ISSN: 0973-7510
E-ISSN: 2581-690X
Immobilization of thermostable lipase from Geobacillus thermoleovorans PPD2 (Lip-A) were carried out on Ni-NTA agarose and carboxymethyl chitosan. Free enzyme was obtained by heterologous expression in Escherichia coli as a host cell. The enzyme showed catalytic activity for transesterification reaction. Transesterification activity of immobilized lipase on Ni-NTA agarose was increased by three fold (75.04% conversion) compared to that the free enzyme (24.65%), while the activity of immobilized lipase on carboxymethyl chitosan was slightly decreased (19.86%).
Geobacillus thermoleovorans PPD2, thermostable lipase, immobilization, Ni-NTA agarose, carboxmethyl chitosan, transesterification
Transesterification of triglycerides with short chain alcohols has an important role on industrial processes, such as biodiesel production or other chemical synthesis. Transesterification reaction is generally performed in non-aqueous media, catalyzed by chemicals or bio-catalyst1. Chemical catalyst provides high transesterification yield, however it has some disadvantages especially on the downstream process: the complexity of side product removal and waste treatment. Biocatalyst is preferable used since it offers clean process and highly specificity of substrates and products2.
Lipases from different sources were reported as biocatalyst for esterification and transesterification reaction3,4,5. Burkholderia cepacia lipase showed high conversion yield (79%) of Jatropha oil into ethyl ester6. Recombinant lipase ITB1.1 from Geobacillus uzenensis local strain, isolated from Manuk hot spring7, was also reported as catalyst for methylation of coconut oil. The product was identified as methyl laurate (47.36%), methyl myristate (19.87%), and methyl caprate (15.96%)8. Lipase from other sources, Pseudomonas fluorescens 26-2, showed 83.8% yield of soybean oil conversion into methyl ester. The enzyme also showed 80% yield conversion from rapeseed and cottonseed oil9. Transesterification assay catalyzed by organic soluble lipase from P. fluorescens AK was reported10. Solubilized enzyme only convert 36% yield of soybean oil conversion into methyl ester, while the native enzyme (insoluble in organic solvent) did not produce the desired product.
Organic solvents used in transesterification reaction show to strip water molecules from the enzyme surface. It may leads to inactivation of most enzymes, including lipases11. In order to overcome this limitation, immobilized enzyme is employed to increase the enzyme stability in organic solvent. Immobilized lipases from Chromobacterium viscosum was reported as catalyst for transesterification reaction of Jatropha oil and ethanol12. The immobilized enzyme on Celite-545 enhanced the product yield to 71%, compared to the free enzyme (62%). Immobilized lipase from Pseudomonas aeruginosa LX1 showed 80% conversion of soybean oil into methyl ester, while the free enzyme showed 71.5% yield13. Immobilized lipase from Mucor meihei on aminated polyethersulfone (PES-NH2) membrane cross-linked by glutaraldehyde, was reported as catalyst for methylation of sunflower oil. The product was identified as methyl palmitate (100%) and methyl stearate (58%) with high quality for biodiesel, according to ASTM standard. Meanwhile, the reaction catalyzed by free lipase produced methyl palmitate (100%) and methyl stearate (45%)14. Several immobilization methods was used on lipase from Talaromyces thermophilus; such as adsorption (CaCO3), ionic binding (Amerlite, Duolite, and DEAE-Sephadex), and covalent binding (chitin, chitosan, and gelatin). Optimum result for transesterification was observed from immobilized lipase on chitosan (activated by glutaraldehyde), with 76% yield conversion from oleic acid into butyl oleate15.
Lipase from local isolate, Geobacillus thermoleovorans PPD2, is potential as biocatalyst in transesterification reaction since it showed tolerance towards short chain alcohols. The enzyme showed optimum temperature at 50 oC and high specificity towards pNP decanoate16. In this paper we described the effect of matrixs bound on the transesterification activity of immobilized lipase. Two matrixs were used to bound the enzyme. One was based on covalent coordination bound at the N-terminal, other based on covalent bound on the random surface of the enzyme.
Chemical and Strains
E. coli BL21(DE3) carries pITBlip2.2 plasmid inserted with PPD2 lipase gene was obtained from our collection. Luria Bertani medium was used for lipase expression, with antibiotic kanamycin (Bio Basic) and IPTG (Thermo Scientific) as marker and inducer. Chitosan powder (Acros Organics), sodium hydroxide (Merck), and monochloroacetic acid were used for carboxymethyl chitosan synthesis. Ni-NTA Agarose (Qiagen, Germany) and carboxymethyl chitosan were used as immobilization matrix. 2,4-nitrophenol (Sigma) was used as the standard for the enzyme assay. 2,4-nitrophenol decanoate (Sigma), n-propanol (Merck), and acetonitrile were used as the substrate for transesterification assay.
Heterologous Expression
Expression of lipase was carried out in Luria Bertani (LB) medium, containing 0.1% kanamycin sulfate. Isopropyl â-D-1-thiogalactopyranoside (IPTG) was added when optical density (OD600) of the medium reached 0.60. Overexpression was conducted at 37 oC for 4 hours, with shaking at 200 rpm. Pellet cell was collected by centrifugation and the overexpressed protein was obtained by two steps, pellet sonication followed by boiling lysis of the debris cell. The supernatant from the second step was filtered by Amicon Ultra-4 Centrifugal Filters (10 kDa), then analyzed by SDS-PAGE on 10% running gel. Freeze dried Lip-A was stored at 4 oC.
Immobilization on Ni-NTA agarose
Ni-NTA agarose matrix was pipetted into column, equilibrated by aqua miliQ and 50 mM K-phosphate buffer pH 8. Soluble protein was then bound into matrix for 30 minutes. The matrix was rinsed with K-phosphate buffer, and the unbound protein was subjected to Bradford assay17. Immobilized Lip-A/Ni-NTA was submersed in K-phosphate buffer and stored at 4 oC.
Carboxymethyl Chitosan Preparation
Carboxymethyl chitosan (CM-chitosan) was prepared using the method described by Chen and Park18. A mixture of 2.0 g of chitosan, 2.7 g of sodium hydroxide, and 20 mL isopropanol:water (4:1) were added into a ûask (100 mL). The mixture was stirred at 50 oC until the polymer swells. Monochloroacetic acid solution (3 g solid dissolved in 4 mL isopropanol) was added dropwise. The final mixture was allowed for 4 hours at 50 oC, then stopped by addition of 40 mL ethanol (70%) into the mixture. The solid product (Na salt of CM-chitosan) was ûltered and rinsed in ethanol, followed by air dried at room temperature. Na-salt CM-chitosan was then suspended in 100 mL ethanol (70%). Hydrochloric acid (37%) was added until the pH reached 3, and the suspension was stirred for 30 min. The solid product (CM-chitosan) was ûltered and rinsed in ethanol, then dried at room temperature.
Immobilization on Carboxymethyl chitosan
A mixture of CM-chitosan (10 mg solid suspended in 2 mL isopropanol) and 1 mL enzyme solution (1 mg enzyme per mL buffer) was stirred at 20 oC for 12 hours. The mixture was centrifuged (3800 g, 30 min), and the pellet was rinsed with K-phosphate buffer. The unbound protein was subjected to Bradford assay, while the freeze dried pellet (immobilized Lip-A/CM-chitosan) was stored at 4oC.
Transesterification Assay
Transesterification assay was conducted by spectrophotometry analysis19 with some modification. Substrates, p-nitrophenyl decanoate (10 mM) and isopropanol (1 M), were mixed in a total volume of 500 µL acetonitrile. Free and immobilized Lip-A (equal to 1 mg protein) were added into each tube to start the reaction. The mixture was incubated at 50 oC with shaking speed at 100 rpm for 2 min. After reaction, 30 µL supernatant was pipetted into another tube and immediately mixed with ethanol (total volume 1.5 mL). The control was carried out using the same condition and protocols but without enzyme addition. The mixture was measured at 315 nm, and the conversion was calculated based on a calibration curve of p-nitrophenol.
Expression and Purification of Lipase
Recombinant lipase used for immobilization was obtained by two steps, resulting in one dominant band on SDS-PAGE electrophoregram (Fig 1). Most of the proteins from host cell were separated in the first step (ultra-sonication lysis), since the heterologous expressed Lip-A formed insoluble and did not appear in the supernatant. The overexpressed lipase was obtained in the second step (boiling lysis) with additional of detergent. Protein solubility was increased by addition of detergent20. Isolation of insoluble thermostable protein by boiling lysis was also reported21, resulting in high purity of the recombinant protein. Detergent contamination on the protein solution was later removed by filtration using Amicon Ultra-4 Centrifugal Filters, and the post-treatment protein was used for immobilization.
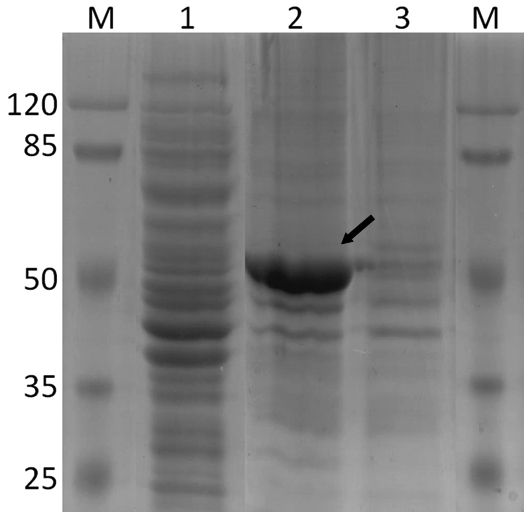
Fig. 1. SDS-PAGE Electrophoregram of PPD2 Lipase through E.coli
[M] protein marker; [1] Ultra-sonication lysate from first step, [2] over expression protein from second step (boiling treatment with addition of detergent), [3] Unbound protein after immobilization process (fluthrough); (→) overexpressed Lip A
Immobilization on Ni-NTA Agarose
Lip-A was successfully immobilized on Ni-NTA agarose after 30 min interaction. Electrophoregram of the unbound protein showed that the matrix showed high affinity towards the enzyme (Fig 1). The interaction between the Ni-NTA and histidine-tag at the N terminus was occurred by covalent coordination bound between the metal ion (Ni2+) and imidazole ring of histidine22. The maximum capacity of matrix was observed at 4 mg protein per 0.4 mL matrix (Fig 2). Immobilized Lip-A do not leach out by washing the matrix with Triton-X (1%) or NaCl (100 mM), while imidazole (100 mM) was effectively removed Lip-A from the matrix (data not shown). When fresh Lip-A was reloaded into the previously used matrix, almost the same amount of protein was bound (data not shown). Other paper also reported that immobilized His-tagged Green Fluorescent Protein (GFP) on Ni-NTA chitosan was specifically removed by imidazole or EDTA23. Re-immobilization of His-tagged GFP on the previously used matrix gave almost the same amount of immobilized GFP. This specific interaction may lead to the possibility of matrix to be reused for immobilization of fresh enzyme.
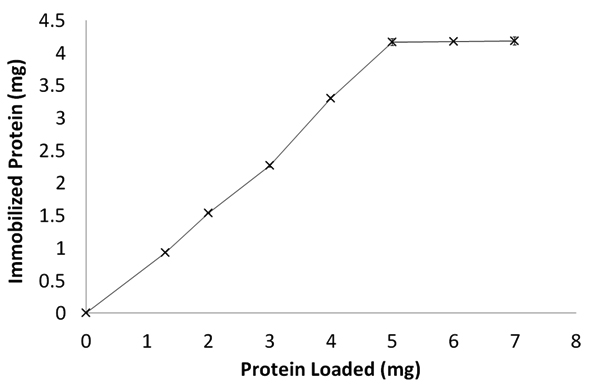
Fig. 2. Immobilization of Lip-A with various amount of protein loaded onto Ni-NTA agarose matrix. The steady state of bound protein was showed at 4mg protein 10/ 4mL of resine
Immobilization on Carboxymethyl Chitosan
Carboxymethyl chitosan (CM-chitosan) was prepared in two steps, synthesis of Na salt CM-chitosan and acidification of the salt. FTIR analysis of these three solids (chitosan, Na CM-chitosan, and CM-chitosan) showed that the CM-chitosan was obtained (Fig 3). The FTIR spectra of chitosan shows the basic characteristic of chitosan at: 3353 cm-1 (O–H stretch), 2871 cm-1 (C–H stretch), 1589 cm-1 (N–H bend), and 1024 cm-1 (C–O stretch). Meanwhile, spectra of CM-chitosan showed peaks at 1724 cm–1 (C=O stretch from –COOH), 1617 and 1520 cm–1 (N–H bend from –NH3+), and 1058 cm–1 (C–O stretch), which fits the characteristic of CM-chitosan as described in previous studies [18,24]. The –COOH peak (at 1724 cm-1) did not show in spectrum of chitosan and Na salt of CM-chitosan, indicated that the carboxylate group was only presented in the final product (Fig 3). Lip-A was successfully immobilized on CM-chitosan after 12 hour reaction. FTIR analysis of CM-chitosan and complex Lip-A/CM-chitosan showed that the C=O peak has shifted from 1724 cm-1 (–COOH) to 1651 cm-1 (–CONH–) (Fig 3). The result suggested that Lip-A was covalently attached to CM chitosan. The amide bond was formed from the carboxylate group in CM-chitosan and the amine group in Lip-A.
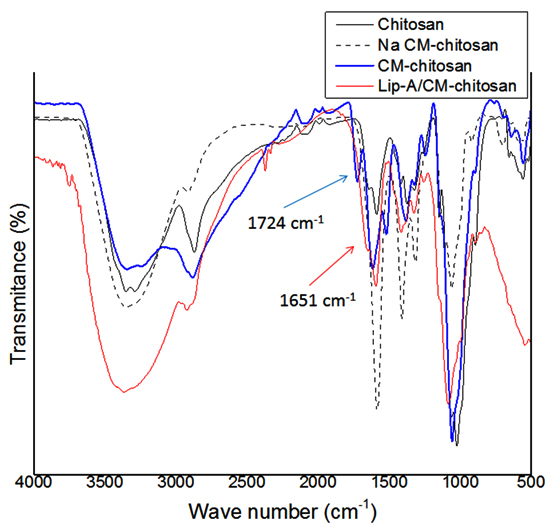
Fig. 3. FTIR spectra of chitosan, Na CM-chitosan, CM-chitosan, and immobilized Lip-A on CM-chitosan (Lip-A/CM-chitosan). Blue arrow showed peak of carbonyl residue from CM-chitosan, meanwhile red arrow showed carbonyl residue (-CONH-) of LipA-CM-chitosan
Transesterification Activity of Immobilized Lipase
Transesterification activity of immobilized Lip-A on Ni-NTA agarose (Lip-A/Ni-NTA) was increased three fold (75.04% conversion) compared to the free enzyme (24.65%). This supports the previews result that immobilized lipase from Burkholderia sp. on alkyl-celite in a packed-bed reactor, also enhanced the activity to 85% conversion, compared to the free enzyme (67%)25. Immobilization of the whole cell of Aspergillus oryzae expressed Fusarium heterosporum lipase, also enhanced the production by approximately three fold26. Meanwhile, the activity of immobilized Lip-A on CM-chitosan (Lip-A/CM-chitosan) was slightly decreased. Lip-A/CM-chitosan showed 19.86% conversion (Table 1), about 80% compared to that the free enzyme activity.
Table (1):
Transesterification activity of free and immobilized Lip-A
Sample |
Transesterification Activity (% conversion) |
|---|---|
Free Lip-A |
24.65 |
Lip-A/Ni-NTA |
75.04 |
Lip-A/CM-chitosan |
19.86 |
Reusability assay of Lip-A/Ni-NTA and Lip-A/CM-chitosan showed that the immobilized enzyme reached half time after fourth usage (41% remained, Fig 4, A) and third usage (30% remained, Fig 4, B), respectively. Activity reduction of immobilized Candida antartica lipase B on octyl agarose was also reported27. The enzyme was catalyzed transesterification reaction between tributyrin and methanol, and the activity was reduced about 10% after fifth usage.
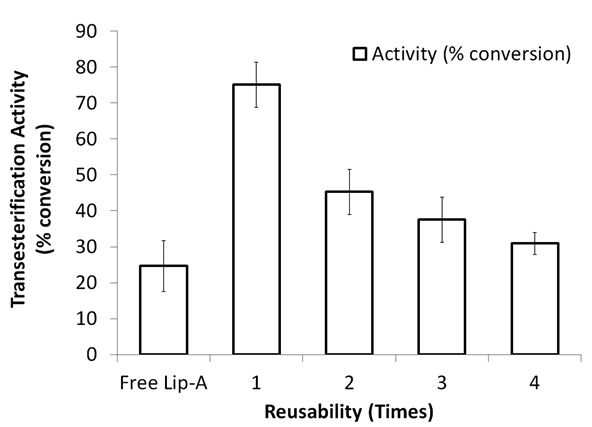
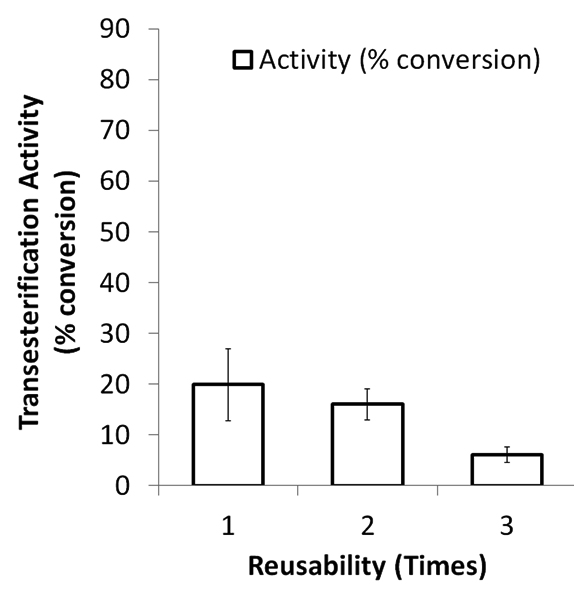
Fig. 4. Transesterification activity and reusability of tree and immobilized Lip-A on Ni-NTA (A), and carbox-ymethyl chitosan (B)
Lip-A/Ni-NTA showed higher activity and reusability compared to that Lip-A/CM-chitosan. This probably due two reason, the first one is interaction between the enzyme to Ni-NTA resulting on covalent coordination bound meanwhile interaction of the enzyme to CM-chitosan resulted the covalent bound. The last interaction is rather stronger compared to that covalent coordination bound. As consequence, the 3D structure of the enzyme is easier to change and hence reducing the activity of the enzyme. The second possibility of loosing activity of the enzyme bound to CM-chitosan is due to the random interaction of the matrix into the surface of the enzyme. There are many possibility that the interaction occurred at the region close to the lid or catalytic site, hence the lid movement would be affected when the catalytic process occured. Meanwhile using Ni-NTA matrix only the N-terminal on the enzyme was bound to the matrix, so that the 3D structure of the enzyme was not influenced by the above interaction resulting on the increasing of the activity of the enzyme. These result suggested that attachement of the enzyme at specific binding and less tight interaction give higher activity and reusability of immobilized of PPD2 lipase.
Lipase of PPD2 was successfully immobilized on Ni-NTA agarose and carboxymethyl chitosan. Enzyme-matrix interaction at specific location (Lip-A/Ni-NTA at the N-terminus) give higher transesterification activity and reusability compared to that nonspecific residues on the enzyme surface (Lip-A/CM-chitosan).
ACKNOWLEDGMENTS
We would like to thank P3MI research program of ITB and PDUPT program, Ministry of Research, Technology and Higher Education, Republic of Indonesia to make this research possible to be carried out.
- Amini, Z., Z. Ilham, H. C. Ong, H. Mazaheri, and W. H. Chen. State of the art and prospective of lipase-catalyzed transesterification reaction for biodiesel production. Energy Conversion and Management. 2017; 141: 339-353.
- Bajaj, A., P. Lohan, P. N. Jha, and R. Mehrotra. Biodiesel production through lipase catalyzed transesterification: an overview. Journal of Molecular Catalysis B: Enzymatic. 2010; 62(1): 9-14.
- Nurhasanah, S. Nurbaiti, F. Madyanti, and Akhmaloka. Diversity of Gene Encoding Thermostable Lipase from Compost Based on Metagenome Analysis. International Journal od Integrative Biology. 2015; 16(1): 07-12.
- Asy’ari, M., P. Aditiawati, R. Hertadi, and Akhmaloka. Cloning and Squence Analysis of Lipase Gene of Halophilic Bacteria Isolated from Mud Crater of Bledug Kuwu, Central Java, Indonesia. Bioscience, Biotechnology Research Asia. 2015; 12(3): 1903-1912.
- Kristia, Y. Y., S. F. Syihab, and Akhmaloka. Cloning and Characterization of Lipase Gene from a Local Isolate of Psedoxanthomonas sp. Biosciences Biotechnology Research Asia. 2017; 14(2): 503-507.
- Shah, S. and M. N. Gupta. The effect of ultrasonic pre-treatment on the catalytic activity of lipases in aqueous and non-aqueous media. Chemistry Central Journal. 2008; 2(1): 1-8.
- Widhiastuty, M. P., Febriani, M. R. Moeis, Akhmaloka, and F. Madayanti. Cloning, homological analysis and expression of Lipase gene from hot spring isolate. International Journal of Integrative Biology. 2011; 11(1): 8-13.
- Brilliantoro, R., R. Zidny, M. P. Widhiastuty, and Akhmaloka. Hydrolytic and transesterification activities of thermostable lipase ITB1.1. Biosciences Biotechnology Research Asia. 2015; 12(1): 01-06.
- Yang, J., B. Zhang, and Y. Yan. Cloning and expression of Pseudomonas fluorescens 26-2 lipase gene in Pichia pastoris and characterizing for transesterification. Applied Biochemistry and Biotechnology. 2009; 159(2): 355-365.
- Zhao, X., B. El-Zahab, R. Brosnahan, J. Perry, and P. Wang. An organic soluble lipase for water-free synthesis of biodiesel. Applied Biochemistry and Biotechnology. 2007; 143(3): 236-243.
- Yang, L., J. S. Dordick, and S. Garde. Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophysical journal. 2004; 87(2): 812-821.
- Shah, S., S. Sharma, and M. N. Gupta. Biodiesel preparation by lipase-catalyzed transesterification of Jatropha oil. Energy and Fuels. 2004; 18(1): 154-159.
- Ji, Q., S. Xiao, B. He, and X. Liu. Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. Journal of Molecular Catalysis B: Enzymatic. 2010; 66(3): 264-269.
- Handayani, N., D. Wahyuningrum, M. A. Zulfikar, S. Nurbaiti, and C. L. Radiman.The synthesis of biodiesel catalyzed by Mucor miehei lipase immobilized onto aminated polyethersulfone membranes. Bioresources and Bioprocessing. 2016; 3(1): 22-26.
- Romdhane, I. B. B., Z. B. Romdhane, A. Gargouri, and H. Belghith. Esterification activity and stability of Talaromyces thermophilus lipase immobilized onto chitosan. Journal of Molecular Catalysis B: Enzymatic. 2011; 68(3): 230-239.
- Permana, A. H., F. M. Warganegara, D. Wahyuningrum, M. P. Widhiastuty, and Akhmaloka. Heterologous expression and characterization of thermostable lipase from Geobacillus thermoleovorans PPD2 through Escherichia coli. Biosciences Biotechnology Research Asia. 2017; 14(3): 1081-1088.
- Bradford, M.M. A rapid and sensitive method for the quantitation of micrograms quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976; 72: 248-254.
- Chen, X. G., and H. J. Park. Chemical characteristics of O-carboxymethyl chitosans related to the preparation conditions. Carbohydrate Polymers. 2003; 53(4): 355-359.
- Fu, X., J. Zheng, X. Ying, H. Yan and Z. Wang. Investigation of Lipozyme TL IM-catalyzed transesterification using ultraviolet spectrophotometric assay. Chinese Journal of Catalysis. 2014; 35(4): 553-559.
- Singh, A., V. Upadhyay, A. K. Upadhyay, S. M. Singh, and A. K. Panda. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microbial cell factories. 2015; 14(1): 41-46.
- Livernois, A. M., D. J. Hnatchuk, E. E. Findlater, and S. P. Graether. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Analytical Biochemistry. 2009; 392(1): 70-76.
- Ward, W. W., & G. Swiatek. Protein purification. Current Analytical Chemistry. 2009; 5(2): 85-105.
- Oshige, M., K. Yumoto, H. Miyata, S. Takahashi, M. Nakada, K. Ito, M. Tamegai, H. Kawaura, and S. Katsura, Immobilization of His-tagged proteins on various solid surfaces using NTA-modified chitosan. Open Journal of Polymer Chemistry. 2013; 3(01): 6-11.
- Fei Liu, X., Y. Lin Guan, D. Zhi Yang, Z. Li, and K. De Yao. Antibacterial action of chitosan and carboxymethylated chitosan. Journal of applied polymer science. 2001; 79(7):1324-1335.
- Tran, D. T., C. L. Chen, and J. S. Chang. Continuous biodiesel conversion via enzymatic transesterification catalyzed by immobilized Burkholderia lipase in a packed-bed bioreactor. Applied Energy. 2016; 168: 340-350.
- Amoah, J., S. H. Ho, S. Hama, A. Yoshida, A. Nakanishi, T. Hasunuma, C. Ogino, and A. Kondo. Converting oils high in phospholipids to biodiesel using immobilized Aspergillus oryzae whole-cell biocatalysts expressing Fusarium heterosporum lipase. Biochemical engineering journal. 2016; 105: 10-15.
- Hirata, D. B., T. L. Albuquerque, N. Rueda, J. J. Virgen-Ortíz, V. G. Tacias-Pascacio, and R. Fernandez-Lafuente. Evaluation of different immobilized lipases in transesterification reactions using tributyrin: Advantages of the heterofunctional octyl agarose beads. Journal of Molecular Catalysis B: Enzymatic. 2016; 133: 117-123.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


