microbiol Rev. 2016;29(2):223-238.
Crossref - Cheng L, Kazmierczak D, Norenhag J, et al. A MicroRNA Gene Panel Predicts the Vaginal Microbiota Composition. mSystems. 2021;6(3):e00175-21.
Crossref - O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402.
Crossref - Kenyon C, Colebunders R, Crucitti T. The global epidemiology of bacterial vaginosis: a systematic review. Ameri J Obstet Gynecol. 2013;209(6):505-523.
Crossref - Redelinghuys MJ, Geldenhuys J, Jung H, Kock MM. Bacterial vaginosis: current diagnostic avenues and future opportunities. Front Cell Infect Microbiol. 2020;10:354.
Crossref - Chao X, Liu Y, Fan Q, Shi H, Wang S, Lang J. The role of the vaginal microbiome in distinguishing female chronic pelvic pain caused by endometriosis/adenomyosis. Ann Translat Med. 2021;9(9):771.
Crossref - Muzny CA, Balkus J, Mitchell C, et al. Diagnosis and Management of Bacterial Vaginosis: Summary of evidence reviewed for the 2021 Centers for Disease Control and Prevention Sexually Transmitted Infections Treatment Guidelines. Clinic Infect Dis. 2022;74(Suppl 2):S144-S151.
Crossref - Chaudhuri AA, So AYL, Sinha N, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187(10):5062-5068.
Crossref - Muzny CA, Schwebke JR. Pathogenesis of Bacterial Vaginosis: Discussion of Current Hypotheses. J Infect Dis. 2016;15(214 Suppl 1):1-5.
Crossref - Ravel J, Moreno I, Simon C, et al. Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am J Obstet Gynecol. 2021;224(3):251-257.
Crossref - George SD, Amerson-Brown MH, Sousa LGV, et al. Investigating Bacterial Vaginosis Pathogenesis Using Peptide Nucleic Acid-Fluorescence In Situ Hybridization With a Focus on the Roles of Gardnerella Species, Prevotella bivia, and Fannyhessea vaginae. Open Forum Infect Dis. 2025;12(9):ofaf556.
Crossref - Deng T, Song X, Liao Q et al. Best among the key molecular diagnostic markers of bacterial vaginosis. AMB Expr. 2025;15(1):1-12.
Crossref - Glascock AL, Jimenez NR, Boundy S, et al. Unique roles of vaginal Megasphaera phylotypes in reproductive health., Microb Genom. 2021;7(12):000526.
Crossref - van Teijlingen NH, Helgers LC, Zijlstra-Willems EM, et al. Vaginal dysbiosis associated-bacteria Megasphaera elsdenii and Prevotella timonensis induce immune activation via dendritic cells. J Reprod Immunol. 2020;137:103072.
Crossref - Chen X, Lu Y, Chen T, Li R. The Female Vaginal Microbiome in Health and Bacterial Vaginosis. Front Cell Infect Microbiol. 2021;11:631972.
Crossref - Erysipelothrix. (n.d.). Taylor & Francis Online. Retrieved from https://taylorandfrancis.com/knowledge/Medicine_and_healthcare/Infectious_diseases/Erysipelothrix/, Accessed 29 september, 2025.
- Staedel C, Darfeuille F. MicroRNAs and bacterial infection. Cell Microbiol. 2013;15(9):1496-1507.
Crossref - Yan XY, Yao JP, Li YQ, et al. Global trends in research on miRNA-microbiome interactions from 2011-2021: A bibliometric analysis. Front Pharmacol. 2022;13:974741.
Crossref - Workowski KA, Bachmann LH, Chan PA, et al. Sexually Transmitted Infections Treatment Guidelines, 2021. MMWR Recomm Rep. 2021;70(4):1-187.
Crossref - Metcalf GAD. MicroRNAs: circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene. 2024;43(28):2135-2142.
Crossref - Mickiewicz J, Mielczarek-Palacz A, Gola JM. MicroRNAs as potential biomarkers in gynecological cancers. Biomedicines. 2023;11(6):1704.
Crossref - Kwon EJ, Kim HJ, Woo BH, Joo JY, Kin YH, Park HR. Profiling of plasma derived exosomal RNA expression in patients with periodontitis: A pilot study. Oral Dis. 2023;29(4):1335-1343.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.