ISSN: 0973-7510
E-ISSN: 2581-690X
The Indonesian Pesticide Regulations state that Malathion and Profenofos have been restricted in their use for agriculture because of is bioaccumulative in ecological systems. Cleaning technology using microorganisms is an effective solution for cleaning pesticide residues. This study aims to identify the bacteria that degrade and the degradation process of Malathion and Profenophos into non-toxic compounds. The research method was experimental, identification of bacteria by 16S-rRNA gene analysis, degradation ability by GC MS. The results of phylogenetic tree analysis showed that the tested bacteria were closely related to Oceanobacillus iheyenis (RPL1) and Exiquobacterium profundum (RPL5) with a similarity level of 87% and 99%. The two bacteria are used as a consortium of test bacteria. The results of degradation based on the observation chromatogram T = 0 showed that the Malathion compound C10H19O6PS2 or butanedioic acid [(dimethoxyphosphinothioyl) thio]) was detected at peak 4, real-time = 19,675, area% = 7.37 and Profenofos compound C11H15BrClO3PSO-(4-Bromo-2-chlorophenyl)o-ethyl s-propyl thiophosphate, peak 8, real-time = 23,957, area% = 6.91. Likewise, the chromatogram results at T = 96 were still detected Malathion ((dimethoxyphosphinothioyl) thio) at peak 14, real-time = 19,675, area% = 2.25, and Profenofos (o- (4-Bromo-2-chlorophenyl)) o – ethyl. s – propyl thiophosphate) peak = 22 real-time = 23,951, area% = 2.2. However, the observation of T = 192 hours, Malathion and Profenofos compounds were not detected. The conclusion showed that the consortium bacteria were able to completely degrade Malathion and Profenophos within 192 hours.
Consortium bacteria, Exiquobacterium profundum, Oceanobacillus iheyenis, Biodegradation, Malathion, Profenofos
Pesticides are widely used to increase agricultural yields, plantations, forestry production, but pesticides can have a negative impact on the non-target environment. Pesticide pollution needs to be controlled because it can damage the ecological balance 1. Malathion and Profenofos are types of pesticides that are widely used by farmers around Lake Rawa Pening in Central Java, Indonesia, even though these pesticides have been banned especially for rice cultivation based on Regulation of the Minister of Agriculture of the Republic of Indonesia Number 39 / Regulation of the Minister of Agriculture / SR.330 / 7 / 2015 2. It is proven that Profenofos is widely used by farmers around Rawa Pening (50%), then Carbamate (16%), Deltamethrin (8%), Imidacloprid (6%), Fentoat (5%), Carbosulfan (5%), Carbofuran (5% ) and Lamda Sihalotrin (4%). The results of the Profenofos residue analysis in the waters of Rawa Pening ranged from 0.021 ppm – 0.08 ppm, the sediments ranged from 0.12 ppm – 0.28 ppm while the Malathion residue in the waters ranged from 0.0366 ppm – 0.0521 ppm and in sediments ; 0.0567 ppm – 0.12 ppm 3, is above the specified threshold e” of 0.01 ppm 4. Therefore, it is necessary to make efforts to clean the residue of Malathion and Profenofos which have long been exposed to the environment. Indigenous bacteria are developed as biological agents in modifying toxic residues into non-toxic compounds 5
The degradation process of Malathion – Diethyl (dimethoxythiophosphorylthio)succinate (C10H19O6PS2) in aquatic systems will be degraded to monocarboxylic acid-dimethyl monocarboxylic acid – dicarboxylic acid – dimethyl dicarboxylic acid- dicarboxylic acid-CO2 6. Malathion fragmentation in the environment will become maloxon (C10H19O7PS), malathion monocarboxylic acid (C8H15O6PS2), and 2-mercaptosuccinic acid (C6H5NO2S) are derivative compounds that are no more toxic than the initial compound, Malathion 7.
Malathion-degrading bacteria produce catabolic enzymes-malathion carboxyl esterase and malathion dicarboxy latoxy reductase which are able to convert malathionic compounds into thiophosphates and phosphates8.
Profenofos pesticides will be broken down by bacteria into mono metabolite compounds and divalent acids through the enzymatic activity of oxidative desulfurization carboxylesterase 6 9 and demethylation processes in mineralization mechanisms that cause minor routes of metabolism, including oxidation, reduction of sulfur and methyl. 10. The result of enzymatic degradation by consortium bacteria is able to degardate Profenofos into simpler compounds, namely 4-Bromo-2-chlorophenol and 1-phenyl-3-hydroxy-1, 2,4-triazole 11
This article discusses critical areas regarding the degradation of Malathion and Profenophos residues contained in the water and sediments of the Rawa Pening lake by a consortium of indigenous bacteria Exiquobacterium profundum – Oceanobacillus iheyenis which are expected to produce simpler and non-toxic final compounds.
Genomic DNA (Promega) Wizard Extraction Kit: EDTA, Lytic enzyme, nuclei lysis solution, RNAase solution, protein precipitation solution, DNA rehydration solution. Bact-FI primer. 5’AGAGTT TGATCMTGGCTCAG3 ‘/UniB1.5’GGTTACSTTGTTACGACTT3′ (Eurogentec AIT), agarose (Vivantis), ethanol 70%, Ethidium bromide, isopropanol, loading dye (Vivantis), marker (Vivantis), and HgCl2. Sediment from Rawa Pening Lake, Profenofos and Malathion Pro Analisys (PA), Sigma Aldrich Laborochemikallen GmbH, Malathion Pestanal Bath SEBC132XV, Profenofos pestanal Bath SZBC132XV, Malathon 96% and Profenofos Curacron 500 EC
Microbial Identification based on 16S-rRNA Gene Analysis
Bacterial identification was carried out using the 16S-rRNA gene analysis method which included DNA extraction, DNA amplification, purification of DNA amplification results, DNA sequencing, and subsequent construction of phylogenetic trees to obtain genetic diversity.
DNA extraction
DNA extraction using the Chelex 100 Kit. Bacterial cells that have been grown for 24 hours are put into a 1.5 ml Eppendorf tube containing 100 µl of aquadest, then add 0.5% saponins and let stand for 24 hours at 4 0 C. The samples were centrifuged at 12,000 rpm for 10 minutes, the supernatant from the centrifuge was discarded. A total of 1 ml of Phosphate Buffer Saline (PBS 1x) was added to the Eppendorf tube, then centrifuged again at 12,000 rpm for 15 minutes, the supernatant was removed, 100 µl aquadest and 50 µl Chelex 100 were added to the tube. The samples were boiled for 10 minutes (samples were vortexed in the first 5 minutes), then centrifuged again at 12,000 rpm for 10 minutes. The DNA containing the supernatant is transferred to a new Eppendorf tube which is ready for the DNA amplification process. 12
DNA amplification
Amplification is a molecular marker using the 16s rDNA Polymerase Chain Reaction (PCR) method. The temperature treatment used in the DNA amplification process is initial denaturation at 95 0C for 3 minutes, then 30 cycles (denaturation at 95 0C for 1 minute, annealing process at 55 0C for 1 minute and extension at 72 0C for 1 minute ), then extension at 72 0C for 7 minutes 13. The primers used for PCR 16S rDNA were universal primers for 27F bacteria (5′-AGAGTTTGATCMTGGCTCAG-3′) and eubacteria specific primers 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) 14. The mixture of materials used were Promega kit (25 µl) primer 270 F (2.5 µl), primer 1492 R (2.5 µl), DNA template (2.5 µl) and aquabides (17.5 µl) so that total volume 50 µl. The ingredients were mixed in a 0.2 ml PCR tube.15
Visualization of DNA Amplification Results
Visualization of the results of DNA amplification was carried out through electrophoresis by inserting 5 µl of PCR products into 1% agarose gel wells. Making 1% agarose gel by dissolving 1 gram of agarose in 100 ml of TAE 1x buffer solution, then heating it in an oven until homogeneous. A total of 5.33 µl Ethidium Bromide was put into the gel solution and shaken so that it was homogeneous. The gel solution is poured into a comb-shaped mold that is placed in an upright position so that it passes through the comb to the desired thickness. Then the gel was allowed to stand for a while until it hardened, then the gel was immersed in a 1x TAE buffer solution, the gel was electrophoresed with a voltage of 100 V for ± 30 minutes. The amplified DNA bands were observed using the Gel Documentation tool. 16
Purification of DNA Amplification Result
Purification was carried out to obtain pure DNA from PCR 16S rDNA amplification. The PCR results were centrifuged at a speed of 12,000 rpm for 7 minutes. The supernatant was removed using a micropipette until the DNA was completely pure. A total of 50 µl of sterile aquadest was added to the DNA pellet and the results of the pure DNA were sequenced to determine the sequence of DNA bases.17
DNA sequencing
Sequencing was carried out according to the PCR sequencing cycle using Big Dye Terminator v.3.1. The formula for sequencing PCR reactions are 2 µl big dye, 2 µl 10x buffer, 4 µl DNA template, 1 µl primer with a concentration of 3.2 pmol, ddH2O to a final volume of 10 µl. DNA amplification carried out by cycles were initial denaturation (96 °C for 2 minutes), denaturation (96 °C for 10 seconds); annealing (50 °C for 5 seconds); and extension (60 °C for 4 minutes) by 25 cycles. PCR results were purified and sequenced using 27F primer. The sequences were analyzed automatically (ABI 3130XL, Applied Biosystem). 18
Phylogenetic Tree Construction
The pesticide-degrading bacteria that had successfully amplified their 16S rRNA gene could be seen from their relationship with other prokaryotes in the database based on their 16S-rRNA gene sequences. The results of partial sequences are edited using the Bioedit program. After obtaining data on the results of nucleotide sequence contigs, the homology will be compared with other prokaryotes in the Gene Bank database. 19 Cluster analysis was carried out using a database from the RDP website (Ribosomal Database Project) with the website (http://www.rdp.com). while making phylogenetic trees using the MEGA 5 program 20
Biodegradation Test of Malathion and Profenofos
The quantitative data analysis was carried out by determining the levels of Malathion and Profenofos which could be obtained based on the area of the chromatogram produced on Gas chromatography-Mass Spectrometry (GC-MS).21 The analysis was performed using a Gas Chromatography-Mass Spectrometry (GS-MS) instrument. The GS-MS conditions at the time of the study were injector temperature 250°C, oven temperature 80°C, column temperature 280°C, detector temperature 250 °C, helium gas flow rate 1ml / min, constant rate, sample constant rate 1 ›l splitless, standard mix 1 ›l 100 ppm.22 To determine the degradation results of the specimens that had been refused were analyzed using GC MS at 0 hours, 96 hours and 192 hours observations.
Identification of indigenous bacteria
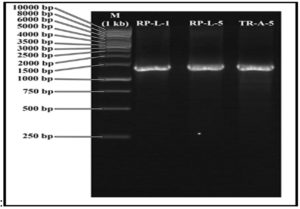
Molecular genetic identification of indigenous bacteria by using genomic-DNA isolation as a template, then the results of genomic-DNA isolation are shown based on the DNA-bands resulting from the 16S-rRNA gene amplification electrophoresis process, shown in the following figure (Figure 1).
Fig. 1. The Results from the Gel Electrophoresis process – 16S-rRNA Amplification. (M) Marker; (1) Bacteria Identification Code = RPL1 and (2) Bacteria Identification Code = RPL5
The species identification by polymerase chain reaction technology (PCR product) using gene-16S rDNA / 16S rRNA (PCR-amplified 16S rRNA) of bacterial species, was carried out using agarose gel electrophoresis method23. The DNA fragments with a size of 50-20,000 bp are the best sizes that agarose gel can separate 24 Analysis using the 16S rDNA/16S rRNA gene has been carried out experimentally in the laboratory because the 16S rDNA/16 rRNA gene is universal and is part of the ribosomal structural RNA which plays an important role in protein synthesis. Therefore the 16 rRNA gene is always present in prokaryotic organisms, is immortal, and almost never is transferred horizontally. This makes the 16S rRNA gene ideal for the reconstruction of the phylogenetic tree and the identification of prokaryotic organisms 25
The isolation process of the tested bacterial genome with the code RPL1 and RPL5 was marked by the formation of one band for each genome of the tested bacteria after being observed using Ultra Violete Transluminator, then it was described by the 16S rRNA gene coding band 1.5 kb, then compared with a Marker (1kb DNA ladder). The results of 16 rRNA DNA amplification were sequenced to obtain the nucleotide sequence and analyzed for similarities using the Gen Bank with the BLAST-N (Basic Local Alignment Search Toll-Nucleotode) program so that the homology and species of bacteria tested could be determined. 26, to determine the phylogeny relationship / relationship with other organisms, the 16S rDNA sequencing results of RPL1 and RPL5 isolates were compared with 16S rDNA sequence data from several species obtained from the data bank. The 16S rDNA sequence data was then synchronized with the ClustalX ver 2.0 program27 The next process is the creation of a phylogenetic tree using the MEGA version 5.03 program with the Neighbor-Joining Tree statistical method, 1000 bootstrap level p-distance models 28 The PCR results of the 16S rDNA gene were shown with a single band on the gel electrophoresis with a size of about 1500 bp.
The results of sequencing using forward and reverse primers to determine the sequence of bacterial nucleotide bases are as follows: (Table 1, fig 2 and fig 3)
Table (1):
Sequencing results (primary forward and reverse).
No |
Code |
Nucleotide base (bp) |
Species name |
Homology |
No accession |
|---|---|---|---|---|---|
1 |
RPL 1 |
1071 |
Oceanobacillus iheyensis |
87 % |
LC10790 |
2 |
RPL 5 |
1238 |
Exuquobacterium profundum |
99 % |
LC19791 |
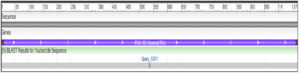
Fig. 2. Exiguobacterium profundum gene for 16S rRNA, partial sequence, strain: RP-L-5 1,238 bp linear DNA GenBank: LC019791.1, species, firmicutes
Fig. 3. Oceanobacillus iheyensis gene for 16S rRNA, partial sequence, strain: RP- L-1 1,071 bp linear DNA. species, firmicutes
Results of 16S-rRNA Gene Sequence of RPL-1 Bacterial GGGGTATTGCATCATAATGCAGTC GAGCGCAGGAAGCTATCTGATCCTCTTTTAGAGGTGACGATAATGGAATGAGCGGCGGACGGGTGAGTAACACGTAGGCAACCTGCCTGTAAGACTGGGATAACTCGTGGAAACGCGAGCTAATACCGGATAACACTTTTCATCTCCTGATGAGAAGTTG AAAGGCGGCTTTTGCTGTCACTTACAGATGGGCCTGCGGCGCATTAGCTAGTTGGTAAGGTAATGGCTTACCAAGGCGACGATGCGTAGCCGACCTGAGAGGGTGATCGGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCGCAATGGAC GAAAGTCTGACGGAGCAACGCCGCGTGAGTGATGAAGGTTTTCGGATCGTAAAACTCTGTTGTTAGGGAAGAACAAGTGCCATAGTAACTATGGCACCTTGACGGTACCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGAATTATTGC GCGTAAAGCGCTCGCAGGCGGTTCTTTAAGTCTGATGTGAAATCTTACGGCTCAACCGTAAACGTGCATTGGAAACTGGGGAACTTGAGTGCAGAAGAGGAGAGTGCAATTCCACGTGTAGCGGTGAAATGCGTATAGATGTGGAGGAACACCAGTGGCGAACGCGACTCTCTGG TCTGTAACTGACGCTGAGTAGCCAAGCGTCGGGAGCGACAGGATTAGATACCCTGGTAGCCCCTGCCGTAGACGATGAGCGCTAGTCGTCAGGGGTTTCCGCCCCTTATGCTGAAGTTACTCATTAAGCACTCCACCTGTGACGTCAGACGCAAGCATCAACTCAAAGGATTTACGCGGAC CACTCAAGCGATGATCACTCGTTTAATTACAGCACCGCGAGAACTTACCAGGCTTGGATTCCTCTGAACATCTAAAATAGCCTTTCCTTCAGGGAAGAGTTCTCCCGACAAAGATTTTTCAACCCANACCTAAATTTCAGTAAGCCCGCACGAAGAAATCTTGA
Results of 16S-rRNA Gene Sequence of RPL-5 Bacterial Samples CAATTGCGCGGCTATAATGCAGTCGAGCGCAGGAAACCGTCTGAACCCTTCGGGGGGACGACGGCGGA ATGAGCGGGGGACGGGTGAGTAACACGTAAAGAACCTGCCCATAGGTCTGGGATAACCACAAGAAATCCGGGCTAATACCGGATGTGTCATCGGACCGCATGGTCCGCTGATGAAAGGGGCTCCGGCGTCTCCCATGGATGGCTTTGCGGTGCATTAGC TAGGTGGTGGGGTAAAGGCCCACCAAGGCGACGATGCATAGCCCAGCTGAGAGGGTGATCGGCCACACTGGGACTGAGACACGGGCCAGACTCCTACGGGAGGGGGCAGTAGGGAATCTTCCCCAATGGACGAAAGTCTGATGGAGCAACG CCGCGTGAACGATGAAAGCTTTCGGGGCGTAAAGTTCTGTTGTAAGGGAAGAACAAGTGCCGCACGCAATGGCGGCGCCTTGACGGTACCTTGCGAGAAAGCCACGGCTAACTACATGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGAATTATTGGG CGTAAAGCGCGCGCAGGCGGCCTCTTAAGTCTGATGTGAAAGCCCCCGGCTCAACCGGGGAGGGCCATTGGAAACTGGGAGGCTTGAGTATATGAGAGAAGAGTGGAATTCCACGTGTAGCGGTGAAATGCGTACAGATGTGAAGGAACACCCTTGTCGAAAGCGACTCTTTGGCCTATA TCTGACGCTGAGGCGCGAAAACGTGGGGAGCAACACGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGAGAGCTAA GTGTTGGAGGGTTCCGCCCTTTGTGCT CAGCTAAGCATTAACACTCCCCTGGGGA GACAGTCGCAGGCTCAACTCAAGGATTG ACGGGACCCCACACCAGTGGAGCATGTGGTTTATTTGAGCACACGGAAAACTTTCCACTCTTGAATCCCCTGACCGGAAAAAATGTACCTTCCCTCTGGGGCAGGGTGACAAGTGTGGATGGTTGCGTCAGCCCCGTCCGAGAGATGCGTTAATCCCCAACAAGGCAACCTTGTCTTTTTTGC ACATTCGTTGGCCCCCTAGGAAATGCCGTGACAACCGAAGAAGGGGGATAACCAAATTCATGCCCTTAAAGTGGGTACACGTGTCAATGGAGGGCAAGGGACCCAACCCCAGTGGACCATCCCAAACGTTTCNTTGGATGGGGGGCACCCCCGTAGACCGAATCTGGCGGGTGC TATACATGCAGTCGAGCGGACAGATGGGAGCTTGCTCCCTGAAGTCAGCGGCGGACGGGTGAGTAACACGTGGGCAACCTGCCTGTAAGACTGGGATAACTCCGGGAAACCGGGGCTAATACCGGATAATTCTTTCCCTCACATGAGGGAAAGCTGAAAGATGGTTTCGGCTATCACTT ACAGATGGGCCCGCGGCGCATTAGCTAGTTGGTGAGGTAACGGCTCACCAAGGCAACGATGCGTAGCCGACCTGAGAGGGTGATCGGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCGCAATGGACGAAAGTCTGACGGAGCA ACGCCGCGTGAGTGATGAAGGTTTTCGGATCGTAAAACTCTGTTGTTAGGGAAGAACAAGTACCGGAGTAACTGCCGGTACCTTGACGGTACCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGAATTATTGGGCGTAAAGCGCGCGCA GGCGGTTCCTTAAGTCTGATGTGAAAGCCCCCGGCTCAACCGGGGAGGGTCATTGGAAACTGGGGAACTTGAGTGCAGAAGAGAAGAGTGGAATTCCACGTGTAGCGGTGAAATGCGTAGAGATGTGGAGGAACACCAGTGGCGAAGGCGACTCTTTGGTC TGTAACTGACGCTGAGGCGCGAAAGCGTGGGGAGCAAACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGAGTGCTAAGTGTTAGAGGGTTTCCGCCCTTTAGTGCTGCAGCAAACGCATTAAGCACTCCGCCTGGGGAGTACGGCCGCAAGGCTGAAACTCAAAGGAATT GACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCTCCTGACAACCCTAGAGATAGGGCGTTCCCCTTCGGGGGACAGGATGACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGT TAAGTCCCGCAACGAGCGCAACCCTTGATCTTAGTTGCCAGCATTCAGTTGGGCACTCTAAGGTGACTGCCGGTGACAAACCGGAAGGAAGGTGGGGGATGACGGTCAAATCATCATGGCCCCTTAAGGACCTGGGGCTAACNCACGTGCTACAATGGGATGGGAACAAAGGGGTTCGAAGACCC GCAAGGTTAANCGGAATCCCCATAAAACATTTTTCAAGTTCNGAATTGCAGGGTTGAAACTCTCCTTGTTTGAAACCCGGATT
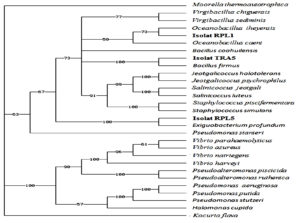
Based on the phylogenetic tree analysis, the test bacterial isolate with code RPL1 has the closest relationship with the Oceanobacillus iheyenis bacteria with a maximum similarity rate of 87%, while the tested bacterial isolates with code RPL5 had the closest relationship with Exiquobacterium profundum with a maximum similarity level of 99%. as follows (Fig 4)
Biodegradation of Profenofos and Malathion by indigenous bacterial consortium
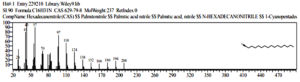
Oceanobacillus iheyenis and Exiquobacterium profundum are indigenous bacteria isolated from the Rawa Pening lake. Both of these bacteria have the ability to degrade against Malathion and profenofos, therefore these bacteria are used as consortium bacteria for research on the biodegradation process of Malathion and Profenofos. The results of Isworo, Purwanto and Sabdono (2016) The results of the test of the degradation ability of selected bacteria in the form of a consortium showed a better ability than the degradation ability of a single isolate. The bacterial consortium Exuquobacterium profundum and Oceanobacillus iheyensis had the best degradation ability of 83.23% while the bacteria consortium Exuquobacterium profundum and bacillus formis had the best degradation ability with a value of 68.75% on the Profenofos substrate 29. The detected biodegradation chemical compounds will be translated into a chromatogram that represents the compound being analyzed. Analysis of the test sample was carried out by observing the retention time and chemical structure of Malathion and Profenofos due to degradation from the bacterial consortium.30 Observations and sampling were carried out at 0 hours, 96 hours, and 192 hours. The chromatogram of chemical compounds biodegradation of the bacterial consortium Exiquobacterium profundum – Oceanobacillus iheyenis at T = 0 hours then detected the malathion compound C10H19O6PS2 or Butanedioic acid, ((dimethoxyphosphinothioyl) thio) -, monoethyl esterer 31, detected on peak 4 with real time = 19,675, area% = 7.37, is follow : (fig 5)
Fig. 5. The chromatogram of the chemical compound Malathion (C10H19O6PS2 or Butanedioic acid, ((dimethoxyphosphinothioyl) thio) -, monoethyl ester
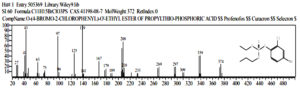
While the cromatogram for the chemical compound Profenofos with the chemical formula C11H15BrClO3PSO- (4-bromo-2-chlorophenyl) o-ethyl s-propyl thiophosphate 32 was detected at peak 8, real time = 23,957, area% = 6.91, as follow : (fig 6)
Fig. 6. Chromatogram and chemical structure of Profenofos (O- (4-Bromo-2-Chlorophenyl) O- Ethyl S- Propyl Thiophosphate))
The complete data on the chemical compounds resulting from degradation is explained based on observations on GC MS with the parameters Peak, Real-Time, Area%, as follows (table 2).
Table (2):
Chemical compounds from the biodegradation of the consortium bacteria (observation t = 0 hours).
Peak |
R.Time |
Area |
Area% |
Height |
Name |
|---|---|---|---|---|---|
1 |
4.034 |
163.635 |
6,7 |
21025 |
(R,S)-2-Butanol, (3R)-3-[(Benzyloxycarbonyl)Amino]- |
2 |
4.240 |
57.279 |
2,4 |
12654 |
T-Butyl (R)-3-(Benzyloxy)-Butanoate |
3 |
7.957 |
59.397 |
2,4 |
14450 |
4,6-Dimethyl-4-Hydroxyhept-5-Enoic Acid |
4 |
13.113 |
98.397 |
4,0 |
39793 |
Pentadecanenitrile(CAS) |
5 |
17.833 |
427.673 |
17,6 |
141327 |
Hexadecanenitrile(CAS) |
6 |
18.384 |
171.142 |
7,0 |
61700 |
Hexadecanoic Acid,Methyl Ester(CAS) |
7 |
19.675 |
179.194 |
7,4 |
61739 |
Malathion E50 |
8 |
21.927 |
145.766 |
6,0 |
45806 |
9-Octadecenal,(Z)-(CAS) |
9 |
22.345 |
85.071 |
3,5 |
30227 |
9-Octadecenoic Acid (Z)-,Methyl Ester(CAS) |
10 |
22.481 |
126.591 |
5,2 |
42298 |
Hexadecanenitrile(CAS) |
11 |
22.958 |
65.369 |
2,7 |
21510 |
Heptadecanoic Acid,16-Methyl-,Methyl Ester(CAS) |
12 |
23.914 |
87.158 |
3,6 |
43370 |
Hexadecanamide(CAS) |
13 |
23.957 |
168.200 |
6,9 |
72947 |
O-(4-Bromo-2-Chlorophenyl)O-Ethyl S-Propyl Thiophosphate # |
14 |
25.481 |
79.450 |
3,3 |
27975 |
9-Octadecenamide(CAS) |
15 |
25.635 |
147.922 |
6,1 |
29660 |
1,1,3,3,5,5,7,7,9,9,11,11,13,13-Tetradecamethylheptasiloxane |
16 |
26.930 |
91.252 |
3,8 |
17443 |
1-Piperazinepropanamide, N-(4-Fluorophenyl)-4-Methyl- |
17 |
27.164 |
60.743 |
2,5 |
17767 |
3-Octadecene-1,2-Diol(CAS) |
18 |
27.258 |
58.346 |
2,4 |
20271 |
14.Alpha-Cheilanth-12-Enic Methyl Ester |
19 |
28.801 |
82.966 |
3,4 |
15704 |
Silicone Grease,Siliconfett |
20 |
29.705 |
77.285 |
3,2 |
19635 |
Silicone Grease,Siliconfett |
2432836 |
100 |
757301 |
In the observation time of t = 0 hours that the tested Malathion and Profenofos compounds were still detected, this indicates that the Malathion and Profenofos compounds have not been completely degraded into simpler compounds.
The chromatogram data of chemical compounds biodegradation results from the bacterial consortium Exiquobacterium profundum-Oceanobacillus iheyenis at T = 96 hours observations, are completely shown in table 3, which is the result of staging on GC MS with parameters peak, real-time, area%, as follow :
Table (3):
Chemical compounds from the biodegradation of the consortium bacteria (observation t = 96 hours).
| Peak | R.Time | Area | Area% | Height | Name | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.975 | 44718 | 1.25 | 9359 | Phenol,3,5-Dimethyl-(CAS) | ||||||
| 2 | 4.477 | 31093 | 0.87 | 10563 | 2-Butenedioic Acid (E)-,Diethyl Ester | ||||||
| 3 | 6.706 | 36954 | 1.04 | 20894 | Phenol,2-methoxy-4-(2-propenyl)-(CAS) | ||||||
| 4 | 7.965 | 72138 | 2.02 | 16400 | 1-(3,3-dimethyl-bicyclo[2.2.1]hept-2-yl)pentan-2-one | ||||||
| 5 | 10.485 | 34744 | 0.97 | 8757 | Pentanedioic acid,2,2-dimethyl-,bis(1-methylpropyl)ester (CAS) | ||||||
| 6 | 12.989 | 54577 | 1.53 | 23184 | Acethydrazide,2-(2-naphthylamino)-N2-(2,6-dichlorobenzylideno)- | ||||||
| 7 | 13.105 | 121792 | 3.42 | 43652 | Dodecanenitrile(CAS) | ||||||
| 8 | 15.995 | 31813 | 0.89 | 13964 | Cyclo(-L-Pro-L-Val-) | ||||||
| 9 | 16.575 | 62669 | 1.76 | 17072 | 1,4-diaza-2,5-dioxo-3-isobutyl bicyclo[4.3.0]nonane | ||||||
| 10 | 17.827 | 401025 | 11.25 | 140342 | Hexadecanenitrile(CAS) | ||||||
| 11 | 18.384 | 139427 | 3.91 | 47978 | Hexadecanoic acid, methyl ester (CAS) | ||||||
| 12 | 18.891 | 72297 | 2.03 | 15832 | 5,10-Diethoxy-2,3,7,8-tetrahydro-1H,6H-dipyrrolo[1,2-a | ||||||
| 13 | 19.493 | 57249 | 1.61 | 18294 | Tetradecanamide | ||||||
| 14 | 19.670 | 80070 | 2.25 | 30221 | Malathion E50 | ||||||
| 15 | 21.914 | 121504 | 3.41 | 37368 | Hexadecenenitrile | ||||||
| 16 | 22.025 | 46680 | 1.31 | 15990 | 9-Octadecenal, (Z)-(Cas) | ||||||
| 17 | 22.336 | 98016 | 2.75 | 34032 | 14-Octadecenoic Acid,Methyl Ester(CAS) | ||||||
| 18 | 22.469 | 150614 | 4.22 | 42639 | Hexadecanenitrile (CAS) | ||||||
| 19 | 22.946 | 75605 | 2.12 | 23171 | Octadecanoic Acid,Methyl Ester(CAS) | ||||||
| 20 | 23.770 | 26632 | 0.75 | 7619 | 4-(4-Bromo-3-Nitro-Benzylidene)-1-(4-Chloro-Phenyl)-Pyrazolidine-3,5-Dione | ||||||
| 21 | 23.913 | 90583 | 2.54 | 42273 | Hexadecanamide(CAS) | ||||||
| 22 | 23.951 | 78877 | 2.21 | 34703 | O-(4-Bromo-2-Chlorophenyl)-O’-Ethyl Ester of Propylthio-Phosphoric Acid | ||||||
| 23 | 24.527 | 41166 | 1.15 | 10099 | Acetamide,N,N’-[(3.beta.)-18-hydroxypregn-5-ene-3,20-diyl]bis- (CAS) | ||||||
| 24 | 24.615 | 33053 | 0.93 | 11041 | 3-(4-Hydroxy-3-methoxyphenyl)-2-isothiocyanatopropionic acid, ethyl ester,TMS | ||||||
| 25 | 25.225 | 65384 | 1.83 | 13189 | 1,3,5,7,9-Pentaethyl-1,9-Dibutoxypentasiloxane | ||||||
| 26 | 25.475 | 28709 | 0.81 | 13917 | 1-(Cyanomethyl)-3-Piperidinecarboxamide | ||||||
| 27 | 25.550 | 34399 | 0.96 | 14017 | N-(2-Adamantan-1-Yl-Ethyl)-4-(Piperidine-1-Sulfonyl)-Benzamide | ||||||
| 28 | 25.620 | 33752 | 0.95 | 7609 | (3E)-4-(1,2-Methoxycarbonylepimino-2,6,6-Trimethylcyclohexyl)-3-Buten-2-One | ||||||
| 29 | 25.863 | 32630 | 0.92 | 20625 | Hexasiloxane, Tetradecamethyl-(CAS) | ||||||
| 30 | 25.936 | 28321 | 0.79 | 14045 | Silikonfett | ||||||
| 31 | 26.010 | 58329 | 1.64 | 19251 | Cyclotetrasiloxane, Octamethyl(CAS) | ||||||
| 32 | 26.055 | 50380 | 1.41 | 19742 | Pentasiloxane,1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||
| 33 | 26.120 | 26494 | 0.74 | 11753 | (2,2-Dibromo-1-Propylcyclopropane)Carboxylic Acid | ||||||
| 34 | 26.150 | 70222 | 1.97 | 17378 | Pentasiloxane, Dodecamethyl- (CAS) | ||||||
| 35 | 26.316 | 58923 | 1.65 | 17078 | 1-Pentene, 1,3-Diphenyl-1-(Trimethylsilyloxy)- | ||||||
| 36 | 26.345 | 86119 | 2.42 | 17965 | 14.Alpha.-Cheilanth-12-Enic Methyl Ester | ||||||
| 37 | 26.430 | 35483 | 1.00 | 16504 | 4-Acetyloxyimino-6,6-Dimethyl-3-Methylsulfanyl-4,5,6,7-Tetrahydro-Benzo | ||||||
| 38 | 26.480 | 63149 | 1.77 | 18989 | Tartronic Acid, 4-(Dimethylethylsilyl)Phenyl-, Dimethyl Ester | ||||||
| 39 | 26.560 | 59074 | 1.66 | 15147 | Cyclotetrasiloxane, Octamethyl- (CAS) | ||||||
| 40 | 26.730 | 34307 | 0.96 | 14017 | Cyclopentasiloxane, Decamethyl- (CAS) | ||||||
| 41 | 26.786 | 31297 | 0.88 | 19358 | Silikonfett | ||||||
| 42 | 26.882 | 51321 | 1.44 | 13561 | Pentasiloxane, 1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||
| 43 | 26.920 | 25776 | 0.72 | 12505 | Sarpagan-17-Ol, 16-[(Acetyloxy)Methyl]-, Acetate (Ester) (CAS) | ||||||
| 44 | 26.985 | 30442 | 0.85 | 12869 | 1,2-Bis(Trimethylsilyl)Benzene | ||||||
| 45 | 27.712 | 47255 | 1.33 | 17719 | Silikonfett | ||||||
| 46 | 27.760 | 30689 | 0.86 | 14958 | Silane, [[4-(2-Isothiocyanatoethyl)-1,2-Phenylene]Bis(Oxy)]Bis[Trimethyl- (CAS) | ||||||
| 47 | 28.085 | 39623 | 1.11 | 20936 | 3-Ethoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsilyloxy)Trisiloxane | ||||||
| 48 | 28.268 | 28903 | 0.81 | 12857 | Tetradecamethylcycloheptasiloxane | ||||||
| 49 | 28.530 | 36573 | 1.03 | 16310 | Silikonfett | ||||||
| 50 | 28.585 | 96854 | 2.72 | 18658 | Silane, Trimethyl[5-Methyl-2-(1-Methylethyl)Phenoxy]- (CAS) | ||||||
| 51 | 28.690 | 40740 | 1.14 | 22872 | Cyclotetrasiloxane, Octamethyl- (CAS) | ||||||
| 52 | 28.747 | 38641 | 1.08 | 23116 | Pentasiloxane, 1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||
| 53 | 28.855 | 53826 | 1.51 | 16658 | 3,4-Isopropylenedioxy-10b(S)-Pancratistatin-1,2-Cyclic Sulfate | ||||||
| 54 | 28.938 | 39942 | 1.12 | 15193 | 3,3-Diethoxy-1,1,1,5,5,5-Hexamethyltrisiloxane | ||||||
| 55 | 29.022 | 35906 | 1.01 | 19894 | Silikonfett | ||||||
| 56 | 29.128 | 28055 | 0.79 | 15987 | Pentasiloxane, 1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||
| 57 | 29.375 | 31829 | 0.89 | 9676 | N-(Cyclohexyl)-3-Ethyl-3-Methyl-1,3-Dihydropyrrol-2-One Alpha.Methyl Ester | ||||||
| 58 | 29.610 | 32848 | 0.92 | 10645 | 14.Alpha.-Cheilanth-12-Enic Methyl Ester | ||||||
| 59 | 29.755 | 29380 | 0.82 | 1E+06 | 1H-Pyrrole-2,4-Dicarboxylic Acid,3,5-Dimethyl-,Diethyl Ester (CAS) | ||||||
| 60 | 29.853 | 45965 | 1.29 | Cyclotetrasiloxane,Octamethyl-(CAS) | |||||||
| 3564836 | 100.00 | ||||||||||
Based on table 3, it shows that the compound Malathion [(Dimethoxy-phosphinothioyl) Thio)] was detected at peak 14, real time = 19.675, area% = 2.25 while Profenofos (O-(4-Bromo-2-Chlorophenyl)O-Ethyl S-Propyl Thiophosphate) detected at peak = 22, real time = 23,951 and area% = 2.2, as follows (Fig 7 and fig 8).
Fig. 7. The chromatogram and chemical structure of Butanedioic Acid Malathion [(Dimethoxyphosphinothioyl) Thio] at observation t = 96 hours
Fig. 8. The chromoatogram and chemical structure of Profenofos[O-(4-Bromo-2-Chlorophenyl) -O’-Ethyl Ester Of Propylthio-Phosphoric Acid] at observation t = 96 hours
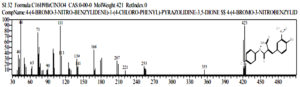
Also detected a chemical compound (4-Bromo-3-Nitro-Benzylidene) -1- (4-Chloro-Phenyl) -Pyrazolidine-3,5-dione which is the result of degradation of the profenofos compound, at peak = 20, real-time = 23,770 , area% = 0.75, as follows: (Fig 9)
Fig. 9. Chemical compounds (4 – Bromo – 3 -Nitro – Benzylidene) -1- (4- Chloro – Phenyl) -Pyrazolidine-3,5-dione
Likewise the chemical compound O- (4-Bromo-2-Chlorophenyl) -O’-Ethyl Ester from Propylthio-Phosphoric Acid resulted from the enzymatic degradation of Profenofos by bacteria, this compound was detected based on a chromatogram at peak = 22, real-time = 24.525 and area % = 2.21, as follow : (Fig 10)
Fig. 10. The chemical compounds O-(4–Bromo–2-Chlorophenyl)-O’-Ethyl Ester of Propylthio-Phosphoric Acid
The chemical compound resulting from the degradation of Profenofos (Profenofos O- (4-Bromo-2-Chlorophenyl) O-Ethyl S-Propyl Thiophosphate) will become a compound of phosphorus and phosphate groups which are degradation compounds that are not toxic. 33
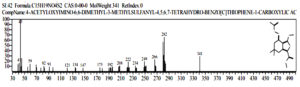
In table 3 also detected compounds resulting from enzymatic malathion degradation by the bacterial consortium, is Chemical compounds of Malathion degraded into Butanedioic Acid, [(Dimethoxyphosphinothioyl) Thio]) detected at peak = 2, real-time = 4,477 , area %. = 0.87 (fig 11).
The chemical compound Butanedioic Acid, [(Dimethoxyphosphinothioyl) Thio] -, Diethyl Ester is a synonym for Malathion Dicarboxylic Acid or Mercapto-O, O-Dimethyl Phosphorodithioate Succinic Acid which is the result of aerobic degradation of Malathion. Butanedioic Acid, [(Dimethoxyphosphinothioyl) Thio] -, Diethyl Ester will be degraded into a compound with this carboxylate group, namely 4-Acetyloxyimino-6,6-Dimethyl-3-Methylsulfanyl-4,5,6,7-Tetra hydro-Benzo [ C] Thiophene-1-Carboxylic Acid. the compound was detected at peak = 37. real time == 26.430, area% = 1.00, as follows: 34 35. (fig 12)
Fig. 12. The chemical compounds 4-Acetyloxyimino-6,6-Dimethyl-3-Methylsulfanyl – 4,5,6,7-Tetra hydro-Benzo [C] Thiophene-1-Carboxylic Acid
Based on these data, Malathion and Profenofos compounds have been degraded into simpler compounds, this can be compared with the decrease in peak values, real time and% area of Malathion and Profenofos compounds.36
The chromatogram of chemical compounds degradation of Malathion and Profenofos by the bacterial consortium Exiquobacterium profundum–Oceanobacillus iheyenis at observation t = 192 hours (table 4), as follows:
Table (4):
Degradation results of chemical compounds (the observation t = 192 hours).
| Peak | R.Time | Area | Area% | Height | Name | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.989 | 48189 | 1.26 | 9539 | Ethanamine,1-(2,4-Cyclopentadien-1-Ylidene)-N,N-Dimethyl-(CAS) | ||||||||||||
| 2 | 5.866 | 63142 | 1.65 | 14298 | 5h-1-Pyrindine | ||||||||||||
| 3 | 8.732 | 64651 | 1.69 | 16861 | 1,3,3-Trideuterio-Endo-6-Hydroxy-9-Oxabicyclo(3.3.1)Nonan-2-One | ||||||||||||
| 4 | 12.985 | 36128 | 0.94 | 14142 | Acethydrazide,2-(2-Naphthylamino)-N2-(2,6-Dichlorobenzylideno)- | ||||||||||||
| 5 | 13.101 | 81069 | 2.11 | 36269 | Tetradecanenitrile | ||||||||||||
| 6 | 15.998 | 57762 | 1.51 | 17709 | 1,4-Diaza-2,5-Dioxo-3-Isobutyl Bicyclo[4.3.0]Nonane | ||||||||||||
| 7 | 17.826 | 353665 | 9.22 | 122031 | Hexadecanenitrile(CAS) | ||||||||||||
| 8 | 18.372 | 98932 | 2.58 | 42608 | Hexadecanoic Acid,Methyl Ester(CAS) | ||||||||||||
| 9 | 18.606 | 35870 | 0.94 | 12399 | 1,4-Diaza-2,5-Dioxo-3-Isobutyl Bicyclo[4.3.0]Nonane | ||||||||||||
| 10 | 19.486 | 38666 | 1.01 | 13905 | 9-Octadecenamide,(Z)-(Cas) | ||||||||||||
| 11 | 21.917 | 119642 | 3.12 | 40310 | Hexadecenenitrile | ||||||||||||
| 12 | 22.02 | 57378 | 1.5 | 17685 | 1H-Fluorene,Dodecahydro-(CAS) | ||||||||||||
| 13 | 22.329 | 57101 | 1.49 | 22640 | 6-Octadecenoic Acid,Methyl Ester,(Z)-(CAS) | ||||||||||||
| 14 | 22.472 | 106596 | 2.78 | 37955 | Hexadecanenitrile(CAS) | ||||||||||||
| 15 | 22.941 | 51171 | 1.33 | 20237 | Tetradecanoic Acid,Methyl Ester(CAS) | ||||||||||||
| 16 | 23.914 | 63356 | 1.65 | 26794 | N-Tetradecanoic Acid Amide | ||||||||||||
| 17 | 24.915 | 30091 | 0.78 | 9222 | Sydnone, 4-Bromo-3-(Dimethylamino)-(CAS) | ||||||||||||
| 18 | 24.975 | 34098 | 0.89 | 12093 | Caprolactone Oxime,(NB)-O-[(Diethylboryloxy)(Ethyl)Boryl]- | ||||||||||||
| 19 | 25.04 | 79169 | 2.06 | 12035 | 3,3-Diethoxy-1,1,1,5,5,5-Hexamethyltrisiloxane | ||||||||||||
| 20 | 25.19 | 50875 | 1.33 | 14698 | 1h-Indole-2-Carboxylic Acid,6-(4-Fluorophenyl)-3-Methyl-4-Oxo-4,5,6,7-Tetra | ||||||||||||
| 21 | 25.28 | 99698 | 2.6 | 20663 | Pentasiloxane,1,1,3,3,5,5,7,7,9,9-Decamethyl | ||||||||||||
| 22 | 25.381 | 46788 | 1.22 | 28603 | 3-Isopropoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsiloxy)Trisiloxane | ||||||||||||
| 23 | 25.415 | 41195 | 1.07 | 22263 | 3,6-Dioxa-2,7-Disilaoctane,2,2,4,7,7-Pentamethyl-(CAS) | ||||||||||||
| 24 | 25.44 | 50152 | 1.31 | 21294 | 1,1,3,3,5,5,7,7,9,9,11,11-Dodecamethyl-Hexasiloxane | ||||||||||||
| 25 | 25.485 | 75004 | 1.96 | 32286 | 1,1,3,3,5,5,7,7,9,9,11,11-Dodecamethyl-Hexasiloxane | ||||||||||||
| 26 | 25.536 | 54901 | 1.43 | 30545 | 3-Ethoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsilyloxy)Trisiloxane | ||||||||||||
| 27 | 25.585 | 90159 | 2.35 | 21884 | 3,7-dibromo-6-ethyl-2-(pent-2′-en-4′-ynyl)octahydropyrano[3,2-b]pyran | ||||||||||||
| 28 | 25.641 | 58196 | 1.52 | 35794 | Phenol,2-(4-diethylaminophenyliminomethyl)- | ||||||||||||
| 29 | 25.715 | 170080 | 4.43 | 29568 | Silicone Grease,Siliconfett | ||||||||||||
| 30 | 25.854 | 130310 | 3.4 | 29240 | (E)-1-[(1′,1′-Dimethylethyl)Diphenylsilyl]-2-(Trimethylsilyl)Ethylene | ||||||||||||
| 31 | 25.896 | 62480 | 1.63 | 26179 | Silikonfett | ||||||||||||
| 32 | 25.947 | 121754 | 3.17 | 23743 | Silikonfett | ||||||||||||
| 33 | 26.134 | 113921 | 2.97 | 20351 | 3-Ethoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsilyloxy)Trisiloxane | ||||||||||||
| 34 | 26.198 | 35139 | 0.92 | 23472 | Silicone Grease,Siliconfett | ||||||||||||
| 35 | 26.303 | 50603 | 1.32 | 21563 | 1,5-Dimethyl-3-(4-Nitrophenyl)-1,3-Dihydro-2,1-Benzisothiazole 2,2-Dioxide | ||||||||||||
| 36 | 26.705 | 56983 | 1.49 | 15843 | 1,1,3,3,5,5,7,7-Octamethyl-Tetrasiloxane | ||||||||||||
| 37 | 27.031 | 55118 | 1.44 | 15602 | Silikonfett | ||||||||||||
| 38 | 27.13 | 75329 | 1.96 | 14331 | Cyclopentasiloxane, Decamethyl-(CAS) | ||||||||||||
| 39 | 27.21 | 39833 | 1.04 | 17390 | Pentasiloxane,1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||||||||
| 40 | 27.378 | 53927 | 1.41 | 15526 | Hydroperoxide,9,10-Dihydro-9,10,10-Triphenyl-9-Anthryl(CAS) | ||||||||||||
| 41 | 27.44 | 32994 | 0.86 | 12840 | Silikonfett | ||||||||||||
| 42 | 27.51 | 45836 | 1.2 | 20107 | Tetracosamethylcyclododecasiloxane | ||||||||||||
| 43 | 27.54 | 41921 | 1.09 | 23431 | 1,3,5,7-Tetraethyl-1-Butoxycyclotetrasiloxane | ||||||||||||
| 44 | 27.674 | 59065 | 1.54 | 14410 | Silikonfett | ||||||||||||
| 45 | 27.73 | 30912 | 0.81 | 13975 | 3-Isopropoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsiloxy)Trisiloxane | ||||||||||||
| 46 | 27.804 | 46229 | 1.21 | 14254 | Silicone Grease,Siliconfett | ||||||||||||
| 47 | 27.858 | 39969 | 1.04 | 17706 | 1,1,3,3,5,5,7,7,9,9,11,11-Dodecamethyl-Hexasiloxane | ||||||||||||
| 48 | 28.599 | 32475 | 0.85 | 14537 | Cyclotrisiloxane,Hexamethyl-(CAS) | ||||||||||||
| 49 | 28.741 | 82232 | 2.14 | 19951 | Silicone Grease, Siliconfett | ||||||||||||
| 50 | 28.83 | 43445 | 1.13 | 19371 | 3-Isopropoxy-1,1,1,5,5,5-Hexamethyl-3-(Trimethylsiloxy)Trisiloxane | ||||||||||||
| 51 | 28.904 | 51224 | 1.34 | 16427 | Tetrakis(Dimethylsilyl)-[18-O]-Dioxide | ||||||||||||
| 52 | 29.064 | 40249 | 1.05 | 19899 | 1h-Pyrrole-3,4-Diacetic Acid, 2-Acetoxymethyl-5-Methoxycarbonyl-,Dimethyl Ester | ||||||||||||
| 53 | 29.14 | 38332 | 1 | 15524 | 2-{4-[2-(4-Methoxymethylphenyl)Vinyl]Phenyl}Propan-2-Ol | ||||||||||||
| 54 | 29.285 | 30528 | 0.8 | 15811 | Anthracene-9-Ol, 9,10-Dihydro-10-(4-Nitrobenzylideno)- | ||||||||||||
| 55 | 29.355 | 38555 | 1.01 | 16368 | 2,5-Dichloro-N,N-Diethyl-Benzenesulfonamide | ||||||||||||
| 56 | 29.44 | 44942 | 1.17 | 13947 | Benzoic Acid, 3-[(Trimethylsilyl)Oxy]-,Trimethylsilyl Ester | ||||||||||||
| 57 | 29.565 | 34341 | 0.9 | 9740 | Silane, Trimethyl[[1-[(Trimethylsilyl) Ethynyl] Cyclohexyl]Oxy] | ||||||||||||
| 58 | 29.69 | 30557 | 0.8 | 12757 | 1,1,1,3,5,7,9,9,9-Nonamethylpentasiloxane18591 Hexasiloxane,Tetradecamethyl-(CAS) | ||||||||||||
| 59 | 29.751 | 30337 | 0.79 | 17289 | Hexasiloxane,Tetradecamethyl-(CAS) | ||||||||||||
| 60 | 29.8 | 31960 | 0.83 | 1306505 | Pentasiloxane,1,1,3,3,5,5,7,7,9,9-Decamethyl- | ||||||||||||
| 3835224 | 100 | ||||||||||||||||
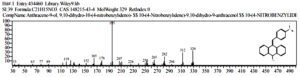
Based on table 4, at the observation t = 92 hours, the chemical compounds of Malathion and Profenofos were not detected. This shows that the concentration disturbance and Profenofos in the sample have broken down completely into simple compounds which are not contaminants. Prediction of Biodegradation of Malathion and Profenofos compounds according to the EAWAG-Biocatalysis and Biodegradation Pathway Prediction System that Malathion and Profenofos compounds will be degraded into simpler compounds, namely Hexadecanenitrile (CAS) chemical compounds Palmitonitrile, Palmitic acid nitrile, N-Hexadecanon 1- Cyanopentadecane which is the result of degradation of Profenofos. these compounds were detected at peak = 7 and real time = 17,826 and Hexadecanenitrile (CAS) at peak = 14, real-time = 22,472 (fig 13), chemical compound Anthracene-9-Ol,9,10-Dihydro-10-(4-Nitrobenzylideno)-(Functional Group-Ol/Alcohol) was detected at peak 54, real time = 29,285 (fig 14), whereas chemical compound 2- {4- [2- (4-Methoxymethylphenyl) vinyl] phenyl} propan-2-ol (functional group-ol / alhohol) with peak = 53 and real time = 29,140 (fig 15) and chemical compounds Acethydrazide compound, 2 – (2-naphthylamino) -N2- (2,6-dichloro benzylideno) is a decomposed benzyl aldehyde group, is a compound resulting from Malathion degradation, detected with peak 4 and real time = 12,985, as follow 33 (fig 16) :
Fig. 13. The chromatogram of chemical compounds Hexadecanenitrile(CAS) Palmitonitrile, Palmitic acid nitrile, N-Hexadecanonitrile, 1-Cyanopentadecane
Fig. 14. Chromatogram of chemical compounds Anthracene -9-Ol, 9,10-Dihydro-10- (4-Nitrobenzylideno) – (Functional Group – Ol / Alcohol)
Fig. 15. The chromatogram of chemical compounds 2- {4-[2-(4-Methoxymethylphenyl) vinyl] phenyl} propan-2-ol (functional group –ol/alhohol)
Fig. 16. The chromatogram of chemical compounds Acethydrazide compound, 2- (2-naphthylamino) – N2 – (2 , 6 – dichloro benzylideno)
At the observation of t = 192 hours, the chemical compounds of Malathion and Profenofos have been degraded into simpler and non-toxic compounds..37
The indigenous bacterial consortium Exiquobacterium profundum – Oceanobacillus iheyenis was able to completely degrade Malathion and Profenofos at observation t = 4 (96 hours observation) based on a decrease in the area % of Malathion from 7.37 to 2.25 and a decrease in area % of Profenofos from 6.91 to 2, 21. At the observation t = 8 (192 hours) Malathion and Profenofos compounds were not detected (area % = 0)
ACKNOWLEDGMENTS
The researcher would like to thank all staff of the Integrated Laboratory of Diponegoro University, Semarang, and all staff of the Dian Nuswantoro University health laboratory, Semarang so that this research can be carried out well.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SI does the research design, wrote the research results, wrote the initial draft of the manuscript. SI and PSO worked together to manage the research analysis. SI manages the literature and makes final draft corrections. Both authors read and approved the manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Meftaul IM, Venkateswarlu K, Dharmarajan R, Annamalai P, Megharaj M. Pesticides in the urban environment: A potential threat that knocks at the door. Sci Total Environ. 2020;711:134612.
Crossref - Mova Al’Afghani M, Paramita D. Regulatory Challenges in the Phasing-Out of Persistent Organic Pollutants in Indonesia. Int Chem Regul Law Rev. 2018;1(1):12-27.
Crossref - Isworo S, Purwanto I, Sabdono A. Impact of pesticide use on organophosphorus and organochlorine concentration in water and sediment of Rawa Pening lake, Indonesia. Res J Environ Sci. 2015;9:233-40.
Crossref - Jensen IM, Whatling P. Malathion: A review of toxicology. Hayes’ Handb Pestic Toxicol. 2010;1527-42. eBook ISBN: 9780080922010
- Siddiqa A, Faisal M. Microbial degradation of organic pollutants using indigenous bacterial strains. In: Handbook of Bioremediation. Elsevier; 2021: 625-637. eBook ISBN: 9780128193839
- Singh B, Kaur J, Singh K. Microbial degradation of an organophosphate pesticide, malathion. Crit Rev Microbiol. 2014;40(2):146-54.
Crossref - Geed SR, Kureel MK, Shukla AK, Singh RS, Rai BN. Biodegradation of malathion and evaluation of kinetic parameters using three bacterial species. Resour Technol. 2016;2:S3-11.
Crossref - Reza M, Fiza J, Hossen F, Ahmed F. Isolation and partial characterization of organophosphate pesticide degrading bacteria from soil sample of Noakhali, Bangladesh. Bangladesh J Microbiol. 2019;36(1):17-22.
Crossref - Cai X, Wang W, Lin L, et al. Autotransporter domain-dependent enzymatic analysis of a novel extremely thermostable carboxylesterase with high biodegradability towards pyrethroid pesticides. Sci Rep. 2017;7(1):3461.
Crossref - Nanda M, Kumar V, Fatima N, et al. Detoxification mechanism of organophosphorus pesticide via carboxylestrase pathway that triggers de novo TAG biosynthesis in oleaginous microalgae. Aquat Toxicol. 2019;209:49-55.
Crossref - E Gonzales-Condori, S Ramírez-Revilla, J Villanueva-Salas. Role of Eisenia foetida in the degradation of profenofos in presence of native bacterial communities. Revista Mexicana De Ingeniería Química. 2020;19(Sup. 1): 45-57.
Crossref - Kang M, Yang JS, Kim Y, Kim K, Choi H, Lee SH. Comparison of DNA extraction methods for drug susceptibility testing by allele-specific primer extension on a microsphere-based platform: Chelex-100 (in-house and commercialized) and MagPurix TB DNA Extraction Kit. J Microbiol Methods. 2018;152:105-8. PMID: 30075237.
Crossref - Ye S, Zeng G, Wu H, et al. Biological technologies for the remediation of co-contaminated soil. Crit Rev Biotechnol. 2017;37(8):1062-76. PMID: 28427272.
Crossref - Santosa GW, Djunaedi A, Susanto AB, Pringgenies D, Ariyanto D. Characteristics of bioactive compounds of Holothuria atra (Jaeger, 1833) associated bacteria. AACL Bioflux. 2020;13(4):2161-2169.
- Gunasegar S, Neela VK. Evaluation of diagnostic accuracy of loop-mediated isothermal amplification method (LAMP) compared with polymerase chain reaction (PCR) for Leptospira spp. in clinical samples: A systematic review and meta-analysis. Diagn Microbiol Infect Dis. 2021;100(3):115369.
Crossref - Lee PLM. DNA amplification in the field: move over PCR, here comes LAMP. Wiley Online Library. 2017;17(2):138-141.
Crossref - Vasiee AR, Mortazavi A, Tabatabaei-yazdi F, Dovom MR. Detection, identification and phylogenetic analysis of lactic acid bacteria isolated from Tarkhineh, Iranian fermented cereal product, by amplifying the 16s rRNA gene with universal primers and differentiation using rep-PCR. Int Food Res J. 2018;25(1):423-432.
- Protopopova M, Pavlichenko V, Gnutikov AA, Chepinoga V. DNA barcoding of Waldsteinia Willd.(Rosaceae) species based on ITS and trnH-psbA nucleotide sequences. In: Information Technologies In the Research Of Biodiversity. 2019:107-115.
Crossref - Kim YG, Choi DH, Hyun S, Cho BC. Oceanobacillus profundus sp. nov., isolated from a deep-sea sediment core. Int J Syst Evol Microbiol. 2007;57:409-413. PMID: 17267988.
- Mello B. Estimating timetrees with MEGA and the TimeTree resource. Mol Biol Evol. 2018;35(9):2334-42. PMID: 29931306.
Crossref - Tony AM, El-Geundi MS, Hussein SM, Abdelwahab MZ. Degradation of malathion in aqueous solutions using advanced oxidation processes and chemical oxidation. Direct Res J Agric Food Sci. 2017;5:174-85. ISSN: 2354-4147.
Crossref - Ozdemir C, Ozdemir S, Oz E, Oz F. Determination of organochlorine pesticide residues in pasteurized and sterilized milk using QuEChERS sample preparation followed by gas chromatography-mass spectrometry. J Food Process Preserv. 2019;43(11):e14173.
Crossref - Tilahun B, Tesfaye A, Muleta D, Bahiru A, Terefework Z, Wessel G. Isolation and molecular identification of lactic acid bacteria using 16s rRNA genes from fermented Teff (Eragrostis tef (Zucc.)) dough. Int J food Sci. 2018;2018:8510620.
Crossref - Green MR, Sambrook J. Analysis of DNA by agarose gel electrophoresis. Cold Spring Harb Protoc. 2019.
Crossref - Quammen D. The tangled tree: a radical new history of life. Simon and Schuster. Nature. 2018;560:26-27.
Crossref - Sarjono PR, Hazrina QH, Saputra A, et al. Isolation, characterization, and identification of endophytic bacteria by 16S rRNA partial sequencing technique from leaves of carica papaya and its potential as an antioxidant. AIP Conference Proceedings. AIP Publishing LLC; 2020:20053.
Crossref - Goncalves LR, Herrera HM, Nantes WAG, et al. Genetic diversity and lack of molecular evidence for hemoplasma cross-species transmission between wild and synanthropic mammals from Central-Western Brazil. Acta Trop. 2020;203:105303.
Crossref - Challa S, Neelapu NRR. Phylogenetic trees: applications, construction, and assessment. Essentials Bioinformatics. 2019;(3):167-92.
Crossref - Newman R, Gilbert MW, Lothridge K. GC-MS guide to ignitable liquids. CRC Press; New York. 2020.
Crossref - Sivakumar S, Anitha P, Ramesh B, Suresh G. Analysis of EAWAG-BBD pathway prediction system for the identification of malathion degrading microbes. Bioinformation. 2017;13(3):73-77.
Crossref - Eawag BH. Swiss Federal Institute of Aquatic Science and Technology. Switzerland, 2014.
- El-Nahhal Y. Toxicity of some aquatic pollutants to fish. Environ Monit Assess. 2018;190:449. PMID: 29974249.
Crossref - Kumar SS, Ghosh P, Malyan SK, Sharma J, Kumar V. A comprehensive review on enzymatic degradation of the organophosphate pesticide malathion in the environment. J Environ Sci Heal Part C. 2019;37(4):288-329.
Crossref - Lozowicka B, Rutkowska E, Jankowska M. Influence of QuEChERS modifications on recovery and matrix effect during the multi-residue pesticide analysis in soil by GC/MS/MS and GC/ECD/NPD. Environ Sci Pollut Res. 2017;24(8):7124-38.
Crossref - Baron S. Medical microbiology. 4th Ed. University of Texas Medical Branch at Galveston; 1996. ISBN-10: 0-9631172-1-1.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.