ISSN: 0973-7510
E-ISSN: 2581-690X
To elucidate the molecular determinants of tetracycline resistance in clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates from the private health sector in KwaZulu-Natal province (KZN), South Africa (SA). Seventy-five clinical MRSA isolates were collected from the private hospital sector in KZN, SA over a one-year period. Susceptibility to antibiotics (tetracycline, doxycycline, minocycline and tigecycline) were determined and tetracycline resistant strains were screened using polymerase chain reaction (PCR) for the presence of four putative tetracycline resistance determinants (tet(K), tet(L), tet(M) and tet(O)). Efflux inhibitors were used to assess the possibility of efflux-mediated resistance.All isolates were mecA gene positive and susceptible to doxycycline, minocycline and tigecycline. Of note, 47 (62.67%) isolates were resistant to tetracycline. Doxycycline exhibited the largest number of intermediate resistance 20 (26.67%) in all the isolates. The tet(M)gene was found in all 47tetracycline-resistant isolates. No tet(L), tet(K) and tet(O) were detected. Efflux inhibitors did not have any significant effect on the sensitivity of tetracycline-resistant isolates suggesting that efflux played a minor role in tetracycline resistance. In conclusion; Tet(M) mainly mediates tetracycline resistance in MRSA in the private health sector in KZN, SA. This report on the prevalence and molecular determinants of tetracycline resistance is the first study on clinical MRSA isolates from the private health sector in SA.
tetracycline; efflux inhibitors;antibiotic resistance; MRSA; South Africa.
Tetracyclines are broad-spectrum antibiotics used in the treatment and prevention of bacterial infections in both humans and animals. Tetracyclines inhibit bacterial 30S ribosome from attaching with the aminoacyl-tRNA, inhibiting protein synthesis1. The discovery of tetracyclines sparked the development of other chemically altered natural antibiotics such as minocycline, doxycycline and recently,the glycylcyclines (tigecycline). Two main mechanisms of resistance to tetracyclines have been identified in Staphylococcus spp.: (i) active efflux resulting from acquisition of the plasmid-located genes, tet(K) and tet(L), and (ii) ribosomal protection mediated by transposon-located or chromosomal tet(M)or tet(O)determinants2.This indicates that tetracycline resistance genes can be transferred amongst the bacterial population.
There have been numerous reports of tetracycline resistance in clinical MRSA isolates in South Africa3–5 owing to the extensive use this antibiotic for various infections in humans and as growth promoters in animal agriculture in the country6. These abusive applications exert a selection pressure on both targeted bacteria and other microflora, leading to resistance in a wide range of bacterial species7. Alarmingly, tetracycline resistance has been reported as 83.4% in the recent largest cohort study of 1236 clinical MRSA isolates from four provinces in South Africa8; however little work has been done to elucidate the genetic determinants of tetracycline resistance in the country. The aim of this study was to ascertain the molecular determinants of tetracycline resistance in methicillin-resistant Staphylococcus aureus (MRSA). To our knowledge, this is the first report describing the genetic basis of tetracycline resistance in clinical MRSA isolates from the private sector in South Africa.
Ethical approval
Permission to carry out this study was granted by the Biomedical Research Ethics Committee (BE394/15) of the University of KwaZulu-Natal (UKZN).
Antibiotics and Efflux pump inhibitors [EPIs]
Antibiotics, namely tetracycline, doxycycline, minocycline, tigecycline as well as efflux pump inhibitors (EPI) including carbonyl cyanide m-chlorophenyl hydrazone (CCCP), thioridazine (TZ), chlorpromazine (CPZ), verapamil (VER) and reserpine (RES) in powder form were purchased from Sigma-Aldrich (St Louis, USA). CCCP is herein used as an experimental protonophore as well as efflux pump inhibitor. RSP (plant-derived efflux pump inhibitor) and VER (Calcium Ca2+ channel blocker) are broad spectrum efflux inhibitors that are commonly used in Gram-positive efflux inhibition experiments. Phenothiazine’s [CPZ and TZ] are mostly used as gram-negative efflux pump inhibitors and their use in Gram-positive bacteria (MRSA) is so far minimal although efflux inhibition has been reported. Solutions of TZ, CPZ, VER were prepared in deionized water; RES was prepared in dimethyl sulfoxide (DMSO) and CCCP in 50% methanol (v/v)9. All solutions were prepared on the day of the experiment and protected from the light.
Bacterial strains
A total of 75 consecutive non-duplicate MRSA isolates were obtained between June 2015 to August 2016, from 21 private healthcare centers in Durban, SA. The isolates were identified using Vitek 2 (bioMerieux, Durham, NC, USA). The cefoxitin disc diffusion test was used to identify potentially MRSA, which was then confirmed by PCR detection of mecA10.
Determination of minimum inhibitory concentrations (MICs)
Cultures were grown in cation-adjusted-Mueller-Hinton broth (Sigma-Aldrich, St Louis, USA) at 37 °C. MICs of antibiotics were determined by the two-fold broth microdilution method and interpreted per the Clinical and Laboratory Standards Institute (CLSI 2016) breakpoints11. Staphylococcus aureus ATCC 29213 was used as a control strain.
MICs in the presence of efflux inhibitors
The inhibitory effect of the efflux inhibitors CCCP, TZ, CPZ, VER and RES onboth tetracycline-resistant and susceptible isolates were evaluated by the microdilution method in medium containing sub-inhibitory EPI concentrations that had no effect on cell viability: 0.25 mg/L CCCP, 8 mg/L TZ, 16 mg/L CPZ, 12.5 mg/L TZ, 200 mg/L VER and 20 mg/L RES12. After an 18 h incubation period at 37°C, the presence of bacterial growth was evaluated visually and the lowest concentration of antimicrobial that presented no visible growth was registered as the MIC. All antibacterial assays were performed in triplicates.
Bacterial DNA extraction and determination of tetracycline resistant genes
NA extraction kit (Gene Elute Bacterial Genomic DNA kit, Sigma-Aldrich) was used to isolate and purify the total DNA from all tetracycline-resistant isolates, as per the manufacturer’s instructions. The presence of genes conferring resistance to tetracycline [tet(K), tet(L), tet(M)and tet(O)] were ascertained by polymerase chain reaction (PCR) as described previously13. All specific primers and programs for detecting antibiotic resistance can be found in Table S1. Nuclease-free water, primers and in-house control strains were included as quality controls in all PCR assays. Amplified products were analyzed by electrophoresis on 1% agarose gel containing 0.5 mg/ml ethidium bromide and photographed under UV illumination. A 10 kb DNA Ladder was used as a reference when running the gel to ensure that the amplimers were within the expected range. Amplimers were further sequenced and BLASTed on NCBI Nucleotide BLAST for confirmation of the genes.
Data and Statistical analysis
The MIC fold changes resulting after the addition of CCCP, TZ, CPZ, RSP and VRP were calculated as the ratio of the MICs of the antibiotic alone to that of the antibiotic plus EPIs. A fold change of ³ 8 was adopted as significant14. MIC50 and MIC90 percentile values, as well as the geometric mean of MIC values were determined. Statistical analysis was undertaken using a One-Way ANOVA followed by the Bonferroni post hoc test using GraphPad Prism 5.0 software; p- values of ³ 0.05 were considered significant.
Susceptibility data of the isolates
A total of seventy-five(75) laboratory confirmed mecA- positive MRSA were recovered from 75 patients from 12 wards, 16 sources in 21 private care centers in KZN, SA (Table 1). The intensive care unit (ICU) recorded the highest number of isolates from the ward; 24 (32.00%) while pus was the specimen with the largest number of isolates; 31 (41.33%). The tetracycline susceptibility pattern of the isolates as well as the MIC distributions of the tested agents are shown in Tables 1, 2, Figure 1 and Table S2. Forty-seven out of the 75 isolates (62.67%) were resistant to tetracycline. All the isolates were susceptible to doxycycline, minocycline and tigecycline.The geometric mean of doxycycline’s MIC (3.69 mg/L) was not significantly different from that of minocycline (2.60 mg/L) it was always a fold higher when MIC50/MIC90 (4/8:2/4 mg/L) and MIC range (³0.25–8: ³0.25–8 mg/L) were compared and had more isolates 20/75 (26.67mg/L) in the intermediate range than minocycline. The MIC50 and MIC90 and geometric mean of MIC values for tetracycline were higher than all the other antibiotics with tigecycline recording the lowest values (Table 2 and Figure 2).
Table 1
Clinical data, minimum inhibitory concentrations (MIC), and results of PCR for 47 tetracycline resistant MRSA isolates
Sample No. |
Isolate |
Clinical data |
MIC (mg/L) |
PCR |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Ribosomal protective proteins |
Efflux proteins |
||||||||||||
Collection date |
Hospital center |
WARD |
SPECIMEN |
TETRACYCLINE |
DOXYCYCLINE |
MINOCYCLINE |
TIGECYCLINE |
TET(M) |
<TET(O) |
TET(L) |
TET(K) |
||
1 |
LNO02 |
15/6/15 |
O |
OPD |
Nasal |
256 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
2 |
LNO03 |
15/6/15 |
B |
ICU |
CVP tip |
128 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
3 |
LNO05 |
18/6/15 |
C |
ICU |
ETT |
128 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
4 |
LNO06 |
24/6/15 |
E |
LW |
Blood |
128 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
5 |
LNO10 |
25/6/15 |
F |
OPD |
Pus |
32 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
6 |
LNO13 |
29/7/15 |
D |
ICU |
Sputum |
16 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
7 |
LNO14 |
29/7/15 |
H |
ICU |
Blood |
64 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
8 |
LNO15 |
30/7/15 |
E |
Surgical |
Pus |
256 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
9 |
LNO18 |
20/8/15 |
A |
Surgical |
Wound |
64 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
10 |
LNO20 |
20/8/15 |
K |
ICU |
Wound |
128 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
11 |
LNO23 |
26/8/15 |
M |
General |
Bone |
64 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
12 |
LNO24 |
03/9/15 |
F |
Trauma |
Cheek |
64 |
2 |
0.5 |
≤0.25 |
+ |
– |
– |
– |
13 |
LNO26 |
03/9/15 |
B |
Surgical |
Head |
32 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
14 |
LNN01 |
08/4/16 |
A |
OPD |
Pus |
32 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
15 |
LNN02 |
08/4/16 |
F |
Surgical |
Urine |
32 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
16 |
LNN06 |
08/4/16 |
Q |
ICU |
Sputum |
64 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
17 |
LNN09 |
08/4/16 |
B |
ICU |
Blood |
32 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
18 |
LNN10 |
20/4/16 |
F |
ICU |
Pus |
64 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
19 |
LNN12 |
20/4/16 |
F |
Surgical |
Pus |
64 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
20 |
LNN14 |
20/4/16 |
L |
ICU |
Blood |
32 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
21 |
LNN15 |
20/4/16 |
L |
Surgical |
Pus |
32 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
22 |
LNN16 |
20/4/16 |
G |
Medical |
Blood |
64 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
23 |
LNN18 |
20/4/16 |
B |
Medical |
Sputum |
64 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
24 |
LNN20 |
03/5/16 |
L |
Surgical |
Pus |
32 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
25 |
LNN22 |
03/5/16 |
M |
Surgical |
Pus |
128 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
26 |
LNN23 |
03/5/16 |
L |
Surgical |
Pus |
256 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
27 |
LNN25 |
03/5/16 |
S |
Pediatric |
Sputum |
64 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
28 |
LNN32 |
03/5/16 |
U |
Pediatric |
ETT |
256 |
8 |
≤0.25 |
≤0.25 |
+ |
– |
– |
– |
29 |
LNN34 |
03/5/16 |
E |
OPD |
Pus |
32 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
30 |
LNN35 |
03/5/16 |
H |
ICU |
Blood |
256 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
31 |
LNN36 |
03/5/16 |
H |
OPD |
Pus |
128 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
32 |
LNN37 |
03/5/16 |
S |
Medical |
Sputum |
128 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
33 |
LNN39 |
30/6/16 |
H |
Surgical |
Tissue |
128 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
34 |
LNN40 |
30/6/16 |
K |
Surgical |
Pus |
64 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
35 |
LNN41 |
06/6/16 |
I |
Theatre |
Pus |
128 |
4 |
2 |
≤0.25 |
+ |
– |
– |
– |
36 |
LNN46 |
20/7/16 |
I |
ICU |
CVP tip |
128 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
37 |
LNN47 |
28/7/16 |
N |
Ortho |
Tissue |
32 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
38 |
LNN49 |
25/8/16 |
M |
Neuro |
Pus |
256 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
39 |
LNN50 |
25/8/16 |
V |
Ortho |
Sputum |
16 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
40 |
LNN51 |
25/8/16 |
N |
ICU |
Pus |
32 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
41 |
LNN52 |
25/8/16 |
N |
ICU |
Blood |
64 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
42 |
LNN53 |
25/8/16 |
G |
ICU |
Pus |
128 |
8 |
8 |
≤0.25 |
+ |
– |
– |
– |
43 |
LNN56 |
31/8/16 |
V |
Medical |
CVP tip |
128 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
44 |
LNN57 |
31/8/16 |
P |
Surgical |
Urine |
256 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
45 |
LNN59 |
31/8/16 |
Q |
ICU |
Pus |
64 |
8 |
4 |
≤0.25 |
+ |
– |
– |
– |
46 |
LNN60 |
31/8/16 |
C |
Surgical |
ETT |
64 |
8 |
2 |
≤0.25 |
+ |
– |
– |
– |
47 |
LNN61 |
31/8/16 |
S |
ICU |
Sputum |
16 |
4 |
4 |
≤0.25 |
+ |
– |
– |
– |
a. ETT, Endotracheal tube; CVP, Central venous catheter; ICU, Intensive/High care unit; LW, Labour ward; OPD, outpatient department; Ortho, Orthopaedic department: Neuro; Neurological department. b.The alphabets A – Z indicates the codes of the hospitals where the MRSA isolates were collected.
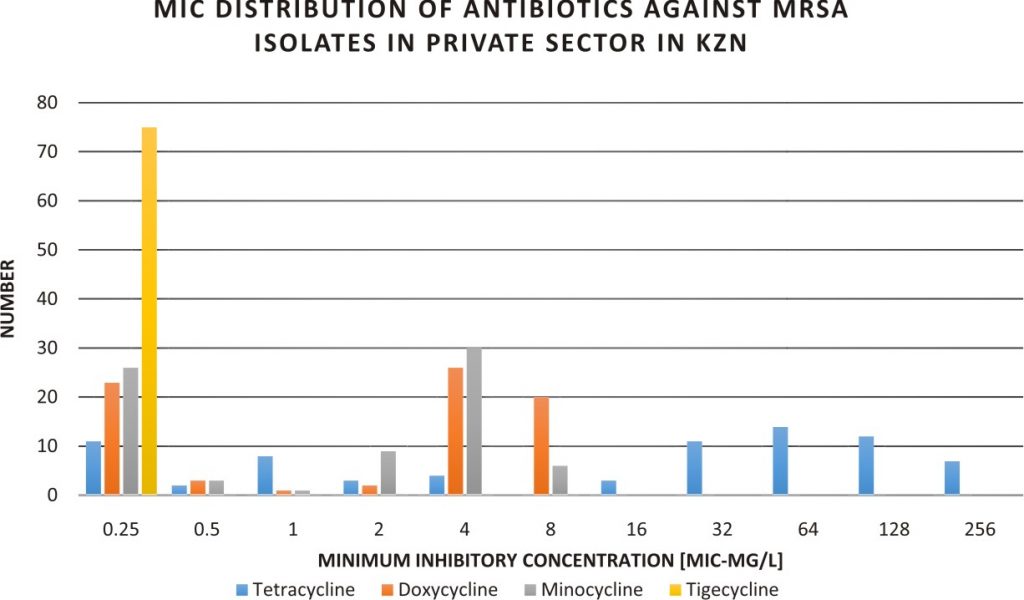 Fig. 1. MIC of tetracycline, doxycycline, minocycline and tigecycline to MRSA isolated from KZN Private Hospitals, SA.
Fig. 1. MIC of tetracycline, doxycycline, minocycline and tigecycline to MRSA isolated from KZN Private Hospitals, SA.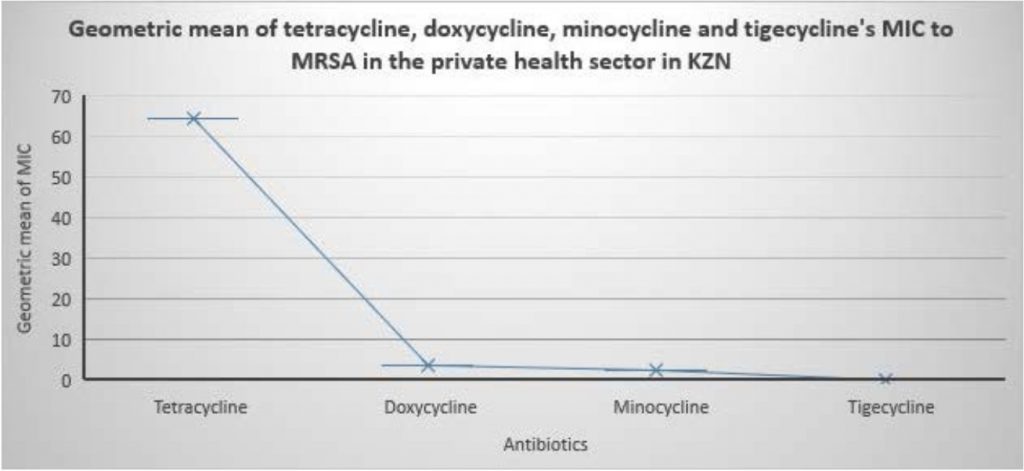 Fig. 2. Geometric mean of tetracycline, doxycycline, minocycline and tigecycline MIC to MRSA in the private health sector in KZN.
Fig. 2. Geometric mean of tetracycline, doxycycline, minocycline and tigecycline MIC to MRSA in the private health sector in KZN.Table 2
Susceptibility overview of the 75 MRSA isolates to tetracyclines
| Antibiotic | Sensitive
N (%) |
Intermediatea
N (%) |
Resistant
N (%) |
MIC (mg/L) | |||
|---|---|---|---|---|---|---|---|
| range | MIC50
|
MIC90
|
geometric mean | ||||
| Tetracycline | 28 (37.33) | 0 (0.00) | 47 (62.67) | ≤0.25 – 256 | 32 | 128 | 58.40 |
| Doxycycline | 55 (73.33) | 20 (26.67) | 0 (0.00) | ≤0.25 – 8 | 4 | 8 | 3.69b |
| Minocycline | 69 (92.00) | 6 (8.00) | 0 (0.00) | ≤0.25 – 8 | 2 | 4 | 2.60b |
| Tigecycline | 75 (100.00) | 0 (0.00) | 0 (0.00) | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25b |
a. An intermediate: Isolate is one that is inhibited in vitro but therapeutic effect is uncertain.
b. P < 0.0001 was observed when the MIC geometric mean of the other antibiotics was compared to tetracycline.
Effects of Efflux pump inhibitors on the isolates
All the efflux pump inhibitors did not significantly reduce the MIC of tetracycline indicating that the ionophore (CCCP),gram-positive efflux pump inhibitors (RES and VER) andphenothiazines (TZ and CPZ)played a minor role in tetracycline resistance (Table S3).
Prevalence of the genetic determinants of tetracycline resistance
The prevalence of the four tetracycline resistance genes [tet(M), tet(O), tet(K) and tet(L)] on the extracted DNA of the all clinical MRSA isolates are shown in Table 1 and Table S2.Tet(M) was found in all the tetracycline resistant isolates. None of the isolates contained tet(L), tet(O) and tet(K).
The rate of antibiotic resistance is increasing faster than the development of new compounds for clinical practice; this is causing a public health crisis. Antibiotic resistance in Gram-positive bacteria is on the rise globally as indicated by the WHO list of high priority pathogens (i.e. MRSA) though much attention has shifted to combating Gram-negative bacteria hence the need to understand the molecular mechanism of resistance to inform the development of appropriate clinical regimen that could be therapeutically effective in curbing this resistance. Evidently research in the public health sector in terms of drug resistance far outweighs that of the public health sector in South Africa and World at large hence the need for research to be broaden to all sectors to elucidate the actual extent of this global menace15.
The demographic data reflected 12 wards and 16 specimen sources from 21 private healthcare centers in KZN, South Africa, representing a wide spectrum of sources within the KZN province. None of the isolates showed resistance to doxycycline, minocycline and tigecycline while 62.67% (47/75) were resistant to tetracycline. The resistance rate of tetracycline (62.67% vs 57.2%) in this study was comparable to another study in South Africa involving 248 confirmed MRSA isolates;4 however, resistance to minocycline (0.0% vs 39.1%) varied considerably in that study than observed herein. This is expected as resistance to minocycline has been reported to be significantly higher in the public than the private sector in South Africa4.
The respective MIC50/MIC90 and geometric mean of MIC clearly indicate the isolates were highly resistant to tetracycline compared to the other antibiotics. Of note, though the geometric mean of doxycycline’s MIC(3.67mg/L)was not significantly different from that of minocycline (2.60mg/L) it was always a fold higher when MIC50/MIC90 (4/8:2/4mg/L) were compared and had more isolates 20/75(26.67mg/L)in the intermediate range than minocycline. This necessitates a cautious prescription of doxycycline as a treatment option for MRSA infections by clinicians as their overuse can cause resistance as a result of selective pressure. All of the isolates were fully susceptible to tigecycline,which was comparable to study conducted on MRSA in South Africa5 and confirms their use as a treatment option for MRSA infections in SA.
The EPIs did not have any significant effect on the MIC when all the different EPIs were added,albeit its resistance was not fully reversed. Thus, efflux may play a minor role in mediating tetracycline resistance. This was contrary to studies by Chovanova et al,16 Hirata et al17 and Stavri et al18 implicating efflux pump inhibitors; CCCP and reserpineas potentiators of tetracycline (aminimum 4-fold reduction in MICs) in Staphylococcus spp.
The tet(M)gene was present in all 47tetracycline-resistantisolates and could be responsible for the higher tetracycline MICs. Tet(M) protects the bacteria from tetracycline by encoding ribosomal protective proteins which inhibits the binding of tetracycline to the ribosome. The tet(O)gene was not detected in any of the isolates, which correlates with studies conducted in Europe19,20 and Pakistan21 indicating that the gene is rare in S. aureus as previously reported by Bismuth et al22. Tet(L) was absent in all the isolates, which is akin to a study conducted in Europe(2000) on 400tetracycline-resistant clinical MRSA isolates in which tet(L) genes were detected in six (1.5%) isolates19. Interestingly, no tet(K)was found in all the isolates screened even though tet(K) has been reported as a common and widely distributed gene in Staphylococcus aureus19–21. However, this was in agreement with a study conducted in South Africa where no tet(K) was found in 27 clinical MRSA isolates5. This is supportive of the assertion that efflux pumps-mediated resistance played a part in inducing resistance to tetracycline in the isolates, tet(M) is the predominant resistance mechanism.
The study indicates that resistance to tetracycline in MRSA in the private health-sector in KZN, South Africa is mainly mediated by tet(M)-encoded ribosomal protection proteins with little or no significant role of efflux pumps. However, strain typing will be needed to provide a better overview of the population clonal structure of MRSA circulating in the private sector niche. To our knowledge, this report on the prevalence and molecular determinants of tetracycline resistance is the first of such study on clinical MRSA isolates from the private health sector in South Africa. This gives credence to how data generated by private sector laboratories can be leveraged in support of public health AMR surveillance in South Africa to inform the development of appropriate clinical regimen that could be therapeutically effective against this menace.
We thank the South African National Research Foundation and the College of Health Sciences, UKZN, for supporting and funding this project. The funders had no role in the study design, data collection and interpretation, nor the decision to submit the work for publication.
Professor S.Y. Essack is a member of the Global Respiratory Infection Partnership sponsored by an unrestricted educational grant from Reckitt and Benckiser. The other authors declare no conflict of interest.
Table S1: PCR primers and cycling parameters for genes presented in this study
Gene |
Primer/sequence |
PCR conditions |
PCR size (bp) |
Reference |
|---|---|---|---|---|
mecA |
F-AACAGGTGAATTATTAGCACTTGTAAG
R-ATTGCTGTTAATATTTTTTGAGTTGAA |
30 s 94 °C, 30 s 55 °C, 60 s 72 °C |
174 |
1 |
tet(M) |
F-GTGGACAAAGGTACAACGAG
R-CGGTAAAGTTCGTCACACAC |
60 s 94 °C, 60 s 55 °C, 90 s 72 °C |
406 |
2 |
tet(O) |
F-AACTTAGGCATTCTGGCTCAC
R-TCCCACTGTTCCATATCGTCA |
60 s 94 °C, 60 s 57 °C, 60 s 72 °C |
514 |
3 |
tet(K) |
F-TCG ATA GGA ACA GCA GTA
R-CAG CAG ATC CTA CTC CTT |
60 s 94 °C, 60 s 55 °C, 90 s 72 °C |
169 |
2 |
tet(L) |
F-TCGTTAGCGTGCTGTCATTC
R-GTATCCCACCAATGTAGCCG |
60 s 94 °C, 60 s 55 °C, 90 s 72 °C |
267 |
2 |
Table S2: Clinical data and minimum inhibitory concentrations (MIC) for 28 tetracycline susceptible MRSA isolates
| No. | Isolate | Clinical data | MIC (mg/l) | PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Collection date | Hospitalbcenter | Ward a | Specimen | Tetracycline | Doxycycline | Minocycline | Tigecycline | Tet(M) | Tet(O) | Tet(L) | Tet(K) | ||
| ATCC 29213C | NA | NA | NA | NA | 0.5 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – | |
| 1 | LNO01 | 10/6/15 | A | ICU | Blood | 2 | 0.25 | 0.5 | ≤0.25 | – | – | – | – |
| 2 | LNO09 | 25/6/15 | F | OPD | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 3 | LNO11 | 24/7/15 | A | ICU | Blood | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 4 | LNO16 | 07/8/15 | A | ICU | Blood | 2 | 0.5 | ≤0.25 | ≤0.25 | – | – | – | – |
| 5 | LNO17 | 20/8/15 | J | ICU | ETT | 2 | 2 | 1 | ≤0.25 | – | – | – | – |
| 6 | LNO19 | 20/8/15 | J | Nursery | Eye | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 7 | LNO21 | 26/8/15 | A | Medical | CVP | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 8 | LNO22 | 26/8/15 | L | General | Humerus | 0.5 | 0.5 | 0.5 | ≤0.25 | – | – | – | – |
| 9 | LNO25 | 03/9/15 | N | Surgical | Head | 0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 10 | LNN03 | 08/4/16 | F | Surgical | Urine | 4 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 11 | LNN04 | 08/4/16 | E | Surgical | Pus | 4 | 1 | ≤0.25 | ≤0.25 | – | – | – | – |
| 12 | LNN05 | 08/4/16 | O | Medical | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 13 | LNN07 | 08/4/16 | R | Medical | Blood | 4 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 14 | LNN08 | 08/4/16 | G | Theatre | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 15 | LNN11 | 20/4/16 | A | Surgical | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 16 | LNN19 | 03/5/16 | G | Medical | Joint fluid | 0.5 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 17 | LNN27 | 03/5/16 | E | ICU | Pus | 4 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 18 | LNN28 | 03/5/16 | K | Medical | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 19 | LNN30 | 03/5/16 | S | Surgical | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 20 | LNN31 | 03/5/16 | T | Surgical | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 21 | LNN33 | 03/5/16 | G | Theatre | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 22 | LNN43 | 20/7/16 | G | ICU | Sputum | 1 | 0.5 | ≤0.25 | ≤0.25 | – | – | – | – |
| 23 | LNN44 | 28/7/16 | B | Medical | Blood | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 24 | LNN45 | 28/7/16 | G | Theatre | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 25 | LNN48 | 03/8/16 | N | ICU | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 26 | LNN54 | 25/8/16 | D | Surgical | Pus | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 27 | LNN55 | 31/8/16 | E | ICU | Sputum | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | – | – | – | – |
| 28 | LNN58 | 31/8/16 | G | Medical | Pus | 1 | ≤0.25 | ≤0.25 | ≤0.25
|
– | – | – | – |
ETT, Endotracheal tube; CVP, Central venous catheter; ICU, Intensive/High care unit; LW, Labour ward; OPD, outpatient department; Ortho, Orthopaedic department: Neuro; Neurological department. b. The alphabets A – Z indicates the codes of the hospitals where the MRSA isolates were collected. c. ATCC 29213- Staphylococcus aureus ATCC 29213
Table S3: Effects of Efflux pump inhibitors on tetracycline resistance on MRSA isolates in KZN
| Isolatec | MIC (mg/L) | |||||
|---|---|---|---|---|---|---|
| Tetracycline | ||||||
| No | + | + | + | + | + | |
| EPI | CCCP [0.25 mg/L] | TZ [8 mg/L] | CPZ [16 mg/L] | VER [200 mg/L] | RES [20 mg/L] | |
| Staphylococcus aureus ATCC 29213 | 0.5 | 0.5 [1]b | 0.25[2] | 0.25[2] | 0.25 [2] | 0.5 [1] |
| Tetracycline-resistant isolates | ||||||
| LNO02 | 256 | 256 [1] | 128 [2] | 128 [2] | 128 [2] | 256 [1] |
| LNO03 | 128 | 128 [1] | 64 [2] | 64 [2] | 64 [2] | 128 [1] |
| LN010 | 32 | 32 [1] | 32 [1] | 32 [1] | 32 [1] | 32 [1] |
| LNN02 | 32 | 32 [1] | 32 [1] | 32 [1] | 32 [1] | 32 [1] |
| LNN06 | 64 | 64 [1] | 64 [1] | 64 [1] | 32 [2] | 64 [1] |
| LNN15 | 32 | 32 [1] | 16 [2] | 16 [2] | 32 [1] | 32 [1] |
| LNN22 | 128 | 128 [1] | 128 [1] | 64 [2] | 128 [1] | 128 [1] |
| LNN32 | 256 | 256 [1] | 256 [1] | 128 [2] | 128 [2] | 256 [1] |
| LNN34 | 32 | 32 [1] | 32 [1] | 16 [2] | 16 [2] | 32 [1] |
| LNN35 | 256 | 128 [2] | 128 [2] | 128 [4] | 64 [4] | 64 [4] |
| LNN49 | 256 | 128 [2] | 128 [2] | 128 [2] | 128 [2] | 256 [1] |
| LNN50 | 16 | 16 [1] | 8 [2] | 8 [2] | 16 [1] | 16 [1] |
| LNN53 | 128 | 128 [1] | 128 [1] | 128 [1] | 128 [1] | 128 [1] |
| LNN60 | 64 | 64 [1] | 64 [1] | 64 [1] | 32 [2] | 64 [1] |
| Tetracycline-susceptible isolates | ||||||
| LNO01 | 2 | 2 [1] | 2 [1] | 1 [2] | 2 [1] | 2 [1] |
| LNO16 | 2 | 2 [1] | 2 [1] | 1 [2] | 2 [1] | 2 [1] |
| LNN03 | 4 | 4 [1] | 2 [2] | 2 [2] | 2 [2] | 4 [1] |
| LNN07 | 4 | 4 [1] | 4 [1] | 2 [2] | 2 [2] | 4 [1] |
| LNN11 | 1 | 1 [1] | 1 [1] | 0.5 [2] | 0.5 [2] | 1 [1] |
| LNN19 | 0.5 | 0.5 [1] | 0.5 [1] | 0. 25 [2] | 0.25 [2] | 0.25 [2] |
| LNN27 | 4 | 4 [1] | 2 [2] | 2 [2] | 2 [2] | 4 [1] |
| LNN58 | 1 | 1 [1] | 1 [1] | 1 [1] | 1 [1] | 1 [1] |
EPI: efflux pump inhibitor; CCCP: carbonyl cyanide m-chlorophenylhydrazone; TZ: thioridazine; CPZ: chlorpromazine; VER: verapamil; RES: reserpine. b. Parenthesis [] indicates the MIC fold change of the isolates after an efflux inhibitor was added. c. Isolate in bold correspond to those which efflux pumps play a major role in tetracycline resistance when efflux inhibitors were added i.e. MIC decrease of ≥ 8-fold change in the presence of the EPIs in comparison to the values with no EPIs.
- Lambert T. Antibiotics that affect the ribosome. Rev sci tech Off int Epiz. 2012; 31(1):57-64.
- Chopra I. New developments in tetracycline antibiotics: Glycylcyclines and tetracycline efflux pump inhibitors. Drug Resist Updat. 2002; 5(3-4):119-125.
- Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis. 2006; 6:125.
- Marais E, Aithma N, Perovic O, Oosthuysen WF, Musenge E, Dusי a G. Antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates from South Africa. S Afr Med J. 2009; 99(3):170-173.
- Amoako DG, Bester LA, Somboro AM, Baijnath S, Govind CN, Essack SY. Plasmid-mediated resistance and virulence mechanisms in the private health sector in KwaZulu-Natal, South Africa: An investigation of methicillin resistant Staphylococcus aureus (MRSA) clinical isolates collected during a three month period. Int J Infect Dis. 2016; 46:38-41.
- Eagar H, Swan G, van Vuuren M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J. S. Afr. Vet. Assoc. 2012; 83(1):16. doi:10.4102/jsava.v83i1.16.
- Holmes AH, Moore LSP, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016; 387(10014):176-187.
- Perovic O, Iyaloo S, Kularatne R, et al. Prevalence and trends of staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: Laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2015; 10(12):1-14.
- Couto I, Costa SS, Viveiros M, Martins M, Amaral L. Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. 2008;(May):504-513.
- Kuntovב L, Pantuטek R, Rבjovב J, et al. Characteristics and distribution of plasmids in a clonally diverse set of methicillin-resistant staphylococcus aureus strains. Arch Microbiol. 2012; 194(7):607-614.
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement M100-S25. USA: CLSI, Wayne, PA, USA; 2016.
- Santos Costa S, Viveiros M, Rosato AE, Melo-Cristino J, Couto I. Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus. BMC Microbiol. 2015; 15(1):232.
- Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. mol cell probes. 2001;15:209-215.
- Osei Sekyere J, Amoako DG. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant enterobacteriaceae. Front Microbiol. 2017; 8:228.
- Gandra S, Merchant AT, Laxminarayan R. A role for private sector laboratories in public health surveillance of antimicrobial resistance. 2016; 11:16-19.
- Chovanovב R, Mezovskב J, Vaverkovב, Mikulב ovב M. The inhibition the Tet(K) efflux pump of tetracycline resistant Staphylococcus epidermidis by essential oils from three Salvia species. Lett Appl Microbiol. 2015; 61(1):58-62.
- Hirakata. NII-Electronic Library Service. Chem Pharm Bull. 1966; 14:369-375.
- Stavri M, Piddock LJ V, Gibbons S. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother. 2007; 59(6):1247-1260.
- Schmitz F-J, Krey A, Sadurski R, Verhoef J, Milatovic D, Fluit AC. Resistance to tetracycline and distribution of tetracycline resistance genes in European Staphylococcus aureus isolates. J. Antimicrob Chemother. 2001; 47:239-240.
- Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000; 45(6):763-770.
- Ullah F, Malik SA, Ahmed J, et al. Investigation of the genetic basis of tetracycline resistance in Staphylococcus aureus from Pakistan. Trop J Pharm Res. 2012; 11(6):925-931.
- Bismuth R, Zilhao R, Sakamoto H, Guesdon JL, Courvalin P. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob Agents Chemother. 1990; 34(8):1611-1614.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


