ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus is the leading cause of milk-borne disease in animals and humans worldwide, and it is often contaminated by enterotoxigenic and antimicrobial-resistant S. aureus strains. The current research work was intended to identify the prevalence of S. aureus from samples of bovine milk from various dairy farms and local vendors of Kamrup Metro District, Assam, India, by phenotypic and genotypic identification along with antibiotic resistance profiling. The conventional aseptic methods were implemented for S. aureus isolation from milk in Baird Parker Agar, supplemented with egg yolk and potassium tellurite. Further, the isolates confirmation was carried out using the automated VITEK system and amplification of the S. aureus specific nuc gene by PCR. Antibiotic susceptibility profiling for variety of 16 antibiotics was obtained through the conventional disc diffusion method. Eighty-five presumptive isolates with jet-black colonies with a white halo on Baird Parker Agar were selected. Thirty-eight isolates were eventually confirmed as S. aureus by the automated method and the detection of nuc gene. Antibiotic profiling revealed about 60.52% of the isolates to be multidrug resistant and 55.26a ± 0.01 mm resistant against Kanamycin. The statistical analysis data expressed correlation between Penicillin G and Ampicillin with 42.10b ± 0.01 mm and correlation among Tetracycline, Methicillin and Streptomycin with 10.52h ± 0.01 mm, respectively. Resistance against Kanamycin, Trimethoprim, Cloxacillin, and Nalidixic acid is concerning as such a resistant pattern has not been extensively reported in bovine milk samples in India, which could indicate the possible emergence of MDR S. aureus strains in the study area.
Bovine Milk Staphylococcus aureus, nuc Gene, Antimicrobial Resistance
Staphylococcus aureus, highly pathogenic gram-positive bacteria that leads to zoonotic diseases. It is considered to cause tissue infections and bacteremia in humans. In dairy animals, S. aureus is a predominant causative agent of mastitis, resulting in enormous monetary losses. In addition to being a frequent cause of mastitis in cows, likewise can might give rise to food poisoning in human. Due to its ability to produce toxins like enterotoxins, alpha-toxin, and other exoenzymes, which promote host cell invasion and spreading, as well as its multidrug resistance. As per WHO, S. aureus is a priority microorganism of global concerns that needs to be studied systematically, understands their antibiotic-resistant profiles and how to manage and minimize health hazards due to it. According to EFSA and the ECDC (2016), this is equivalent nearly half of all food-borne outbreaks linked to bacterial poisons in 2015. Approximately 240,000 instances of staphylococcal food poisoning are reported to the Centre’s for Disease Control each year in the United States, resulting in 1,000 hospitalizations and six fatalities.1
S. aureus produced Staphylococcal enterotoxins (SE) act on particular emetic receptors present in the intestinal wall, causing food poisoning. Enterotoxins are short extracellular proteins that dissolve in water. Based on their antigenicity, 23 unique SE has been recognized till date. S. aureus has the astounding ability to resist antimicrobial drugs and elude the human immunity. S. aureus potential to combat particular antimicrobial stressors is influenced by both intrinsic and acquired resistance. It may evolve resistance to numerous antimicrobial agents by conveying different resistance attributes on plasmids or transposons. It also possesses a number of intrinsic features that hinder the efficiency of antimicrobial agent. The propensity of S. aureus strains to produce food poisoning were investigated using molecular biology approaches. Several studies suggested that stressors, like the pH and temperature of milk, could affect the expression of genes that regulate the synthesis of enterotoxins. Methicillin-resistant S. aureus (MRSA) in particular is causing S. aureus infections, are emerging as a major public health problem. Ever since the first isolate of MRSA was identified in England in 1961, MRSA has emerged as a leading cause of nosocomial infections worldwide. The spike in S. aureus infection and antibiotic-resistant strains in low and middle-income countries are quite alarming and of great concern.1-7 Although antimicrobial therapy is crucial for combating mastitis; however, S. aureus has poor response to antimicrobial drug therapy.
Acquired antimicrobial resistance has the potential to be transmitted to humans. Overuse of antibiotics causes resistance through the introduction of foreign resistance genes or the emergence of point mutations that alter the antimicrobial target and trigger the antimicrobial to degrade or reduce the internal concentration of antimicrobials within the cell. Over 35,000 individuals in the USA are estimated to die while receiving treatment for resistant bacteria, out of the 2.8 million patients treated annually (CDC, 2019). Antimicrobial resistance and their pathogenicity in S. aureus have been researched in mastitis affected bovine milk with mastitis from the middle east China, Iran, Brazil, India and Turkey.2-4, 7-9
India ranked first position in the world for milk production, which is accounted for 196.18 million tonnes.10 In reference to Assam, as per to the report of National Dairy Development Board statistics, 2017, the dairy sector in Assam produces the per capita availability of milk is 69 g/day. According to the study of Lindahl et al., more than 90% of Guwahati dairy farms, rely primarily on milk production for their revenue. Hence, increasing in milk production would have a significant impact on farmer’s economies, in addition to increase consumer access to milk.11, 12 India is one of the world’s top consumers of antibiotics and has a high prevalence of infectious diseases. Studies has shown a high level of multidrug resistance in S. aureus and major drug-resistance along with major virulence gene that were detected in humans and livestock population in Assam.13,14 Geographically, North and East India have a higher hospital associated (HA)-MRSA form burden than West and South India, according to studies.13,14 Studies has shown milk in Assam had a greater percentage (29.41%) of livestock- MRSA and it was found that the MRSA prevalence in animal meat ranged from 28.6% to 48.57%. The study also revealed that prevalence of MRSA and S. aureus in Guwahati was significantly higher than the national average.14-16
In Assam, Kamrup Metro District is known as a major hub for the commercial production of milk. Few studies,14,17-19 have been reported from Assam and regardless of being the major milk production centre, Kamrup (M) district lacks sufficient studies. Hence, this study aims to scrutinizes the prevalence of antibiotic-resistant S. aureus in samples of bovine milk obtained from different dairy farms, local vendors, and commercial vendors in Kamrup (M) Districts, Assam, India, using both conventional and molecular approaches.
Sampling
Two hundred and twenty samples (approximately 10 mL) were obtained in sterile vials from different dairy farms, and local and commercial vendors of Kamrup (M) Districts (26.1701°N, 91.9490°E) of Assam, India, during the months between January-October 2023 (Figure 1). The study region was split into 5 major zones as per zones reported in www.mapsofindia.com and random sampling method was employed with the initial target of 50 samples from each zone. However, as it was observed availability of cattle farms were not uniform accordingly in few areas less than 50 samples were collected. So, 50 samples were collected from Panikhaiti (26.2006°N, 91.8757°E); 50 samples from Sonapur (26.1172°N, 91.9802°E); 47 samples from Jorabat (ASSAM) (26.0989°N, 91.8623°E); 30 samples from Rani (25.7905°N, 91.2626°E) and 43 samples Ulubari (26.1683°N, 91.7541°E). The samples were shipped to the laboratory in a chilled container and further processed for S. aureus isolation within 24 hours by employing standard procedures.
Figure 1. Primer locations for sample collections (location identified using Google Map application)
Isolation and preliminary screening using Automated VITEK system
With some minor modifications, the protocol outlined by Sharma et al. was employed to isolate S. aureus.20 Milk samples were enriched in Tryptic soy broth (TSB) (M011, HiMedia, Mumbai, India) by inoculating 1 mL sample with 9 mL sterile enrichment TSB and kept at 37°C for 24 hours. After 24 hours, a loopful of broth was streaked on Baird Parker Agar (BPA) (M043B, HiMedia, Mumbai, India), supplemented with egg yolk and potassium tellurite for screening the samples for the presence of Staphylococcus species and incubated for 48 hours at 37°C. The colonies with jet-black colonies with a white halo appearance were selected and maintained as pure culture for further validations of Staphylococcus aureus through Automated VITEK system (VITEK MS, Biomerieux), morphological and biochemical tests. The presumptive Staphylococcus isolates were validated by amplification of nuc gene with PCR.
Morphological and biochemical characterisation of the isolates
The presumptive positive S. aureus isolates confirmed through Automated VITEK system (VITEK MS, Biomerieux) were further analysed for morphological and biochemical characterisation. The morphological characterization was performed by Christian Gram’s staining method. The isolates were subjected to Sulphide, Indole and Motility test, Oxidase test, Coagulase test, Catalase test, Methyl-red test, and Voges-Proskauer test.
Molecular identification of isolates by detection of nuc gene
The Genomic DNA was extracted from freshly grown S. aureus using the Hot-cold lysis method. S. aureus specific nuc gene (279 bp) was screened by PCR from the isolates by using primers as – NUC-F: 5-’ F: GCGAT GATGGTGATACGGTT-3’ and NUC-R: 5’AGCCAAGCCTTGACGAACTAAAGC-3’ for coding a 279 bp nuc gene.21 The reaction was sought in a final volume of 20 µL consisting of a 10 µL PCR Master Mix composed of a thermostable DNA polymerase, dNTPs and MgCl2, 2 µL of DNA template and 10 pmol of each primer. The PCR cycling setups included an initial denaturation of 95°C for 3 min; followed by 35 cycles of denaturation 94°C for 30 sec, annealing 52.2°C for 30 sec, extension 72°C for 60 sec and final extension at 72°C for 8 min. PCR output were visualized on 1.5% agarose gel with the ethidium bromide dye on a UV transilluminator.
Antimicrobial resistance profiling of the isolates
The isolates that were confirmed by biochemical characterization and Automated VITEK system as S. aureus (n = 38) were subjected to antimicrobial susceptibility test by standard Kirby-Bauer method disc diffusion method on Muller-Hinton agar (M173, HiMedia, Mumbai, India). A 24 hours overnight broth culture of S. aureus isolates was standardized by diluting to 0.5 McFarland’s standard, and inoculated by spreading on the surface of prepared Mueller-Hinton agar plates. The antibiotics tested against the isolates were Streptomycin (10 mcg) (SD031, HiMedia, Nashik, India); Vancomycin (30 mcg) (SD045, HiMedia, Nashik, India); Penicillin-G (10 mcg) (SD028, HiMedia, Nashik, India); Trimethoprim (5 mcg) (SD039, HiMedia, Nashik, India); Methicillin (10 mcg) (SD136, HiMedia, Nashik, India); Ciprofloxacin (5 mcg) (SD060, HiMedia, Nashik, India); Cephalothin (30 mcg) (SD050, HiMedia, Nashik, India); Rifampicin (5mcg) (SD030, HiMedia, Nashik, India); Gentamycin (10 mcg) (SD016, HiMedia, Nashik, India); Nalidixic acid (30 mcg) (SD021, HiMedia, Nashik, India); Cloxacillin (20 mcg) (SD284, HiMedia, Nashik, India); Chloramphenicol (30 mcg) (SD0006, HiMedia, Nashik, India); Erythromycin (15 mcg) (SD013, HiMedia, Nashik, India); Ampicillin (10 mcg) (SD002, HiMedia, Nashik, India); Kanamycin (5 mcg) (SD223, HiMedia, Nashik, India); Tetracycline (30 mcg) (SD037, HiMedia, Nashik, India) was used to evaluate the susceptibility patterns of selected isolates. The inhibitory zones were taken as a standard norm as given in the CLSI, USA, to test pathogenic microorganisms that might lead to zoonotic diseases (As per Clinical and Laboratory Standards Institute, 2021).
Statistical analysis
The statistical evaluations were executed using Microsoft Excel and SPSS. The correlation was applied to ascertain the existence of any association between the antibiotic sensitivity patterns of the antibiotics. The level of significance of differences in positive S. aureus strains in terms of nucgene was determined (p < 0.05).
Isolation and preliminary screening using VITEK Automated VITEK system
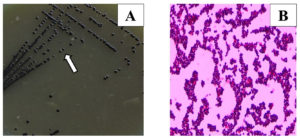
Two hundred and twenty samples of bovine milk were screened on Baird Parker Agar (BPA) (HiMedia), supplemented with potassium tellurite and egg yolk to detect the Staphylococcus species. From 220 samples, 350 isolates were initially cross-examined for confirmation of S. aureus via the Automated VITEK system (VITEK MS, Biomerieux). Out of 350 isolates, 38 were confirmed as positive S. aureus. Further, 38 positive S. aureus strains were screened for morphological and biochemical characterization and molecular identification (Figure 2; Table 1).
Figure 2. (A) Staphylococcus aureus in Baird Parker Media; (B) Gram’s staining of S. aureus (100X) grapes like gram-positive cocci
Table (1):
Morphological Characteristics of S. aureus from Bovine Milk Samples
| No. | Location of the Isolates | Isolated ID | Gram staining | Culture characteristics on selective media | Automated Identification of S. aureus by VITEK |
|---|---|---|---|---|---|
| 1 | Panikhaiti | P2 | Gram-positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
| 2 | P7 | Gram-positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 3 | P25 | Gram-positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 4 | P10 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 5 | P24 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 6 | P22 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 7 | P11 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 8 | P21 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 9 | Sonapur | S32 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
| 10 | S36 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 11 | S1 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 12 | S15 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 13 | S21 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 14 | S7BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 15 | Jorabat
(ASSAM) |
J15BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
| 16 | J21 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 17 | J4BNS | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 18 | J17 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 19 | J4BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 20 | J18BBL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 21 | J9BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 22 | J10BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 23 | J16BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 24 | J7 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 25 | J9 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 26 | J2BLB | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 27 | J13BL | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 28 | J5 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 29 | J4 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 30 | Rani | G5 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
| 31 | R1 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 32 | R7 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 33 | R12 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 34 | R13 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 35 | R18 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 36 | R24 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + | |
| 37 | Ulubari | U3 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
| 38 | U15 | Gram- positive cocci (in clusters) | BPA: Typical jet-black colonies surrounded by halo zone | + |
Morphological and biochemical characterization
There was no variation observed in the morphological and biochemical characterization in all the 38 S. aureus isolates. Gram’s staining showed the evidence of irregularly arranged gram-positive bunches of cocci mimicked to a bunch of grapes on all the 38 samples. Biochemical analysis revealed that 38 isolates yield positive for the catalase, coagulase, methyl-red, and Voges-Proskauer test, and were negative in Motility, H2S Production, Indole, and oxidase test (Figure 2; Table 1 and Table 2).
Table (2):
Biochemical Characterization of S. aureus isolated from Bovine milk
No. |
Isolates ID |
Motility test |
H2S Production test |
Indole test |
Coagulase test |
Catalase test |
Oxidase test |
Methyl-red test |
Voges-Proskauer test |
|---|---|---|---|---|---|---|---|---|---|
1 |
P2 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
2 |
P7 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
3 |
P25 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
4 |
P10 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
5 |
P24 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
6 |
P22 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
7 |
P11 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
8 |
P21 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
9 |
S32 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
10 |
S36 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
11 |
S1 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
12 |
S15 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
13 |
S21 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
14 |
S7BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
15 |
J15BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
16 |
J21 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
17 |
J4BNS |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
18 |
J17 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
19 |
J4BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
20 |
J18BBL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
21 |
J9BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
22 |
J10BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
23 |
J16BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
24 |
J7 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
25 |
J9 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
26 |
J2BLB |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
27 |
J13BL |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
28 |
J5 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
29 |
J4 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
30 |
G5 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
31 |
R1 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
32 |
R7 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
33 |
R12 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
34 |
R13 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
35 |
R18 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
36 |
R24 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
37 |
U3 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
38 |
U15 |
– |
– |
+ |
+ |
+ |
– |
+ |
+ |
Molecular identification of the isolates
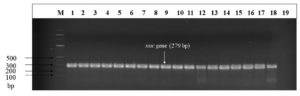
All the 38 isolates shortlisted for genomic detection of the nuc gene (S. aureus specific target gene) were positive at 279 bp visualized on 1.5% agarose gel subjected with ethidium bromide dye under a UV transilluminator (Figure 3). In the current investigation, no discernible variations were observed between the different positive S. aureus strains in terms of nuc gene (p < 0.05).
Figure 3. Agarose gel electrophoresis for visualization of the nuc (279 bp). M: molecular weight marker (100 bp); lanes 1-18 nuc amplicon; lane 19 negative control
Antibiotic resistance profiling
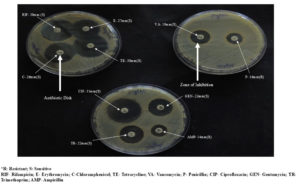
In the current research study, the following antimicrobial agents as Streptomycin (10 mcg); Vancomycin (30 mcg); Penicillin-G (10 mcg); Trimethoprim (5 mcg); Methicillin (10 mcg); Ciprofloxacin (5 mcg); Cephalothin (30 mcg); Rifampicin (5 mcg); Gentamycin (10 mcg); Nalidixic acid (30 mcg); Cloxacillin (20 mcg); Chloramphenicol (30 mcg); Erythromycin (15 mcg); Ampicillin (10 mcg); Kanamycin (5 mcg); Tetracycline (30 mcg) were used to check the antibiotic susceptibility patterns of 38 S. aureus isolates (Figure 4).
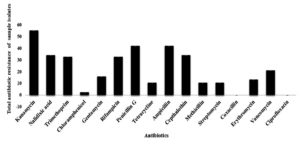
In the current research findings, the highest antibiotic resistance against the S. aureus isolates was exhibited by the Nalidixic acid (13.162 mm), followed by Kanamycin (14.202 mm), Trimethoprim (15.606 mm), Vancomycin (18.512 mm), Streptomycin (20.254 mm), Methicillin (23.452 mm), Ampicillin (25.096 mm), Penicillin (25.43 mm), Rifampicin (26.794 mm), Erythromycin (28.088 mm), Cephalothin (29.272 mm) and, Ciprofloxacin (31.382 mm) and the lowest resistance activity was exhibited by Cloxacillin (32.978 mm). The lowest zone of inhibition indicated the highest resistivity of the antibiotics. The values given along with the antibiotics represent the zone of inhibition expressed by the S. aureus isolates (Table 3).
Table (3):
Average zone of inhibition by antibiotics (mm) of the study area
| No. | Sample Area | Antibiotics (mcg) | Average of antibiotic activity area wise (%) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEP (30) | MET (10) | K (5) | S (10) | COX (200) | NA (30) | TR (5) | E (15) | C (30) | VA(30) | CIP (5) | GEN (10) | RIF (5) | P (10) | TE (30) | AMP (10) | ||||
| 1 | Panikhaiti | 35.75 | 33.12 | 0 | 23.87 | 41.25 | 17.75 | 25 | 32.37 | 30.25 | 22.5 | 37 | 22.37 | 31.37 | 29.87 | 34.87 | 30 | 27.959 | |
| 2 | Sonapur | 31.16 | 27.5 | 16.83 | 19.83 | 38.66 | 14.83 | 18.83 | 25.5 | 27 | 18.16 | 28.5 | 23.66 | 29.5 | 32 | 31.5 | 29.33 | 25.799 | |
| 3 | Rani | 34.42 | 25.71 | 19.85 | 18.14 | 34.42 | 17 | 15.14 | 29.14 | 27.14 | 18.14 | 30.85 | 25.28 | 27.57 | 32.28 | 27.71 | 29.42 | 25.763 | |
| 4 | Jorabat | 30.53 | 30.93 | 16.33 | 19.93 | 37.06 | 11.73 | 19.06 | 23.93 | 26.53 | 19.26 | 30.06 | 23.73 | 30.53 | 31 | 30.4 | 28.73 | 25.609 | |
| 5 | Ulubari | 14.5 | 0 | 18 | 19.5 | 13.5 | 4.5 | 0 | 29.5 | 24.5 | 14.5 | 30.5 | 27 | 15 | 2 | 22.5 | 8 | 15.219 | |
| Average zone of inhibition by antibiotics (mm) | |||||||||||||||||||
| 6 | 29.272 | 23.452 | 14.202 | 20.254 | 32.978 | 13.162 | 15.606 | 28.088 | 27.084 | 18.512 | 31.382 | 24.408 | 26.794 | 25.43 | 29.396 | 25.096 | |||
However, the total antibiotic-resistant activity against the S. aureus was revealed by the antibiotic Kanamycin with the value of 55.26a ± 0.01 mm, and the least resistance activity was expressed by the antibiotic Cloxacillin (0.00j ± 0.01 mm) and Ciprofloxacin (0.00j ± 0.01 mm). Variance analysis at the significance value of p ≤ 0.05 showed that there no significant variations in its activity between the antibiotics Penicillin (42.10b ± 0.01 mm) and Ampicillin (42.10b ± 0.01 mm; Nalidixic acid (34.21c ± 0.01 mm) and Cephalothin (34.21c ± 0.01 mm); Trimethoprim (32.75d ± 1.74 mm) and Rifampicin (32.89d ± 2.30 mm); Tetracycline (10.52h ± 0.01 mm), Methicillin (10.52h ± 0.01 mm) and Streptomycin (10.52h ± 0.01 mm) (Table 4; Figure 5).
Table (4):
Overall antibiotic-resistant activity of the isolates (mm)
No. |
Antibiotics |
Total Antibiotic-Resistant activity/Pattern of the isolates (mm) |
|---|---|---|
1 |
Kanamycin |
55.26a ± 0.01 |
2 |
Nalidixic acid |
34.21c ± 0.01 |
3 |
Trimethoprim |
32.75d ± 1.74 |
4 |
Chloramphenicol |
2.36i ± 0.01 |
5 |
Gentamycin |
15.78f ± 0.01 |
6 |
Rifampicin |
32.89d ± 2.30 |
7 |
Penicillin G |
42.10b ± 0.01 |
8 |
Tetracycline |
10.52h ± 0.01 |
9 |
Ampicillin |
42.10b ± 0.01 |
10 |
Cephalothin |
34.21c ± 0.01 |
11 |
Methicillin |
10.52h ± 0.01 |
12 |
Streptomycin |
10.52h ± 0.01 |
13 |
Cloxacillin |
0.00j ± 0.01 |
14 |
Erythromycin |
13.15g ± 0.01 |
15 |
Vancomycin |
21.05e ± 0.01 |
16 |
Ciprofloxacin |
0.00j ± 0.01 |
CV=3.228 CD (0.05) = 1.2 |
Antimicrobial resistance is becoming a significant threat globally with the driving up of antibiotic-resistant organisms. Globally, antibiotic resistance shows no hints of diminishing, diseases associated microbes that were previously eradicated by antibiotics are revamping themselves because of antibiotic-resistant drugs.22 The origin of antibiotic resistance is multidimensional, and its consequences impact worldwide.23 The mainspring of antimicrobial resistance includes poverty, filthy conditions, improper use of antibiotics, overseas travel, and increasing healthcare services.24 Organizations, like the WHO, CDC, the World Economic Forum, and the Infectious Diseases Society of America have declared antibiotic resistance to be “global public health concern”.23 The WHO stated, the pathogenic S. aureus exhibits a massive threat to human health globally.25 Infections triggered by antibiotic-resistant S. aureus can lead to numerous clinical disorders, not included in Global Burden of Disease (GBD) estimates to date. The main cause of mortality and morbidity by antibiotic-resistant S. aureus has become notoriously tricky to define and has encountered definitional alterations leading to great concern for public health.24, 26
India’s state of Assam is well-known for its contrasting warm and cool weather with significant humidity. Assam is located amid the “pre-humid” and “humid” climate zones, according to Thornthwaite’s categorization. Assam endures a high annual humidity (80-85) % and temperature variations ranging from 8°C-10°C in the winter to up to 36°C in the summer. Overall, this area has a humid mesothermal Gangetic climate (Koppen classification), with the exception of the high mountain barriers. Sweating and hydration of the stratum corneum of the skin is stimulated by high temperature and relative humidity. Particularly during the summer, both of these attributes render the ideal habitat for S. aureus to thrive on the epidermis. A higher frequency of MRSA and Staphylococcus aureus infections was proven to be correlated with high humidity.14,27,28
Assam, the second most enormous state in India’s North Eastern (NE) region, is connected to all the six other NE States and also to neighbouring countries like Bhutan and Bangladesh. The city Guwahati, which is regarded as the gateway of NE states falls within the Kamrup (M) district. Through this gateway, in a day-to-day life bulk of trade and commerce takes place, where milk serve as the commodity that is traded across the different regions for consumption. The present study, therefore, aims to grasp the prevalence scenario of antibiotic-resistant S. aureus from samples of bovine milk obtained from different dairy ranches, local and commercial vendors of Kamrup (M) Districts, Assam, India, as it is considered as a busiest commercial hub for handling out trades and services with the other NE states, and nearby countries. The unnoticed consumption might lead to global health challenges (antibiotic resistance) in humans, from any kind of endemic breakthrough to epidemic, leading to pandemic.
Therefore, in this study, the isolation, identification and antibiotic susceptibility characterization of S. aureus from bovine milk picked up from different dairy farms and local vendors of Kamrup Metro Districts, Assam, has been elaborated. The present research analysis witnessed the S. aureus prevalence to be 17.27% (38/220) in the samples accumulated from different dairy farms and local vendors of Kamrup Metro Districts of Assam, India based on conventional phenotypic methods including colony morphology, Gram’s staining, biochemical characteristics and Vitek automated system. 38 isolates shortlisted for S. aureus genomic identification by the nuc gene were positive. The heat-resistant nuclease (nuc) gene function as a specific marker, tightly associated with the synthesis of enterotoxin in S. aureus.29 Staphylococcal toxins are very resistant to heat. Enterotoxins continues to exist some biological activity in milk even after pasteurization or heating at 121°C for 28 min.30
Studies have reported that antibiotics such as Ampicillin, Amoxicillin, Azithromycin, Amikacin, Cefotaxime, Nalidixic acid, Gatifloxacin, Tetracycline, Chloramphenicol, Penicillin G, Oxacillin, Clavulanic acid, Methicillin, Erythromycin, Gentamicin, Ciprofloxacin, Clindamycin, Chloramphenicol, Trimethoprim/sulfomethoxazole, Ceftaroline, Cefoxitin, Rifampicin, Lincomycin, Minocycline, Ofloxacin, Kanamycin, Amoxycillin+ Sulbactam, Streptomycin, Cotrimoxazole, Sulphafurazole, Vancomycin, Cloxacillin, Ceftriaxone were used to administer against the S. aureus infection in animal.20,31-34
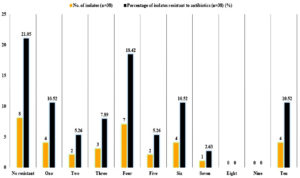
In the current study, variance analysis of the total antibiotic resistant activity of the isolates revealed that the antibiotic Kanamycin was able to inhibit the isolates at the highest concentration at 55.26a ± 0.01 mm, followed by Penicillin G and Ampicillin at 42.10b ± 0.01 mm, followed by Nalidixic acid and Cepthalothin at 34.21c ± 0.01 mm, followed by Rifampicin at 32.89d ± 2.30 mm and Trimethoprim at 32.75d ± 1.74 mm, Vancomycin at 21.05e ± 0.01 mm followed by Gentamycin at 15.78f ± 0.01 mm, followed by Erythromycin 13.15g ± 0.01 mm, followed by Chloramphenicol at 2.36i ± 0.01 mm and least concentration of Coxacillinand Ciprofloxacin at 0.00j ± 0.01 mm and lowest concentration at Tetracycline, Methicillin, Streptomycin at 10.52h ± 0.01 mm, (Figure 4). Of 38 positive isolates of S. aureus, 23 (60.52%) isolates showed resistant towards two or more antibiotics (Figure 6).
Antimicrobial resistance patterns to Penicillin, Ampicillin, Trimethoprim and Vancomycin were found in studies reported from cow’s milk in the Hawassa area, South Ethiopia.35 Akindolire et al., revealed resistance against Penicillin G, Ampicillin, Vancomycin, Gentamicin, Kanamycin, and Methicillin from milk samples aggregated from retail outlets of the North-West Province, South Africa.36 Sudhanthiramani et al., revealed resistance against Penicillin, Ampicillin, Methicillin, Cephalothin, Tetracycline and Gentamicin from milk samples acquired from the local vendors of Tirupathi region, India.37 Antibiotic resistance towards Nalidixic acid, Cloxacillin, Erythromycin, Kanamycin and Vancomycin collected from various places in the Meerut region and animal husbandry department dairy farms of Babugarh cantt (India).20
The isolates from Panikhaiti showed antibiotic resistance to Kanamycin, Gentamicin, Rifampicin, Penicillin G and Ampicillin. Those from Sonapur showed resistance to Vancomycin, Cephalothin, Kanamycin, Penicillin G, Ampicillin and Rifampicin. Isolates from Jorabat (Assam) showed resistance towards Cephalothin, Kanamycin, Nalidixic acid, Trimethoprim, Erythromycin, Rifampicin, Tetracycline, Penicillin G and Ampicillin. Isolates from Rani showed resistance to Cephalothin, Methicillin, Kanamycin, Cloxacillin, Nalidixic acid, Trimethoprim, Vancomycin, Rifampicin, Penicillin G and Ampicillin. Isolates from Ulubari were found to be resistant towards Cephalothin, Methicillin, Nalidixic acid, Trimethoprim, Oxacillin, Vancomycin, Rifampicin, Penicillin G and Ampicillin.
Overall, in all the study areas, resistance against Kanamycin was widely prevalent. Resistance against penicillin G and Ampicillin were prevalent in four out of five study areas. Vancomycin resistance was predominant in the Sonapur, Rani, and Ulubari areas and Methicillin resistance was found only in Rani and Ulubari. Rifampicin and Nalidixic acid were prominent in three out of five studies. Trimethoprim was found prevalent in three out of five study areas. Resistance towards Penicillin G and Ampicillin has been commonly reported worldwide against S. aureus from samples of bovine milk.20, 35-37
Insufficient studies have been reported from India showing antibiotic resistance towards Kanamycin, Nalidixic acid, Rifampicin, trimethoprim and Coxacillin particularly from bovine milk samples. Resistance towards Kanamycin, Nalidixic acid, and Cloxacillin has been only reported from Meerut (India).20 S. aureus resistance towards Trimethoprim and Rifampicin has not been reported from India, particularly from bovine milk samples previously. However, antibiotic resistance towards these antibiotics has been reported from South Ethiopia35 and South Africa.36
In the current study penicillin and ampicillin expressed highest correlation with 42.10b ± 0.01 mm. These antibiotics belongs to the b-lactam antibiotic class which effectively hinders the catalytic activity of bacterial transpeptidases. Based on the primary sequence analog and catalytic mechanism, the enzymes are categorized into 4 classes namely Penicillin, Cephalosporins, Carbapenems and Monobactams. Penicillin exhibit resistance against enzyme Class B and ampicillin shows resistance against Class C of b-lactamase, however in the present statistical correlation it was noted that penicillin and ampicillin showed the highest level of correlation at 42.10b ± 0.01 mm. Pajohesh et al. reported the existence of correlation between antibiotics penicillin and ampicillin from bovine milk. It indicates the S. aureus isolates are able to adopt such pathways of resistance which can help the bacteria resistant against two or more antibiotics.38
Another notable finding of the study is there was correlation among tetracycline, streptomycin and methicillin with a correlation value of 10.52h ± 0.01 mm. Tetracycline belongs to the broad-spectrum antibiotics while the streptomycin belongs to aminoglycoside antibiotic whereas, methicillin belongs to the narrow-spectrum b-lactam antibiotic of the penicillin class. Tetracycline and streptomycin (Aminoglycoside) are known to confer resistance through different forms of pathway mechanisms including efflux pump, enzymatic modification and target site modification via chromosomal mutation or an enzyme whereas methicillin confer resistance through enzymatic modification. The common mechanism of conferring resistance among these three antibiotics could be through enzymatic modification. Hence it might be possible the bacteria may adopt a strategy in such a fashion that it modifies the common enzyme targets that can bestow resistance contrary to these three antibiotics at the same time.
Panikhaiti, one of commercial milk producing and distribution areas of Kamrup (M), has the the highest prevalence antibiotic resistant S. aureus which could be due to existence of various industries such as mining industries, refinery, brick manufacturing industry, automobile industry, food processing industries which involve the transportation of goods and other raw material from other states, possibly bringing along with it antibiotic resistant strains which along with other hazardous pollutants makes it way to soil, water and grazing land of the area on which cattle rearers are dependent upon enhancing the chances of passing of antibiotic resistant strains to bovine body and milk. The areas is one of the hotspot areas for picnic, hence migration of people from the commercial area and other adjoining area further elevates the chances of prevalence of antibiotic resistant strains. Sonapur is at the junction of two cities of Guwahati and Nagaon, a common transit area for goods and animals moving to upper Assam from other states, hence antibiotic-resistant strains might as well contaminate the soil and water sample in such transit zones. Likewise, Jorabat is another junction area between Assam and Meghalaya, which in turn allow the commercial movement of animals and goods from other states as well as from Indo Bangladesh Border through Dawki. Rani located adjacent to the airport zone and its state border with Meghalaya (Tura); the location might contribute to the existence of antibiotic resistance as in this area, transboundary trafficking of the animals is common and human movement is also prevalent. Reports suggest such human activities among cross border animal transportation could be one of the leading causes of antibiotic-resistant strains spreading widely.39
Further, animals in such border/transit zone could be prone to higher level of infection ultimately causing cattle rearers to use high amount of antibiotics to inhibit bacterial growth promotion and disease prevention without proper diagnosis, thus aggravating the situation as there is always high number of antibiotic residues in the body of animals. Further severity of antibiotic resistance S. aureus could be directly correlated with the unhygienic condition of maintaining of the animals, along with the improper mode of collection of the milk. The existence of antibiotic resistance S. aureus also indicates the sub-clinical level of S. aureus that may lead to developing infection and may eventually spoil the milk samples. The subclinical level of S. aureus might also indicate the administration of antimicrobial agents, which is being used to control the bacterial count to prevent infection.
The present research work could be one of first detailed reports of prevalence of antibiotic-resistant strains from the milk samples of Assam and how the cross boundary inter-state as well as international areas is widely prone to antibiotic resistant strains. Further, the study also discloses that the bacteria adopt the resistance strategy in such a frame that it is able to exhibit inhibition against three antibiotics by manoeuvring a single pathway. Such emerging trend of adopting resistance mechanisms against antibiotics by bacteria through modification of common genes, proteins and/or pathways, may eventually result in the inheritance of deadly antibiotic resistance patterns in S. aureus.
S. aureus was resistant to multiple antibiotic classes which might end up to severe health conditions. The current research work discloses that cattle breeding and trash management face critical role in the dispersion of antibiotic-resistant pathogens from animals to humans and the environment. Production of raw milk of high compositional and hygienic quality by farmers is crucial to milk processing business entities, milk consumers and the farmers themselves. For consumers, consumption of milk tainted with pathogenic bacteria might rise to diseases. The resistant strains can infect the human population through inappropriate farming practices and negligent handling of the milk equipment during milking which can be the reason for infection in human beings. The antibiotic-resistant microorganisms have emerged as a consequence of overuse of antibiotics during farm practices. Implementing of stringent biosecurity norms on dairy ranches will enhance sanitation and well-being, while overseeing antibiotic utilization may limit the threats to human through restricting antibiotic remnants in the products. Considering the present findings of this study, further molecular typing of these antibiotic-resistant S. aureus along with phylogenetic analysis with other S. aureus strains of South east Asian countries would give us an idea regarding origin of antibiotic-resistant pattern.
ACKNOWLEDGMENTS
The authors are greatly thankful to the Department of Biosciences, Assam Don Bosco University, Assam, India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Grispoldi L, Massetti L, Sechi P, et al. Characterization of enterotoxin-producing Staphylococcus aureus isolated from mastitic cows. J Dairy Sci. 2019;102(2):1059-1065.
Crossref - Vaez H, Ghazi-Saeidi K, Moradi A, et al. Antibiotic resistance pattern of methicillin resistant S. aureus isolated from Health-educational centers of Gorgan, Iran, 2008-2009. Iran J Med Microbiol. 2010;3(4):31-36.
- Lan XY, Zhao SG, Zheng N, et al. Microbiological quality of raw cow milk and its association with herd management practices in Northern China. J Dairy Sci. 2017;100(6):4294-4299.
Crossref - Khan A, Firyal S, Khan I, et al. Phenotypic and genotypic characterization of beta-lactams resistant Staphylococcus aureus isolates from bovine mastitis and its zoonotic implications. Pak Vet J. 2020;40(4):523-526.
Crossref - Timsina R, Shrestha U, Singh A, Timalsina B. Inducible clindamycin resistance and erm genes in Staphylococcus aureus in school children in Kathmandu, Nepal. Future Sci OA. 2020;7(1):FSO361.
Crossref - Houri H, Samadpanah M, Tayebi Z, Norouzzadeh R, Malekabad ES, Dadashi AR. Investigating the toxin profiles and clinically relevant antibiotic resistance genes among Staphylococcus aureus isolates using multiplex-PCR assay in Tehran, Iran. Gene Reports. 2020;19:100660.
Crossref - Mahros MA, Abd-Elghany SM, Sallam KI. Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health concern. Int J Food Microbiol. 2021;346:109165.
Crossref - Akanbi OE, Njom HA, Fri J, Otigbu AC, Clarke, AM. Antimicrobial Susceptibility of Staphylococcus aureus Isolated from Recreational Waters and Beach Sand in Eastern Cape Province of South Africa. Int J Environ Res Public Health. 2017;14(9):1001.
Crossref - Bakheet AA, Amen O, Habaty SHAL, Darwish SF. Prevalence of Staphylococcus aureus in broiler chickens with special reference to beta-lactam resistance genes in the isolated strains. Alex J Vet Sci. 2018;57(2):25-33.
Crossref - Mishra P, Matuka A, Abotaleb MSA, Weerasinghe WPMCN, Karakaya K, Das SS. Modeling and forecasting of milk production in the SAARC countries and China. Modeling Earth Systems and Environment. 2021;8:1-13.
Crossref - NDDB, 2017. National Dairy Development Board statistics [WWW Document]. http://www.nddb.org/information/stats/. (Accessed 7 March 2017)
- Lindahl JF, Deka RP, Melin D, et al. An inclusive and participatory approach to changing policies and practices for improved milk safety in Assam, northeast India. Global Food Security. 2018;17:9-13.
Crossref - Kaur J, Singh H, Sethi T. Emerging trends in antimicrobial resistance in bloodstream infections: multicentric longitudinal study in India (2017-2022). Lancet Reg Health-Southeast Asia. 2024:26:100412.
Crossref - Deka NK, Handique PJ, Borah P, et al. Antimicrobial Resistance and Major Virulence Gene Detection in Methicillin Resistant Staphylococcus aureus in Humans and Livestock Animals of Assam: A North Eastern State of India. J Pure Appl Microbiol. 2023;17(2):951-965.
Crossref - Das P, Mazumder PB. Prevalence of Staphylococcus in raw meat samples in Southern Assam, India. J Agric Vet Sci. 2016;9(1):23-29.
- Kumar N, Sharma G, Leahy E, et al. Understanding antibiotic usage on small-scale dairy farms in the Indian states of Assam and Haryana using a mixed-methods approach-Outcomes and challenges. Antibiotics. 2021;10(9):1124.
Crossref - Sharma I, Brinty A. Isolation and identification of Staphylococcus aureus from bovine mastitis milk and their drug resistance patterns in Silchar town dairy farms, NE India. Online International Interdisciplinary Research Journal. 2014;4:256-260.
- Mohantana A, Mazumder PB, Bhattacharjee A, Deb B. Identification of Enterotoxigenic coagulase negative Staphylococcus spp. isolated from bovine milk and milk products of southern Assam, India. J Pure Appl Microbiol. 2015;9(1):49-56.
- Kakati S, Talukdar A, Hazarika RA, et al. Bacteriological quality of raw milk marketed in and around Guwahati city, Assam, India. Vet World. 2021;14(3):656.
Crossref - Sharma D, Sharma PK, Malik A. Prevalence and antimicrobial susceptibility of drug resistant Staphylococcus aureus in raw milk of dairy cattle. Int Res J Microbiol. 2011;2(11):466-470.
- Badua AT, Boonyayatra S, Awaiwanont N, Gaban PBV, Mingala CN. Antibiotic resistance and genotyping of mecA-positive methicillin-resistant Staphylococcus aureus (MRSA) from milk and nasal carriage of dairy water buffaloes (Bubalus bubalis) in the Philippines. JAdv Vet Anim Res. 2020;7(3):397-406.
Crossref - Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10(12):S122-S129.
Crossref - Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645-1658.
Crossref - Dunachie SJ, Day NP, Dolecek C. The challenges of estimating the human global burden of disease of antimicrobial resistant bacteria. Curr Opin Microbiol. 2020;57:95-101.
Crossref - Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens. 2021;10(10):1310.
Crossref - Serwecinska L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water. 2020;12(12):3313.
Crossref - Singh VP, Sharma N, Ojha CSP. (Eds.). The Brahmaputra basin water resources (Vol. 47). Springer Science & Business Media. 2013.
- Mermel LA, Machan JT, Parenteau S. Seasonality of MRSA infections. PloS One. 2011;6(3):e17925.
Crossref - Karimzadeh R, Ghassab RK. Identification of nuc nuclease and sea enterotoxin genes in Staphylococcus aureus isolates from nasal mucosa of burn hospital staff: a cross-sectional study. New Microbes New Infect. 2022;47:100992.
Crossref - Yehia HM, Ismail EA, Hassan ZK, Al-Masoud AH, Al-Dagal MM. Heat resistance and presence of genes encoding staphylococcal enterotoxins evaluated by multiplex-PCR of Staphylococcus aureus isolated from pasteurized camel milk. Biosci Rep. 2019;39(11):BSR20191225.
Crossref - Bantawa K, Sah SN, Limbu DS, Subba P, Ghimire A. Antibiotic resistance patterns of Staphylococcus aureus, Escherichia coli, Salmonella, Shigella and Vibrio isolated from chicken, pork, buffalo and goat meat in eastern Nepal. BMC Res Notes. 2019;12(1):1-6.
Crossref - Pekana A, Green E. Antimicrobial resistance profiles of Staphylococcus aureus isolated from meat carcasses and bovine milk in abattoirs and dairy farms of the Eastern Cape, South Africa. Int J Environ Res Public Health. 2018;15(10):2223.
Crossref - Mashouf RY, Hosseini SM, Mousavi SM, Arabestani MR. Prevalence of enterotoxin genes and antibacterial susceptibility pattern of Staphylococcus aureus strains isolated from animal originated foods in West of Iran. Oman Med J. 2015;30(4):283.
Crossref - Hasanpour-Dehkordi A, Khaji L, Sakhaei-Shahreza MH, et al. One-year prevalence of antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from raw meat. Trop Biomed. 2017;34(2):396-404.
- Daka D, Gsilassie S, Yihdego D. Antibiotic-resistance Staphylococcus aureus isolated from cow’s milk in the Hawassa area, South Ethiopia. Annals of Clinical Microbiology and Antimicrobials. 2012;11:1-6.
Crossref - Akindolire MA, Babalola OO, Ateba CN. Detection of antibiotic resistant Staphylococcus aureus from milk: A public health implication. Int J Environ Res Public Health. 2015;12(9):10254-10275.
Crossref - Sudhanthiramani S, Swetha CS, Bharathy S. Prevalence of antibiotic-resistant Staphylococcus aureus from raw milk samples collected from the local vendors in the region of Tirupathi, India. Veterinary World. 2015;8(4):478.
Crossref - Pajohesh R, Tajbakhsh E, Momtaz H, Rahimi E Relationship between biofilm formation and antibiotic resistance and adherence genes in Staphylococcus aureus strains isolated from raw cow milk in Shahrekord, Iran. Int J Microbiol. 2022;(1):1-10.
Crossref - van Uhm D, South N, Wyatt T. Connections between trades and trafficking in wildlife and drugs. Trends Organ Crime. 2021;24(4):425-446.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.