ISSN: 0973-7510
E-ISSN: 2581-690X
One of the most important problems to the fish in aquaculture is pathogenic bacteria. Therefore is a serious necessity in aquaculture to upgrade microbial hegemony strategies. The present study focused on isolation, screening, biochemical and molecular level characterizations of potential probiotic bacteria from the fish pond in Basra city southern Iraq. Isolated bacteria were characterized based on their morphological and biochemical traits and were identified using automated instrument (Vitek II) and 16S rRNA gene sequencing. As a result, bacteria were identified as Pediococcus pentosaceus. In order to be used in aquaculture as probiotics, bacteria must have good probiotic properties such as the ability to live in the presence of bile salts and low pH. Hence, bacteria were incubated with different concentrations of bile salts and pH values for different periods of time. In addition to that, bacteria were subjected to additional tests, such as tolerance to survive in simulated human gastrointestinal tract conditions and antibiotic susceptibility. Results of all these tests indicated that Pediococcus pentosaceus has good probiotic properties. The pathogenic bacteria (Salmonella sp.) were identified using the automated instrument for bacterial identification (Vitek II). The antagonistic ability of P. pentosaceus toward the pathogenic bacteria (Salmonella sp.) was tested using the agar sawing method. The result indicated that P. pentosaceus has good antagonistic ability.
Pediococcus pentosaceus, Bile salt, pH, antagonistic activity.
Recently the production of fish in the world has been grown, according to reduce the outputting in fishing – with increasing the fish demanding at a mediocre yearly average of 3.2%, over stepping the growth of the inhabitance of the world to 1.6%. The individual consumption of fish in the world rose from 9.9 kg in the 1960s to 19.2 kg in 2012. Depending on the FAO’s last numeration, fish farming has grown to 90.4 million tons in the 2012, which comprise 66.6 million tons of fish and 23.8 million tons of aquatic algae1.
Fish deaths, especially those caused in the larval phase; resulting from both pathogenic and opportunistic bacteria, are among the major problems of fish farming in the present, leading to large economic losses. Despite the development of sterilization and sanitation techniques, food preparation, reprocessing and storage still carry the risk of food contamination by unwanted microorganisms. Environmental conditions and microbial water quality at fishing sites are the main reason for the presence of foodborne pathogens as well as the contamination of ponds with animal, human and agricultural waste2. The excessive use of antibiotics to solve this problem, especially those that are not biodegradable and characterized by long-term survival in the water has led to the spread and increase of antibiotic-resistant bacteria in the environment, an increase of antibiotic resistance in fish pathogens, transfer of these resistance determinants to bacteria and then land animals and finally become human pathogens along with alterations of the bacterial flora both in sediments and in the water column3. Concomitantly, probiotics have widely been suggested as eco-friendly alternatives to antibiotics. However, the way in which probiotics are applied in aquaculture is a key factor in their favorable performance. Probiotics as an alternative to chemicals and antibiotics have proven to be effective in promoting successful aquaculture, as they have the potential to improve water quality, increase tolerance to stress, generate high-quality livestock4.
Hence, the current study was designed to isolate Pediococcus pentosaceus from the fish pond in Basra city southern Iraq, and to evaluate its probiotic properties as well as their antimicrobial mode of action against fresh water fish pathogens, Salmonella sp.
Collection of samples
Nine water samples were collected from three Cyprinus carpio (Comman carp) fish ponds located in Marine Science Center- Basra University, Al-Garma campus (Fig. 1), with coordinates as represented in the Table (1), using 500 ml glass bottles, during April 2018. Samples were put in the icebox and transferred to the laboratory where they were saved in cooling (4°C) till be used. The feeding system applied in these ponds does not contain any bacterial additives. Physical and chemical properties like temperature, pH and salinity of water in the ponds during sampling were 26.4°C, 7.1 and 2.1 ms/cm respectively.
Table (1):
Coordinations of sampling stations.
Sampling stations |
Latitude |
Longitude |
|---|---|---|
Fish pond no.1 |
30°33’39.91″N |
47°44’28.34″E |
Fish pond no.2 |
30°33’35.37″N |
47°44’25.31″E |
Fish pond no.3 |
30°33’38.47″N |
47°44’30.20″E |
Isolation and enumeration of bacteria from water of the fish pond
A series of dilutions were performed under severe sterilization for all water samples collected, then 0.1 ml from each dilution were spread on the surface of de Mann Rogosa Sharpe agar medium (MRS, Hi media), and incubated aerobically at 37°C for 48h. The clear bacterial colonies were taken from the culture medium to produce pure culture.
Characterization of bacteria
Morphological and biochemical tests were carried out for diagnosis the probable type of bacterial colony from MRS agar5 and for accurate identification, Vitek II (Biomיrieux, USA) has been used.
Molecular level characterization: 16S rRNA gene sequencing
Extraction of the total genomic DNA of the bacteria was carried out using 2 ml samples of overnight cultures grown in MRS broth at 37°C, following the manufacturer’s instructions for Gram positive bacteria Presto™ Mini gDNA Bacteria (Geneaid Biotech Ltd., Taiwan). The extracted DNA was stored in sterilized vials at – 20°C until used as PCR templates.
PCR amplification of 16S rRNA gene
Extracted DNA was used to amplify the 16S rRNA gene. For PCR reaction, universal primers of 27 F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492 R (5′- GGTTACCTTGTTACGACTT-3′) have been used according to6.
The total volume of PCR reaction fraction was 50µl consisting on: 20 ng/µl as DNA concentration, 25µl of the Master Mix, 1µl of each primer (10 pmol). PCR process was done during 35 cycles and take 3 h at 25 min using a PCR device (Macrogen thermal Block, Bioneer, Korea). The PCR conditions were carried out according to7. The PCR product was analyzed by 1% agarose gel8 using electrophoresis apparatus (Fisher Scientific- USA) and 100 bp DNA marker (Gene Rule-Fermentas). The 16S rRNA sequence was performed by the macrogen factor (Korea).
Probiotic properties of isolates
Probiotic properties of thirty five isolates, such as survival in acid, tolerance against bile salts, and tolerance to gastrointestinal juices have been studied as follows.
Acid tolerance test
The acid tolerance test has been done using MRS broth with different pH (3, 5, 7, and 9), which prepared using HCl 1% (J. T. Baker) and NaOH 1 N (Hi media) in addition to control flask9, then autoclaved at 121°C for 15 min (triplicate has been used for each test). 0.1 ml from overnight cultured bacteria in MRS broth has been used to inoculate each flask, which were then incubated at 30°C. Optical density (OD) as the bacterial growth rate was measured using the spectrophotometer (Shimadzu, UV-1800, Japan) at wave length of 600 nm next 3 and 24 h of incubation.
Bile tolerance (Oxgall treatment)
For this test, MRS broth with different concentrations (0.0, 0.15, 0.25, and 0.5% (w/v)) of Oxgall bile salts (Difco) has been prepared. Then, each concentration was inoculated with 0.1 ml from overnight cultured bacteria and incubated at 30°C. The bacterial growth rate in each concentration has been measured using a spectrophotometer (Shimadzu, UV-1800, Japan) at the wave length of 600 nm after incubation at 37°C for 4 and 24 h. Triplicate has been used for each test.
Survival to simulated human gastrointestinal tract
To provide in vitro situation as those found in the digestive tract, the preparation of gastric and pancreatic juices was done as following: 3 mg.ml-1 from pepsin (Sigma) and 1 mg.ml-1 from pancreatin USP (Sigma- Aldrich) were dissolved in sterile solution of sodium chloride (0.5%, w/v). pH was adjusted to 3 and 8 using HCl (3 mol.L-1) and NaOH (1 mol.L-1). 0.2 ml of the overnight bacterial rinse in the saline buffer of phosphate (pH 7.0) were injected with 1.0 ml of simulated gastric or pancreatic juice and 0.3 ml NaCl (0.5%, w/v), then incubated at 37°C. Colony forming unit (CFU/ml) was counted after incubation for 180 min for gastric tolerance, and 240 min for basic pH tolerance. The initial viable count (CFU/ml) of the washed cell suspension from each probiotic tested was determined prior to the transit tolerance assay, and was used to calculate loss of viability10 using the following formula; Survival rate (%)=[Log CFU after treatment /Log CFU before treatment]׳100.
Antibacterial test
Fresh water fish pathogens, Salmonella sp. was used to study the antibacterial ability of probiotic bacteria, using the well diffusion techniques11. Shortly, 1 ml from overnight culture of Salmonella sp. in Tryptone soya broth (TSB, Hi media) was distributed through the sterile loop on Tryptone soya agar (TSA, Hi media). A probiotic suspension used as an antibacterial agent was the supernatant obtained from a 24 h bacterial culture after being centrifuged at 3000 rpm for 5 minutes.
Antibiotic susceptibility test
Antibiotic susceptibility was tested using Vitek II, in which antimicrobial sensitivity tests are performed in the same manner as antimicrobial dilution cards to determine the deposition point: minimum inhibitory concentration (MIC) against organisms.
Statistical analysis
Statistical analysis was carried out using one way analysis of variance (ANOVA) to compare means and significantly different means were separated using LSD; with post test if P<0.05 and using SPSS ver.10 software.
Isolation and identification of Pediococcus pentosaceus
During the current study, 18 isolates which gave positive reactions to Gram staining and negative reaction to catalase were selected for further characterization. The isolates have been identified as Pediococcus pentosaceus based on their phenotype, cultural characteristics and several biochemical tests. The colonies on MRS agar appeared as illustrated in (Table 2). And to emphasize the diagnosis, Vitek II has been used and gave the result with a confidence degree of 95%.
Table (2):
Morphological and biochemical properties of the bacteria.
| Colony Morphology | |
|---|---|
| Configuration | Round |
| Cell shape | Round |
| Motility | Non motile |
| Pigment White | Creamy |
| Gram reaction | + |
| Surface Mucoid | Mucoid |
| Biochemical Tests | |
| H2S formation | – |
| Catalase | – |
| Nitrate reduction | – |
| Urease | – |
| Indole Production | + |
| Methyl red test | + |
| V P Reaction | + |
| Citrate utilization | + |
| Acid Production from | |
| Glucose | + |
| Lactose | – |
Molecular level characterization: 16S rRNA sequencing
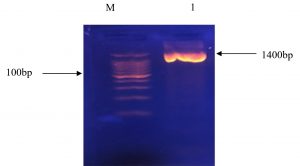
The bacteria were exposed to the RNA sequence analysis of the 16S rRNA. Approximately 1400 bp-band size of 16S rRNA gene was detected on the agarose gel as shown in Figure 2, as a result of the amplification. The 16S rRNA sequence was presented in the 16S ribosomal RNA sequence for Blast in NCBI Gene Bank website (www.ncbi.nlm.nih.gov/blast). The highest sequence similarity of the bacteria was 100%, and the accession number of the deposited sequence was NR-042058.1.
Fig. 2. Bands of agarose gel electrophoresis for 16S rRNA gene PCR product; M: 100 bp DNA marker, 1:the band size of the sample is about 1400 bp.
Acid tolerance
The tested isolates gave good results to selection probiotic criteria (pH test). From Table 3, the isolates have activity at pH 3 and 5 after 3 h of incubation and this activity decrease at 24 h of incubation. Meanwhile the activity of growth at pH 7 and 9 were greater after 24 h of incubation. The results of the statistical analysis showed that there were significant differences in the bacterial optical density among the studied pH values in the two studied time, and between the studied time, and at the significant level P ³ 0.05.
Table (3):
The effects of both pH and concentration of bile salts on the survival of Pediococcus pentosaceus, the number of bacteria in gastric and intestinal juices, and the antibiotic sensitivity pattern of various antibiotics.
Medium pH |
Optical density (600 nm) (3 h.) |
Optical density (600 nm) (24 h.) |
|---|---|---|
3 |
0.564 |
0.327 |
5 |
0.886 |
0.639 |
7 |
0.629 |
1.687 |
9 |
1.511 |
1.645 |
Bile salts % |
Optical density (600 nm) (3 h.) |
Optical density (600 nm)(24 h.) |
Zero |
1.686 |
1.557 |
0.15 |
0.125 |
0.239 |
0.25 |
0.132 |
0.398 |
0.5 |
0.161 |
0.568 |
Isolate |
Survival rate % in (pH 3) (240 min) |
Survival rate % in (pH 8) (180min) |
1 |
75 |
89 |
2 |
71 |
85 |
3 |
66 |
88 |
4 |
64 |
80 |
5 |
54 |
75 |
Interpretation |
MIC |
Antibiotic |
Interpretation |
MIC |
Antibiotic |
|---|---|---|---|---|---|
S |
≤0.12 |
Erythromycin |
I |
1 |
Benzylpencillin |
S |
≤0.25 |
Clindamycin |
I |
0.5 |
Ampicillin |
S |
≤2 |
Linezolid |
S |
0.5 |
Cefotaxime |
S |
0.25 |
Vancomycin |
S |
1 |
Ceftriaxone |
S |
0.5 |
Tetracycline |
S |
2 |
Levofloxacin |
R |
Inducible Clinadamycin |
Bile salt tolerance
The isolates were screened for their ability to tolerate the bile salts under different concentrations (0.15, 0.25 and 0.5%). Results (Table 3) show activity (P < 0.05) in all of the concentrations after 3 h incubation, and this increase with the time of incubation (24h).
Probiotic survival in gastrointestinal juices
The probiotic retention rate in both stomach and pancreatin juices was as shown in Table 3. Results show good survive rate in both juices (pH 3 and pH 8) during 24 h, with efficient survival rate in pH8 than in pH 3 with presence significant (P < 0.05) among the isolate.
Antibiotic susceptibility test
Table 3 shows the MIC values obtained for the different antibiotics tested for the studied bacteria. To determine whether an organism is sensitive, mild opponent or opponents to antimicrobials, the break value of MIC is adopted. From results we can conclude that the understudying bacteria were sensitive to the most tested antibiotics with different MIC values, whereas it was resistance to only inducible Clindamycin.
Extracellular antimicrobial activity
Fourteen (77.77%) of the 18 P. pentosaceus exhibited extracellular antimicrobial activity in their supernatants against the growth of Salmonella sp. The diameter’s range of the inhibition zone was within 22-30 mm.
The undesirable effects of bacterial diseases are a major concern for the aquaculture industry, especially, for economically viable species such as Cyprinus carpio. As a result of increased fish mortality and low yields, farmers have used chemotherapy and antibiotics to protect their investments. The extensive use of antibiotics has a significant impact on public health in the environment and on the development of pathogens resistant to antibiotics. Therefore, probiotic organisms have been proposed12.
Throughout the current study, from all of the isolates getting from the water samples of fishponds, only twenty isolates, which give Gram-positive and catalase-negative reaction, were chosen for further characterization. Only 18 isolates of Pediococcus pentosaceus were identified according to. In addition to that, the bacteria were identified using Vitek II with confidence degree of 95%. These results are in agreement with those recorded by9. Identification of bacteria using 16S rRNA is considered more dependent and accurate than traditional process. Different feature related to good probiotic properties of bacteria has been studied. Good probiotic sources must at least stay alive at pH 313, because of the high acidity in the stomach. For this, bacteria have been tested to withstand acidity during different time periods. The growth rate (observed through optical density) of lactic acid bacteria (LAB) showed that, P. pentosaceus were capable of living in acidic and basal circumstances. Some researchers14 found that the top action of probiotic achieved at pH 7. Meanwhile, in the present study, the isolates have the ability to survive and grow at different pH values and the highest activity was at pH 9. These outcomes harmonize with the results reported by15 and16, and can be proved that the studied bacteria have one of the most selective standards for probiotics17. Acid tolerance in probiotic bacteria has been also reported in other studies6; reported the viability of LAB (92.61% of Lactobacillus plantarum at pH 3 after 90 min). Kim and Austin18 notified that, the survival of the isolates (Carnobacterial) obtained from the rainbow trout intestine appeared at acidity range from 5 to 1019 represented that The LAB strains have a survival rate above 50% at low pH after 4h exposure.
The study of resistance to bile salts by probiotic bacteria is considered extremely important, because bile salts can act as antibacterial agents towards normal flora20. From the results obtained in the present study, the isolates of P. pentosaceus have the ability to tolerate different concentrations of bile salts in two incubation periods (3 and 24 h). This tolerance may be necessary for probiotic bacteria to growth and survive in the fish gut as mentioned by21,14b,22 found that the probiotic bacteria have the ability to survive and grow in different concentrations of bile salts16a reported that the all eight isolated they gated can be grown in different concentrations of bile salts (0.017, 0.014, 0.014, 0.012, 0.012%)(w/v) with different resistances to these bile salts concentrations exhibited by the tested isolates23 observed strain-dependent tolerance responses when subjected to different bile salts, in addition to the effect of the source of isolates in their resistance ability24. Therefore, the advantages of probiotic bacteria which can tolerate a wide range of pH and bile salts are not only their ability to live in the stomach and intestine, but also their ability to grow and tolerate any stress condition.
The present study revealed that the supernatant of P. pentosaceus suppressed the growth of Salmonella sp.25 revealed the ability of Pediococcus sp. to inhibit the growth of S. aureus, P. aeruginosa and E. coli, due to bacteriocin production in early stage of death phase. Probiotic bacteria may display a broad antimicrobial spectrum against fish pathogens, through different antimicrobial metabolites such as lactic acid, hydrogen peroxide, diacetyl, acetaldehyde, and/or bacteriocins26. These antimicrobials may play as potent outer membrane-disintegrating agents which are known to hydrolyze and damage peptidoglycan, a cell wall component of Gram-negative bacteria26b,27. Moreover, probiotic bacteria may reduce the growth of pathogenic bacteria by competition for space or attachment surface or by competition for nutrients28. In addition, the production of acids by probiotics leads to lowers the pH of the digestive tract and inhibits the growth of pathogenic microorganisms25. Furthermore, many researchers29 reported that, the dietary enhancement of Pediococcus sp. has positively influenced the body composition and increases intestinal microflora.
The antibiotic sensitivity pattern of P. pentosaceus against various antibiotics has been studied and results indicated that the bacteria were sensitive to the most tested antibiotics with differences in MIC values, whereas they were resistant to only inducible Clindamycin. In this respect, Klare, et al.30 have identified the MICs from 16 antibiotics for 473 isolates of LAB, including Lactobacillus, Pediococcus and Lactococcus. Their results proved that the majority of LAB was sensitive towards Penicillin, Ampicillin, Ampicillin/Sulbactam, Quinupristin/Dalfopristin, Chloramphenicol and Linezolid, whereas, three probiotic strains were resistant to Streptomycin. Another study has found that Lactobacillus species are affected by many inhibitors of cell wall synthesis such as Penicillin and Ampicillin31. reported that P. pentosaceus LPP32, LPM83 and B5 were Clindamycin resistant33 reported that among the different lactobacilli tested, they found high prevalence of resistances to Tetracycline (68% resistant isolates), Lincomycin (64.5%), Enrofloxacin (60%) and Ampicillin (50%)34 founded that among 29 Lactobacillus strains only S4 (L. reuteri), S5 (L. plantarum), S8a (L. rhamnosus) and S8b (L. acidophilus) displayed resistance to Ampicillin and Penicillin.
Depending on the present results, it can be concluded that the Pediococcus pentosaceus have perfect probiotic properties suggesting their application in the fish ponds as good, environment friendly and economic additives serving thus in: 1) prevention or reduction of pathogenic infection in fish via their antimicrobial activity, 2) increase of fish yield by improving the vitality of fish larvae through preventive colonization with selected beneficial bacteria, and 3) protection of the environment through elimination of the use of antibiotics reducing thus the appearance of antibiotic-resistant bacterial strains. And for future application of these probiotics in fish pond, we recommended to add other safety assays such as the investigation for virulence factors using molecular biology, the haemolysin and gelatinase productions and the bile salt hydrolase activity.
ACKNOWLEDGMENTS
The authors are thankful to Marine Science Center, special bacterial lab for all support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RS and AM designed the experiment and wrote paper. FN and BM coverage the technical aspects. All authors read and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Cajas-Cano, C. M. M. a. L., Blue Growth: The 2014 FAO State of World Fisheries and Aquaculture. Fisheries, 2014; 39(11): 552–553.
Crossref - Kluga, A.; Kacaniova, M.; Kantor, A.; Kovalenko, K.; Terentjeva, M. In Identification of microflora of freshwater fish caught in the Driksna river and pond in Latvia, 11th Baltic Conference on Food Science and Technology” Food science and technology in a changing world” FOODBALT 2017, Jelgava, Latvia, 27-28 April 2017, Latvia University of Agriculture: 2017; pp 164-168.
- Manage, P.M., Heavy use of antibiotics in aquaculture: Emerging human and animal health problems – A review. Sri Lanka Journal of Aquatic Sciences, 2018; 23(1): 13-27.
Crossref - Esteban, L.J. a. M.ֱ., Administration of Probiotics in the Water in Finfish Aquaculture Systems: A Review. Fishes, 2018; 3(33): 1-13.
Crossref - Holt, J.G.; Krieg, N., Bergey’s manual of systematic bacteriology, vol. 1. The Williams and Wilkins Co., Baltimore, 1984; 1-1388.
- Damayanti, E.; Yusiati, L.M.; Dinoto, A., 16s rRNA Identification of Pediococcus spp. from broiler and studies of adherence ability on immobilized mucus. Indonesian Journal of Biotechnology, 2012; 17(2): 96-106.
- Al Jassim, R.A.; Scott, P.T.; Trebbin, A.L.; Trott, D.; Pollitt, C.C., The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiology Letters, 2005; 248(1): 75-81.
Crossref - Harisha, S., Biotechnology procedures and experiments handbook. Laxmi Publications, Ltd.: 2008.
- Ayo-Olalusi, C. I., Isolation and Identification of Probiotics Pediococcus pentosaceus2 and Pediococcus pentosaceus1 from the Gut of Tilapia Guineensis for Use in Aquaculture Production International Journal of Research Studies in Microbiology and Biotechnology (IJRSMB), 2017; 3(1): 15-20.
Crossref - Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K., Selective detection, enumeration and identification of potentially probiotic Lactobacillus and Bifidobacterium species in mixed bacterial populations. International Journal of Food Microbiology, 1997; 35(1): 1-27.
Crossref - Balcazar, J.L.; Vendrell, D.; de Blas, I.; Ruiz-Zarzuela, I.; Muzquiz, J.L.; Girones, O., Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture, 2008; 278(1-4): 188-191.
Crossref - Charles Drago Kato, Ruth Kabarozi, Samuel Majalija, Andrew Tamale, Nathan Lubowa Musisi; Sengooba, a. A., Isolation and identification of potential probiotic bacteria on surfaces of Oreochromis niloticus and Clarias gariepinus from around Kampala, Uganda. African Journal of Microbiology Research, 2016; 10(36): 1523-1530.
Crossref - Kumar, B.V.U., K.; Reddy, O, Isolation and Identification of Pediococcus pentosaceus from Cow’s Milk Curd and Its Use in Grape Juices Fermentation. Journal of Advances in Biology & Biotechnology, 2017; 13(4): 1-11.
Crossref - (a) Allameh, S.K.; Daud, H.; Yusoff, F.M.; Saad, C.R.; Ideris, A., Isolation, identification and characterization of Leuconostoc mesenteroides as a new probiotic from intestine of snakehead fish (Channa striatus). African Journal of Biotechnology, 2012; 11(16): 3810-3816; (b) Muthukumar, P.; Kandeepan, C., Isolation, identification and characterization of probiotic organisms from intestine of fresh water fishes. International Journal of Current Microbiology and Applied Sciences, 2015: 4: 607-616.
- Davati, N.; Yazdi, F.T.; Zibaee, S.; Shahidi, F.; Edalatian, M.R., Study of lactic acid bacteria community from raw milk of iranian one humped camel and evaluation of their probiotic properties. Jundishapur Journal of Microbiology, 2015; 8(5): 1-6.
- (a) Lavanya, K.; Dayakar, Y., Isolation and characterization of probiotic bacteria from the soil samples of the coastal areas of (Gudur division, Nellore Dt.) for utilization in Shrimp farming. International Journal of Fisheries and Aquatic Studies, 2017; 5(4): 371-376; (b) Davati, N.; Yazdi, F. T.; Zibaee, S.; Shahidi, F.; Edalatian, M. R., Study of lactic acid bacteria community from raw milk of iranian one humped camel and evaluation of their probiotic properties. Jundishapur Journal of Microbiology, 2015; 8(5).
- Cakir, I. Determination of some probiotic properties on Lactobacilli and Bifidobacteria. Ankara Ankara University 2003.
- Kim, D. H.; Austin, B., Characterization of probiotic carnobacteria isolated from rainbow trout (Oncorhynchus mykiss) intestine. Letters in Applied Microbiology, 2008; 47(3): 141-147.
Crossref - Somashekaraiah R.S.B., Deepthi B.V. and Sreenivasa M.Y., Probiotic Properties of Lactic Acid Bacteria Isolated From Neera: A Naturally Fermenting Coconut Palm Nectar. Frontiers in Microbiology, 2019; 10(1382): 1-11.
Crossref - Fontana, L.; Bermudez-Brito, M.; Plaza-Diaz, J.; Munoz-Quezada, S.; Gil, A., Sources, isolation, characterisation and evaluation of probiotics. British Journal of Nutrition, 2013; 109(S2): S35-S50.
Crossref - Salminen, S.V.W., Atte, Lactic acid bacteria: microbiological and functional aspects. CRC Press: 2004.
Crossref - Cebeci, A.; G rakan, C., Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiology, 2003; 20(5): 511-518.
Crossref - Liong, M.; Shah, N., Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. Journal of Dairy Science, 2005; 88(1): 55-66.
Crossref - Sahadeva, R.; Leong, S.; Chua, K.; Tan, C.; Chan, H.; Tong, E.; Wong, S.; Chan, H., Survival of commercial probiotic strains to pH and bile. International Food Research Journal, 2011; 18(4): 1515-1522.
- Sukumar, G.; Ghosh, A.R., Pediococcus spp. a potential probiotic isolated from Khadi (an Indian fermented food) and identified by 16S rDNA sequence analysis. African Journal of Food Science, 2010; 4(9): 597-602.
- (a) Araujo, C.; Munoz-Atienza, E.; Poeta, P.; Igrejas, G.; Hernandez, P. E.; Herranz, C.; Cintas, L. M., Characterization of Pediococcus acidilactici strains isolated from rainbow trout (Oncorhynchus mykiss) feed and larvae: safety, DNA fingerprinting, and bacteriocinogenicity. Diseases of aquatic organisms, 2016; 119(2): 129-143; https://doi.org/10.3354/dao02992 (b) Huang, J.-B.; Wu, Y.-C.; Chi, S.-C., Dietary supplementation of Pediococcus pentosaceus enhances innate immunity, physiological health and resistance to Vibrio anguillarum in orange-spotted grouper (Epinephelus coioides). Fish & Shellfish Immunology, 2014; 39(2): 196-205.
Crossref - (a) Xing, C.F.; Hu, H.H.; Huang, J.B.; Fang, H.C.; Kai, Y.H.; Wu, Y.C.; Chi, S.C., Diet supplementation of Pediococcus pentosaceus in cobia (Rachycentron canadum) enhances growth rate, respiratory burst and resistance against photobacteriosis. Fish & Shellfish Immunology, 2013; 35(4): 1122-1128;
Crossref
(b) Alakomi, H.L.; Skytta, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I., Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Applied and Environmental Microbiology, 2000; 66(5): 2001-2005;
Crossref
(c) Jang, S.; Lee, D.; Jang, I. S.; Choi, H.-S.; Suh, H. J., The culture of Pediococcus pentosaceus T1 inhibits Listeria proliferation in Salmon fillets and controls maturation of kimchi. Food Technology and Biotechnology, 2015; 53(1): 29. - Sanchez-Ortiz, A.C.; Luna-Gonzalez, A.; Campa-Cordova, I.Escamilla-Montes, R.; del Carmen Flores-Miranda, M.; Mazon-Suastegui, J.M., Isolation and characterization of potential probiotic bacteria from pustulose ark (Anadara tuberculosa) suitable for shrimp farming. Latin American Journal of Aquatic Research, 2015; 43(1): 123-136.
Crossref - (a) O’sullivan, M.; Thornton, G.; O’sullivan, G.; Collins, J., Probiotic bacteria: myth or reality? Trends in Food Science & Technology, 1992; 3: 309-314;
Crossref
(b) Ferguson, R.; Merrifield, D. L.; Harper, G. M.; Rawling, M. D.; Mustafa, S.; Picchietti, S.; Balcazar,, J. L.; Davies, S. J., The effect of Pediococcus acidilactici on the gut microbiota and immune status of on growing red tilapia (Oreochromis niloticus). Journal of Applied Microbiology, 2010; 109(3): 851-862;
Crossref
(c) Merrifield, D.; Bradley, G.; Harper, G.; Baker, R.; Munn, C.; Davies, S., Assessment of the effects of vegetative and lyophilized Pediococcus acidilactici on growth, feed utilization, intestinal colonization and health parameters of rainbow trout (Oncorhynchus mykiss Walbaum). Aquaculture Nutrition, 2011, 17(1): 73-79;
Crossref
(d) Moslehi, F.; Sattari, M.; Masouleh, A. S., Effects of Pediococcus pentosaceus as a probiotic on intestinal microbiota and body composition of Siberian sturgeon, Acipenser baerii Brandt, 1869. International Journal of Aquatic Biology, 2016; 4(1): 11-16. - Klare, I.; Konstabel, C.; Werner, G.; Huys, G.; Vankerckhoven, V.; Kahlmeter, G.; Hildebrandt, B.; M ller-Bertling, S.; Witte, W.; Goossens, H., Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. Journal of Antimicrobial Chemotherapy, 2007; 59(5): 900-912.
Crossref - Gueimonde, M.; Sanchez, B.; de los Reyes-Gavilan, C. G.; Margolles, A., Antibiotic resistance in probiotic bacteria. Frontiers in Microbiology, 2013; 4(202): 1-6.
Crossref - Munoz-Atienza, E.; Gomez-Sala, B.; Araujo, C.; Campanero, C.; del Campo, R.; Hernandez, P. E.; Herranz, C.; Cintas, L. M., Antimicrobial activity, antibiotic susceptibility and virulence factors of Lactic Acid Bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiology, 2013; 13(1): 15.
Crossref - Dec, M.; Nowaczek, A.; Stipied-Pyniak, D.; Wawrzykowski, J.; Urban-Chmiel, R., Identification and antibiotic susceptibility of Lactobacilli isolated from turkeys. BMC Microbiology, 2018; 18(1): 168.
Crossref - Feckaninova A, K. J., Mudronova D, Schusterova P, Marunokova IC, Popelka P, Characterization of two novel lactic acid bacteria isolated from the intestine of rainbow trout (Oncorhynchus mykiss, Walbaum) in Slovakia. Aquaculture, 2019; 15(506): 294-301.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.