ISSN: 0973-7510
E-ISSN: 2581-690X
Monkeypox is a zoonotic viral infection caused by monkeypox virus which belongs to the Poxviridae family of genus Orthopoxvirus. Usually, the virus transmission happens when the individual comes in contact with the infected person through body fluids, animal lesions, respiratory droplets or through virus contaminated materials. Clinical presentation of the monkeypox has shown significant resemblance to that of smallpox and chickenpox, belonging to the same orthopoxvirus genus but were eradicated during 1980s globally. Monkeypox may lead to a range of medical complications including clinical symptoms like fever, rashes, headaches, back pain, myodynia and swollen lymph nodes. As far as the treatment modalities are concerned, the antiviral therapeutic agents developed for the smallpox treatment, were also permitted to be used for the monkeypox treatment. However, there is no proven treatment for human monkeypox. In the current study, we have focused on designing of a best probable ligand against the target MPXVgp158 (Monkeypox virus protein). Since Tecovirimat is an FDA approved compound known as an antipoxviral drug, the study aimed to develop a Monkeypox virus protein MPXVgp158 inhibitor which is bioavailable and biocompatible as well through drug designing using computational tools. Molecular docking (MD) analysis displayed Tecovirimat with lesser binding energy, higher non-bonded interaction capability, and more stability against MPXVgp158, with efficient binding mode of interactions. Hence, Tecovirimat was adjudged to be the potential candidate against MPXVgp158 inhibition.
Monkeypox Virus, Molecular Docking, Molecular Dynamic Simulations, Computer-Aided Drug Designing
World Health Organization (WHO) declared the outbreak of Monkeypox as a global public health emergency on July 23, 2022 (WHO, 2022). The disease is known to affect various tissues and organs including skin, mucous linings, tonsils, lymphatic nodes, spleen, eyes, with majority of symptoms bearing similarity to that of smallpox. 1,2 Monkeypox virus can reach the brain parenchyma and display neuroinvasive characteristics through two different routes: olfactory epithelium and hematogenous penetration.3 The very first report of isolation of monkeypox virus was reported from cynomolgus monkeys during the smallpox like disease outbreaks during the year 1958 in Copenhagen. The Democratic Republic of the Congo was the first country to report a monkeypox case of a nine-months old baby.4 Since then, the virus has spread throughout Africa, splitting into clades in Central and West Africa.5,6 Therefore, the West African clade, bringing the monkeypox outbreak during the year 2003 in nations like United Kingdom, Israel, and Singapore, is blamed for the present outbreak of monkeypox.7,8 More than 82000 monkeypox cases with laboratory confirmations were recorded between 1st January 2022 and December 2022 in 110 countries and territories with 65 deaths.9-12 The virus is regarded as a possible biological weapon due to its ability to spread quickly among humans.13 Monkeypox virus is a zoonotic Poxviridae family Orthopoxvirus belonging to Chordopoxviridae subfamily and is related to the viruses that cause cowpox, mousepox, and smallpox.14 Notably, with almost 200 different genes, Orthopoxvirus genomes are among the biggest known ones for animal viruses.15,16 As a result, many researches have concentrated on the genomic, proteomics, structural, and morphogenetic organization of the poxviruses.17 Poxviruses comprising of double-stranded DNA result in infections that multiply in the cytoplasm of the host cell.18 The terminal sections of the monkeypox genome encode virulence and host range factors, while the core portion encodes structural proteins and necessary enzymes. The genome of the smallpox-causing variola virus shares 96.3% similarities with that of the monkeypox virus.19 The proteomes of the monkeypox and vaccinia viruses also have a lot in common. The vaccinia virus serves as a model poxvirus in experimental conditions and is utilised in the smallpox vaccine.17 According to reports, immunization against smallpox were also reported to be potentially effective against monkeypox as well with about 85% of efficacy. 20 Smallpox immunisation programmes have been discontinued after the disease was proclaimed eradicated in 1980. Although it is advised to use smallpox medications against monkeypox,21 their safety and effectiveness in human beings has not yet been determined.22 JYNNEOS and ACAM2000 are the only two vaccines currently recommended and approved for human use by the USFDA.9,23 The increase in the hospitalisation rate among patients with monkeypox indicates that appropriate therapy is essential that too within a short period of time.24 Therefore, the present resurgence of monkeypox shows that medications and therapies tailored specifically for this disease are urgently needed. The process of discovery and testing of new drugs typically take ten years, since they require numerous steps, including the identification of early lead, animal studies, and clinical trials.
Therefore, in the context of an epidemic, drug repurposing is a tempting choice. One advantage of the drug repurposing strategy is the substantial reduction in testing time. Particularly, medications that were once licenced for use against different illnesses have undergone comprehensive toxicity testing and can therefore be given to the general public without risk.25 Drug repurposing research may be essential to combating the disease as reported cases of monkeypox rise globally. In order to forecast authorised medications that can be utilised against monkeypox, we offer an in silico molecular docking study in this direction. We focussed on the mpxvgp158 protein from the monkeypox virus that is known to be crucial for the viral replication cycle. In a recent study, Abduljalil and Elfiky concluded that Norov-29 and bemnifosbuvir, two antiviral drugs, can bind to the active site of DNA dependent RNA polymerase (DdRp) and help in fighting against human monkeypox virus (HMP)26 demonstrated that eight drugs namely NMCT and rutaecarpine for A48R, fosdagrocorat and lixivaptan for I7L, simeprevir for D13L, nilotinib for A50R, and hypericin and naldemedine for F13L can be potential targets for the inhibition of HMP so that fatalities can be decreased.27 D9 decapping enzyme and thymidylate kinase (TMPK) enzymes of viruses may also make potent antiviral drugs (Doxorubicin, Cefiderocol, Tipranavir, and Dolutegravir) targets, according to earlier research. Drug development pipelines now include computational tools that allow screening of libraries of small molecules in order to identify lead compounds that can be further refined to create candidate medications for clinical trials.28 Researchers recently showed that Tecovirimat’s efficiency against monkeyviruses was due to its prospective binding pocket and potential binding mode with F13, a phospholipase D present in poxviruses.29 F13 made a significant contribution to the improvement and infectiousness of extracellular viruses. The effectiveness of Tecovirimat has been demonstrated both in vitro and animal studies.22 Therefore, in order to find possible inhibitors for the selected drug target, we docked the Tecovirimat drug against mpxvgp158.
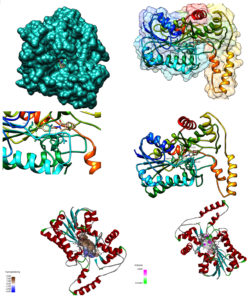
The design of a best probable ligand has been performed following the standard approach for designing of drug molecules based on the optimized structure using the Monkeypox virus protein MPXgp158 as the target. The target protein was aligned for similar proteins using BLAST search against the PDB database, revealing the best template for our target with structural similarities. Based upon the best similar PDB structures, ITASSER was used for the homology modeling.30 PROCHECK and PROSA were used for the validation of the structures which were designed. Active site searches were established by the literature support. To find out the investigation site for our study, we have selected Tecovirimat as a ligand molecule which has been reported as the first antipoxviral drug approved for MPX disease in US, having an inhibitory effect on MPX.31,32 The same ligand has been explored through PubChem. In the present study, we performed docking using AutoDock (parameter version 4.2), in which preparation of the protein and ligands PDBQT files along with the creation of grid-box was accomplished using GUI AutoDock Tools.33 The prepared file in PDBQT format was saved using AutoGrid, and the grid box was created with Grid Point Spacing of 0.375 Ao with an even number of Grid Points. The coordinates of the central grid point of maps were recorded as 65.756, 65.966, 65.707, respectively. In the docking process, both ligand/protein are considered rigid (Figure 1). Lamarckian Genetic algorithm was used to perform Docking studies along with Local Search default parameters with ΔG values refering to the significant binding among the developed ligands.34
The complex generated with the lowest binding affinity was extracted for MD simulation process. The top docked complex of Mpox-tecovirimat, which was showing the highest binding affinity potential, was further subjected to MD simulations using the GROMACS 2020.2 package in order to study the structural deviations in a dynamic environment for a time scale of 100 nanoseconds. The MD simulations were subsequently examined based on various parameters including energy of interaction, solvation free energy (DGsolv), root mean square fluctuation (RMSF), solvent accessible surface area (SASA), angle distribution, Radius of gyration (Rg), and root mean square deviation (RMSD) values as a function of time.
The compound Tecovirimat is an antiviral drug which has been reported to have a promising activity against orthopoxviruses. In order to combat the growing threat of zoonotic diseases, further research into pharmacopoeia is urgently needed. Among other poxviruses, monkeypox is now posing a threat to human survivability. Tecovirimat is more effective and safer than other licenced medications like cidofovir and brincidofovir since cidofovir has dose-limiting nephrotoxicity and brincidofovir has gastrointestinal and hepatocellular toxicity. As opposed to the other medications, Tecovirimat has better overall tolerability.22,35,36 Tecovirimat, however, is not approved for use during pregnancy.37 Few recent studies demonstrated the effectiveness of Tecovirimat in treating monkeypox patients. The monkeypox virus was detected in a 37-year-old immunosuppressed male who received 600 mg of Tecovirimat twice daily for a 2-week treatment along with doxycycline, ceftriaxone, and valacyclovir. The results showed no discernible harmful effects, and the skin flaws vanished right away.38 The safety of oral Tecovirimat doses during monkeypox infection was recently evaluated by one study, and it revealed minimal side effects, such as fatigue, headache, nausea, itching, and diarrhea.39 Only a few clinical trials on the use of Tecovirimat to treat monkeypox disease are available (NCT05534984, NCT02080767). Trials are still ongoing. In trials, Tecovirimat oral doses were given to patients based on their body weight. Confirmed monkeypox patients were given oral Tecovirimat twice daily for 14 days at the prescribed dose of 600 mg in three doses of 200mg each. As a result, lesions gradually disappeared and no deaths were noted. Each participant in the study completed the full course of Tecovirimat therapy regimen. This report has limitations due to the small number of enrolled patients.40 So, additional large-scale trials are required to determine Tecovirimat antiviral activity, dosage, and adverse effects. Even while animal studies can be convincing, human efficacy in later clinical trials is not always precisely correlated with efficacy seen in animals. Tecovirimat safety data could be gathered from people who have monkeypox rather than only healthy participants. Thus, human research with monkeypox are both necessary and feasible.
In our study, we have tried to explore the same compound for developing the best potential bioavailable inhibitor against MPXVgp158 protein. The highest alignment score revealed among the query and the database segments which is in agreement with earlier reports as well.41 Computational drug designing approach was used to develop MPXVgp158 protein inhibitor. With MPXVgp158 sequence as a query, sequence, BLAST search was carried out against protein database. ITASSER was used further to perform homology modeling of the subject and the template. This was followed by the use of PROCHECK and PROSA for the model evaluation. As a result, the C-Score, Z-Score, and G-Factor values of all the models were retrieved. Residual Plot score which is reflective of the quality of the local model was further assessed by generating a plot of energies as a function of the residue sequence position using PROSA. The model score values obtained bear consistency and are in agreement with the earlier studies.42,43 Model V5 was adjudged as the best model having 89.8% core value, with a goodness factor (G-factor) value of -0.79 (shown in Table 1)
Table (1):
ITASSER generated energy refined models.
S.No |
Model |
C-Score |
Core Value |
G-factors |
Z-Score |
|---|---|---|---|---|---|
1 |
V1 |
-1.43 |
80.0% |
-0.64 |
-8.7 |
2 |
V2 |
-2.10 |
75.8% |
-0.91 |
-9.32 |
3 |
V3 |
-3.96 |
68.1% |
-0.73 |
-9.53 |
4 |
V4 |
-4.26 |
67.1% |
-0.73 |
-9.35 |
5 |
V5 |
-5.00 |
89.8% |
-0.79 |
-12.48 |
The energy refined models of the query protein (MPXVgp158) were generated using ITASSER, which were ranked based on the size of the cluster. Significantly higher C Score values with better quality are further testimony to the finalized models which are in agreement with earlier reports.44
From literature support and in the docked complex, the following amino acids i.e. THR at 75, 116, 147, VAL 79, ASP 80, GLU 120, and LYS 151 positions, have been revealed as the best binding pocket of the target protein.
The seed molecule from PubChem “Tecovirimat” is capable enough of binding as well as inhibiting the MPOX virus protein. The selected lead molecule with the target protein has been docked using AutoDock4.2 (Figure 1). Ligand 51 displayed best binding affinity with a ΔG value of -5.91 kcal/mole (Table 2). The study is in agreement with previous reports as well.45
Table (2):
Bioinformatics analysis of the selected ligand molecules.
S.No |
Ligand Nomenclature |
Best Conformation |
ΔG (kcal/mol) |
RMSD Vales |
Inhibition Constant (millimolar) |
|---|---|---|---|---|---|
1 |
Lig15 |
9 |
-5.01 |
4.58 |
428.48 |
2 |
Lig28 |
6 |
-4.94 |
4.66 |
392.42 |
3 |
Lig33 |
8 |
-4.77 |
2.92 |
401.24 |
4 |
Lig38 |
6 |
-4.27 |
2.92 |
348.77 |
5 |
Lig51 |
9 |
-5.91 |
4.7 |
474. 80 |
MD simulation
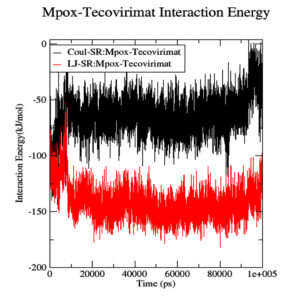
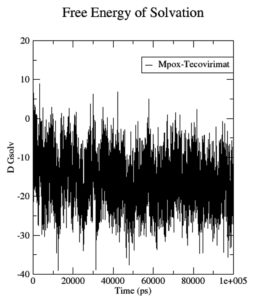
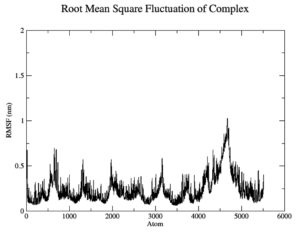
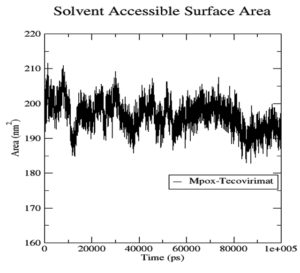
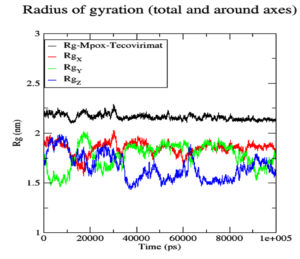
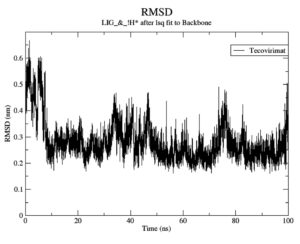
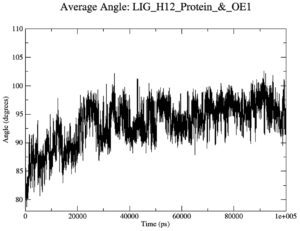
Although the Mpoxgp158-Tecovirimat complex fluctuated at the beginning of the simulation based on the average Coulomb’s short-range (Coul-SR) values but became stable after ten nanoseconds with an average value of -63.58 KJ/mol (Figure 2). The average Lennard-Jones short-range (LJ-SR) value was reportedly higher at the beginning but remained static after 10 ns with an average value of -141.93 KJ/mol.46 The solvation-free energy of the complex was reported to be static with an average value of -15 DGsolv (Figure 3). The RMSF values of the Mpoxgp158-Tecovirimat complex were calculated to understand the local fluctuations taking place for assessing the flexibility of the atoms. The RMSF values for the complex remain under 0.5 nm (Figure 4). Theoretically, the solvent-accessible surface area (SASA) is indicative of the solvent accessibility of the protein. Our results are quite consistent with other studies as well.47 Throughout the simulations, SASA fluctuated around 180 nm2. SASA value for the Mpoxgp158-Tecovirimat complex remained low in the middle of simulations (Figure 5). Based on the above observations, we can conclude that the ligand, Tecovirimat interacts well with the MPXVgp158 protein. Upon analysis of the plot of the radius of gyration (Rg) which was spanning over 100 nanoseconds, the data was indicative of the compactness of the protein during MD simulations. Throughout simulations, the radius of gyration for the Mpoxgp158-Tecovirimat complex remained stable, having a value of 2.25 nm (Figure 6). The ligands’ root mean square deviation (RMSD) was also recorded in order to observe ligand stability during simulations. The RMSD value for Tecovirimat remained higher till eight nanoseconds but afterwards remained stable, with a value of 0.3 nm, indicating the stability of Tecovirimat during MD simulations (Figure 7). The RMSD values observed are quite in agreement with earlier studies in the literature.48 The angle of Tecovirimat in the binding pocket of Mpox protein was also recorded and found stable with an average value of 93.41°, indicating the stability of Tecovirimat in the binding pocket of Mpox protein (Figure 8).
In the present study, we have molecularly docked Tecovirimat with Mpox protein. Computational-based drug designing method is both time and cost-effective to select compounds as the best probable drugs for further studies. Tecovirimat has shown the highest binding energy. Tecovirimat was found to have lesser binding energy, higher non-bonded interaction capability, and more stability against Mpox protein, with good binding mode of interactions. Hence, Tecovirimat was adjudged to be the potential candidate against MPXVgp158 inhibition. However, there is a need to further analyze the target for further investigations so as to reduce fatalities caused by the Monkeypox virus.
ACKNOWLEDGMENTS
The authors would like to thank Maharishi Markandeshwar (Deemed to be University), Mullana (Ambala) Haryana for providing the requisite platform to carry out this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Huhn GD, Bauer AM, Yorita K, et al. Clinical characteristics of human monkeypox, and risk factors for severe disease. Clinical infectious Diseases. 2005;41(12):1742-1751.
Crossref - Ly-Yang F, Miranda-Sánchez A, Burgos-Blasco B, Fernández-Vigo JI, Gegúndez-Fernández JA, Díaz-Valle D. Conjunctivitis in an Individual With Monkeypox. JAMA Ophthalmology. 140(10):1022-1024.
Crossref - Sepehrinezhad A, Ashayeri Ahmadabad R, Sahab-Negah S. Monkeypox virus from neurological complications to neuroinvasive properties: current status and future perspectives. Journal of Neurology. 2022:1-8.
Crossref - Ladnyj I, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bulletin of the World Health Organization. 1972;46(5):593.
- Likos AM, Sammons SA, Olson VA, et al. A tale of two clades: monkeypox viruses. Journal of General Virology. 2005;86(10):2661-2672.
Crossref - Isidro J, Borges V, Pinto M, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nature Medicine. 28, 1569–1572.
Crossref - Forni D, Molteni C, Cagliani R, Sironi M. Geographic structuring and divergence time frame of monkeypox virus in the endemic region. The Journal of Infectious Diseases. 2022;
Crossref - Otu A, Ebenso B, Walley J, Barceló JM, Ochu CL. Global human monkeypox outbreak: atypical presentation demanding urgent public health action. The Lancet Microbe. 3(8); e554-e555;
Crossref - CDC, 2022, https://www.cdc.gov/poxvirus/monkeypox/vaccines/index.html.
- Zhao H, Wang W, Zhao L, et al. The First Imported Case of Monkeypox in the Mainland of China—Chongqing Municipality, China, September 16, 2022. China CDC Weekly. 2022;4(38):847-848.
- Chandran D, Dhama K,MK Aslam M, et al. Monkeypox: An Update on Current Knowledge and Research Advances. J Exp Biol Agric Sci. 2022;10(4):679-688.
Crossref - Gessain A, Nakoune E, Yazdan Y. Monkeypox. N Engl J Med. 2022;387(19):1783-1793.
Crossref - Lane HC, Montagne JL, Fauci AS. Bioterrorism: a clear and present danger. Nature Medicine. 2001;7(12):1271-1273.
Crossref - Parker S, Nuara A, Buller RML, Schultz DA. Human monkeypox: an emerging zoonotic disease. Future Medicine, 2007; 2(1).
Crossref - Harrison SC, Alberts B, Ehrenfeld E, et al. Discovery of antivirals against smallpox. Proceedings of the National Academy of Sciences. 2004;101(31):11178-11192
- Bauer D. A history of the discovery and clinical application of antiviral drugs. British Medical Bulletin. 1985;41(4):309-314
Crossref - Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31-124.
Crossref - Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol 5: a010199. 2013.
Crossref - Shchelkunov SN, Totmenin AV, Babkin IV, et al. Human monkeypox and smallpox viruses: genomic comparison. FEBS Letters. 2001;509(1):66-70.
Crossref - Fine P, Jezek Z, Grab B, Dixon H. The transmission potential of monkeypox virus in human populations. International Journal of Epidemiology. 1988;17(3):643-650.
Crossref - Belongia EA, Naleway AL. Smallpox vaccine: the good, the bad, and the ugly. Clinical Medicine & Research. 2003;1(2):87-92.
Crossref - Sherwat A, Brooks JT, Birnkrant D, Kim P. Tecovirimat and the treatment of monkeypox—past, present, and future considerations. New England Journal of Medicine. 2022;387(7):579-581.
Crossref - Chakraborty S, Mohapatra RK, Chandra D, et al. Monkeypox vaccines and vaccination strategies: Current knowledge and advances. An update- Correspondence. Int J Surg. 2022;105:106869-106869.
Crossref - Moschese D, Giacomelli A, Beltrami M, et al. Hospitalisation for monkeypox in Milan, Italy. Travel Medicine and Infectious Disease. 2022;49:102417.
Crossref - Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nature Reviews Drug Discovery. 2019;18(1):41-58.
Crossref - Abduljalil JM, Elfiky AA. Repurposing antiviral drugs against the human monkeypox virus DNA-dependent RNA polymerase; in silico perspective. Journal of Infection. 2022; 85 (6),702-769.
Crossref - Lam HYI, Guan JS, Mu Y. In silico repurposed drugs against monkeypox virus. Molecules. 2022;27(16):5277.
Crossref - Malviya R, Sharma A. Applications of Computational Methods and Modeling in Drug Delivery. Machine Learning and Analytics in Healthcare Systems. CRC Press; 2021:163-190.
- Li D, Liu Y, Li K, Zhang L. Targeting F13 from monkeypox virus and variola virus by tecovirimat: molecular simulation analysis. Journal of Infection. 2022;85(4):e99-e101.
Crossref - Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):1-8.
Crossref - Rathjen NA, Shahbodaghi SD. Bioterrorism. American Family Physician. 2021;104(4):376-385.
- Al-Musa A, Chou J, LaBere B. The resurgence of a neglected orthopoxvirus: Immunologic and clinical aspects of monkeypox virus infections over the past six decades. Clinical Immunology. 2022:109108.
Crossref - Huey R, Morris GM. Using AutoDock 4 with AutoDocktools: a tutorial. The Scripps Research Institute, USA. 2008;8:54-56.
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. Journal of Molecular Recognition. 1996;9(1):1-5.
Crossref - Chittick G, Morrison M, Brundage T, Nichols WG. Short-term clinical safety profile of brincidofovir: A favorable benefit–risk proposition in the treatment of smallpox. Antiviral Research. 2017;143:269-277.
Crossref - Yu J, Raj SM. Efficacy of three key antiviral drugs used to treat orthopoxvirus infections: a systematic review. Global Biosecurity. 2019;1(1).
Crossref - Khalil A, Samara A, O’Brien P, Ladhani S. Call for a unified approach to Monkeypox infection in pregnancy: Lessons from the COVID-19 pandemic. Nature Communications. 2022;13(1):1-4.
Crossref - Hernandez LE, Jadoo A, Kirsner RS. Human monkeypox virus infection in an immunocompromised man: trial with tecovirimat. The Lancet. 2022;400(10355):e8.
Crossref - Desai AN, Thompson GR, Neumeister SM, Arutyunova AM, Trigg K, Cohen SH. Compassionate use of tecovirimat for the treatment of monkeypox infection. JAMA. 2022; 328(13):1348–1350.
Crossref - Mbrenga F, Nakoune E, Malaka C, et al. Monkeypox treatment with tecovirimat in the Central African Republic under an Expanded Access Programme. MedRxiv. 2022.
Crossref - Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Research. 2006;34(suppl_2):W6-W9.
Crossref - Laskowski R, MacArthur M, Thornton J. PROCHECK: validation of protein-structure coordinates. 2006; 25(2),722-725.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 2007;35(2):W407-W410.
Crossref - Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12(1):7-8.
Crossref - Morris GM, Huey R, Olson AJ. Using autodock for ligand-receptor docking. Current protocols in Bioinformatics. 2008;24(1):8.14. 1-8.14. 40.
Crossref - Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: long-range electrostatic effects. Annual Review of Biophysics and Biomolecular Structure. 1999;28(1):155-179.
- Chen H, Panagiotopoulos AZ. Molecular modeling of surfactant micellization using solvent-accessible surface area. Langmuir. 2019;35(6):2443-2450.
Crossref - Fukutani T, Miyazawa K, Iwata S, Satoh H. G-RMSD: Root mean square deviation based method for three-dimensional molecular similarity determination. Bulletin of the Chemical Society of Japan. 2021;94(2):655-665.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.