ISSN: 0973-7510
E-ISSN: 2581-690X
Klebsiella pneumoniae (K. pneumoniae) is an important pathogen associated with various infections. The emergence of antibiotic resistances, such as quinolone resistance and those due to extended-spectrum beta-lactamases (ESBL), reduces the available choices for treatment. The objectives of the current study include the evaluation of the prevalence of the plasmid-mediated quinolone resistance genes qepA, acrA, acrB, and aac(6’)-Ib-cr by polymerase chain reaction (PCR) in K. pneumoniae and the determination of the mechanism relating these genes to the ESBL phenotype and resistance to other groups of antibiotics. In total, 300 clinical isolates of K. pneumoniae were included in the study. Isolates were subjected to antibiotic sensitivity tests using the disc diffusion method. Quinolone resistance by the minimum inhibitory concentration method and detection of ESBL resistance by the double disc diffusion method were also determined. PCR analyses revealed the prevalence of acrA, aac(6’)-Ib-cr, acrB, and qepA in 74.3%, 73.7%, 71%, and 6.7% of the isolates, respectively. Quinolone-resistant isolates positive for plasmid-encoded genes represented 82.7% of K. pneumoniae isolates positive for ESBL activity. The results also showed that the isolates of K. pneumoniae carrying plasmid-encoded quinolone resistance genes had significantly increased resistance to amikacin, amoxicillin/clavulanate, gentamicin, and cefoxitin than those isolates without quinolone resistance genes. Therefore, there was a high prevalence of acrA, acrB, and aac(6’)-Ib-cr among K. pneumoniae and the prevalence of quinolone resistance was significantly associated with the ESBL resistance phenotype. Moreover, the presence of quinolone resistance genes was associated with resistance to aminoglycosides, namely amikacin and gentamicin.
Antibiotic resistance, K. pneumoniae, quinolone resistance genes, gram-negative bacilli
Klebsiella pneumoniae (K. pneumoniae) are non-motile, encapsulated gram-negative bacilli that may be found in the environment, such as soil and water, and on medical devices.1,2 K. pneumoniae is associated with various human infections, such as urinary tract, respiratory tract, wound, and blood stream infections. Antibiotic therapies against K. pneumoniae include cephalosporins, aminoglycosides, and quinolones. However, resistance to commonly used antibiotics has emerged.3,4
Fluoroquinolone resistance is among the antibiotic resistances identified in K. pneumoniae in the last few years. Fluoroquinolone resistance can be mediated via two different mechanisms: through mutations in the chromosomally encoded genes for gyrase and topoisomerase IV enzymes or through plasmid-mediated quinolone resistance (PMQR) determinants, namely, Qnr determinants, QepA and AcrAB efflux pumps, and aminoglycoside-acetyltransferase Ib-cr enzymes.5 QepA efflux pumps extrude antibiotics to the extracellular environment, leading to decreased antibiotic activity. The other efflux system, AcrAB, acts as a chemical transporter that transports various antibiotics to the extracellular environment, leading to antibiotic resistance.6,7 AcrAB consists of an outer membrane component TolC, an inner membrane transporter AcrB, and a periplasmic protein AcrA that bridges the two integrated membrane components. aac(6’)-Ib-cr encodes an acetyltransferase AAC(6’)-Ib variant with substitutions in tryptophan 102 to arginine and in aspartic acid 179 to tyrosine. These substitutions lead to fluoroquinolone acetylation and to a reduction in susceptibility to fluoroquinolones and aminoglycosides, such as tobramycin, kanamycin, and amikacin.8 Moreover, there are reports about the association between the presence of quinolone resistance and extended-spectrum beta-lactamases (ESBL) in K. pneumoniae.9,10
The objectives of the current study were to evaluate the prevalence of the PMQR genes qepA, acrA, acrB, and aac(6’)-Ib-cr by polymerase chain reaction (PCR) in K. pneumoniae and to determine their relationship with the ESBL phenotype and with other antibiotic resistances.
Bacterial Strains
The analysis included 300 isolates of K. pneumoniae obtained from different clinical samples (100 blood, 80 urine, 70 wound, and 50 sputum cultures) from King Fahd Hospital, Jazan, Saudi Arabia, from January 2018 to May 2020. K. pneumoniae isolates were identified using standard microbiological techniques, including Gram staining and manual biochemical identification methods (according to the Clinical and Laboratory Standards Institute [CLSI] guidelines).11 Biochemical identification included the use of triple sugar iron agar, a motility test, the Simmons citrate test, and ornithine and lysine decarboxylation tests (Oxoid-Thermo Fisher Scientific, USA). Escherichia coli (E. coli) ATCC 25922 was used as the positive control for all biochemical reactions.
Antibiotic Susceptibility Tests by the Disc Diffusion Method
Antibiotic susceptibility tests were performed according to the CLSI guidelines using the disc diffusion method.11 Briefly, pure colonies of K. pneumoniae were used for the preparation of cultures with a turbidity equivalent of 0.5 McFarland standards; cultures were spread over Mueller–Hinton agar. Discs were either impregnated with 5 μg of cefoxitin, 10 μg of imipenem, 20 or 10 μg of amoxicillin/clavulanate, or 30 μg of ceftazidime, tetracycline, cefotaxime, trimethoprim/sulfamethoxazole, gentamicin, amikacin, or cefepime (Oxoid-Thermo Fisher Scientific,). For antibiotic susceptibility testing, Escherichia coli strain ATCC 25922 was used as the quality control strain.
Minimum Inhibitory Concentrations (MICs) of the Quinolone Ciprofloxacin
The microdilution method (according to the CLSI guidelines) was used to determine the MIC of ciprofloxacin for the isolates.11 The results were interpreted according to the CLSI guidelines; isolates were classified as resistant to ciprofloxacin at MICs ≥ 4.0 μg/mL, as ciprofloxacin sensitive at MICs ≤ 1.0 μg/mL, and as ciprofloxacin intermediates at MICs between 1 and 2.0 μg/mL. E. coli ATCC 25922 was used as the quality control strain.
Determination of Extended-Spectrum Beta-Lactamase (ESBL) activity in K. pneumoniae
The double disc diffusion method was used to determine the ESBL activity for isolates of K. pneumoniae resistant to ceftazidime and cefotaxime according to the CLSI guidelines. The double disc method was performed using discs impregnated with 30 μg of cefotaxime and 30 μg of ceftazidime alone or in combination with 10 μg of clavulanic acid. An increase of 5 mm or more in the inhibition zone around the discs after combining the beta-lactam antibiotic with clavulanic acid was considered positive for ESBL activity.11 The ESBL-positive and negative control strains used in this study were E. coli ATCC 25922 and K. pneumoniae ATCC 700603, respectively.
Determination of PMQR genes in K. pneumoniae
DNA Extraction
DNA from isolated colonies of K. pneumoniae was extracted using a Qiagen DNA extraction kit (Qiagen, Hampshire, United Kingdom). Extracted DNA was kept frozen at -20°C till amplification.
Molecular identification of K. pneumoniae
Table 1 shows the results from the molecular identification of K. pneumoniae isolates by PCR using universal 16S rRNA bacterial primers (16S forward and reverse primers). The PCR was carried out in a total volume of 50 μL using Dream Tag Green Master Mix (Thermo Fisher Scientific) containing 25 μL of master mix ready to use, 1 μL (10 pmol) of each reverse and forward primers, 5 μL of DNA template, and 18 μL of nuclease-free water. The mixture was subjected to an initial denaturation process at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 56°C, and primer extension at 72°C for 1 min, and a final extension cycle at 72°C for 5 min. Negative and positive controls were included in each experiment. PCR products were analyzed by gel electrophoresis using 1.5% (wt/vol) agarose gels in Tris-Acetate EDTA buffer stained with ethidium bromide and visualized using a Gel Doc XR imaging system (Bio-Rad).
Table (1):
Genes, primers and the amplified base pair (bp).
Gene |
Sequence of the primer |
bp |
|---|---|---|
16S |
5′- AGA GTT TGA TCC TGG CTC AG – 3′ 5′- ACG GCT ACC TTG TTA CGA CTT – 3′ |
1500 |
QepA |
5’- CTGCAGGTACTGCGTCATG -3’ 5’- CGTGTTGCTGGAGTTCTTC -3’ |
403 |
acrA |
5’- TCTGATCGACGGTGACATCC -3’ 5’- TCGAGCAATGATTTCCTGCG -3’ |
157 |
acrB |
5’- CAATACGGAAGAGTTTGGCA -3’ 5’- CAGACGAACCTGGGAACC -3’ |
64 |
Aac(6’)-Ib |
5’- TTGCGATGCTCTATGAGTGGCTA -3’ 5’- CTCGAATGCCTGGCGTGTTT -3’ |
611 |
Standard Sequencing and Data Analysis
The PCR products were subjected to a high purification process. An ABI PRISM® 3730XL Analyzer with 96 capillary type using a BigDyeTM Terminator Cycle Sequencing Kit with AmpliTaq® DNA polymerase (FS enzyme) (Applied Biosystems) was used to perform sequencing reactions according to the manufacturer’s protocols. Single-pass sequencing was performed on each template using 16S F primers. Gel elution was performed using the MG Gel Extraction SV (MD007) kit (MGmed), following the manufacturer’s instructions.
The sequence similarity was examined and compared to the reference sequences through the Basic local Alignment Search Tool (BLAST) (www.ncbi.nlm.nih.gov/BLAST/) and GenBank (www.ncbi.nlm.nih.gov/genbank/).12
An evolutionary tree was constructed for the sequenced samples using molecular evolutionary genetic analysis (MEGA 5).
PCR for PMQR gene detection in K. pneumoniae
The amplification of qepA, acrA, aac(6’)-Ib-cr, and acrB was carried out using the primers listed in Table 1. A Qiagen (ready to use) amplification mixture (25 µL) was used for the entire process.
The amplification process was performed as follows: an initial denaturation step at 94°C for 5 min, 36 amplification cycles consisting of 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C, and a final extension step at 72°C for 5 min.8
Gel electrophoresis of the amplification products was carried out on a 1% agarose gel stained with ethidium bromide.
Positive PCR products were purified, and direct sequencing was performed to confirm the positive results (Thermo Fisher Scientific).
Statistical Analysis
The data were analyzed using the Statistical Package for the Social Sciences (SPPS) 22 and were stated as number and percentage. The results were compared using the chi-square test, P values < 0.05 were considered as statistically significant.
Molecular identification
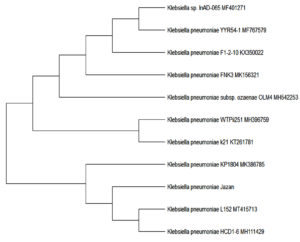
To confirm the biochemical identification results, approximately 1500 bp of the 16S gene of each isolate were amplified (Fig. 1) and partial sequencing of the PCR products using the 16S forward primer was performed. BLAST search results showed that the 16S sequences from the isolates from the Jazan region aligned with many K. pneumoniae sequences published in GenBank (Table 2). Sequences were also used to determine the evolutionary relationship between K. pneumoniae isolates from the Jazan region and other isolates from GenBank (Fig. 2).
Table (2):
Similarity to some K. pneumoniae isolates from Gen Bank.
Gen Bank sequence ID |
Country |
|---|---|
MH396759.1 |
Nigeria |
MT415713.1 |
China |
MH542253.1 |
India |
MK156321.1 |
Turkey |
Fig. 1. Agarose gel electrophoresis image for 16S gene of K. pneumoniae using universal primers. Lane1; 100bp ladder, lane 2; positive control, lanes 3,4,5,7 isolate samples and lane 6 negative control.
Quinolone resistance, as determined by ciprofloxacin MICs ≥ 4.0 μg/mL, was found in 240 (80%) of the K. pneumoniae isolates, and ESBL was determined in 197 (65.7%) of the isolates as well (Table 3).
Table (3):
Prevalence of ESBL and quinolone resistance among isolated K. pneumoniae.
Phenotypic resistance |
No. of isolates |
% |
|---|---|---|
ESBL |
197 |
65.7 |
Quinolone resistance |
240 |
80 |
A PCR study searching for PMQR genes was positive in 233 of the isolates. acrA was found in 74.3% of the isolates, whereas aac(6’)-Ib-cr, acrB, and qepA were found in 73.7%, 71%, and 6.7% of the K. pneumoniae isolates, respectively (Table 4).
Table (4):
Frequency of acA, acrB, qebA and aac (6’)-Ib-cr in K. pneumoniae (n=300).
Gene |
No. |
% |
|---|---|---|
acrA |
223 |
74.3 |
acrB |
213 |
71 |
qebA |
20 |
6.7 |
aac (6’)-Ib-cr |
221 |
73.7 |
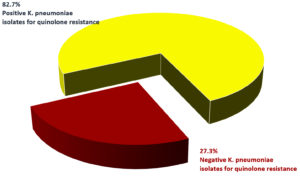
K. pneumoniae positive for PMQR genes represented 82.7% of the isolates that were also positive for ESBL, as determined by the double disc method (Fig. 3).
Fig. 3. Positive K. pneumoniae isolates for quinolone resistance genes among isolates positive for ESBL. P=0.01
Antibiotic resistance was compared between PMQR-positive and negative K. pneumoniae isolates (Table 5). The results showed that the isolates of K. pneumoniae positive for PMQR genes had significantly increased resistance to amikacin (P = 0.0001), amoxicillin/ clavulanate (P = 0.0001), gentamicin (P = 0.001), and cefoxitin (P = 0.002).
Table (5):
Comparison of antibiotics resistance between positive versus negative K. pneumoniae for the studied genes.
Antibiotics |
Positive K.pneumoniae isolates for genes (n=233) No. % |
Negative K.pneumoniae isolates for genes (n=67) No. % |
P |
|---|---|---|---|
Amikacin |
101 |
13 |
P=0.0001 |
Tetracycline |
89 |
22 |
P=0.3 |
amoxicillin–clavulanate |
156 |
26 |
P=0.0001 |
chloramphenicol |
169 |
49 |
P=0.5 |
Cefotaxime |
158 |
55 |
P=0.01 |
Ceftazidime |
155 |
50 |
P=0.5 |
Gentamicin |
103 |
15 |
P=0.001 |
Trimethoprim sulfamethoxazole |
92 |
23 |
P=0.7 |
Cefepime |
106 |
34 |
P=0.7 |
Cefoxitin |
122 |
50 |
P=0.002 |
Imipenem |
106 |
29 |
P=0.4 |
Recently, there has been an increase in worldwide reports on PMQR genes in K. pneumoniae. Most of these studies report a higher prevalence of PMQR genes in K. pneumoniae compared than in E. coli.13,14 However, the rates of prevalence vary considerably with the geographic area.13,15
In the present study, 80% of the K. pneumoniae isolates were found to be resistant to quinolones, according to the MIC study. A high frequency of resistance to quinolones has been reported previously, ranging from 41% to 89%.16-18 Resistance to fluoroquinolone may be attributed to improper use of quinolone, especially in urinary tract infections, leading to the transference of resistance genes to other susceptible Enterobacteriaceae by horizontal plasmid transfer.
A significant association between ESBL production and the presence of PMQR genes was found (P = 0.01). This phenomenon has also been shown in previous studies, with differences in prevalence rates ranging from 5% to 48%.19-22 Moreover, a recent study reported that all K. pneumoniae strains with quinolone resistance genes were also ESBL strains.10 The co-resistance to fluoroquinolones and beta-lactams was explained by the presence of quinolone-resistant genes, including ESBL determinants, in identical mobile genetic elements.10
The molecular investigation done in the present study revealed that the acrA and acrB frequencies in K. pneumoniae were 74.3% and 71%, respectively. These results are in line with a previous study by Heidary et al.8 in which it was determined that efflux systems mediated antibiotic resistance; the AcrAB efflux pump, which plays a vital role in the resistance of K. pneumoniae to quinolone was included.23-25
The second most prevalent gene was aac(6’)-Ib-cr. Previous studies also reported a high frequency of aac(6’)-Ib-cr associated with quinolone resistance.8 Aac(6′)-Ib-cr (ciprofloxacin resistance) is a variant of Aac(6’)-Ib; it is responsible for the resistance to tobramycin, amikacin, and kanamycin due to two amino acid substitutions involved in acetylation and subsequent reduction reactions Thus, Aac(6′)-Ib-cr may be responsible for diminishing the activity of several antibiotics, such as norfloxacin and ciprofloxacin subsequently reduced.26-28 Therefore, in addition to quinolone resistance, PMQR determinants can play a significant role in resistance to other antibiotics, particularly β-lactams and aminoglycosides. This hypothesis supports the finding of increased resistance to amikacin and gentamicin among isolates positive for quinolone resistance genes.
The least frequently determined gene was qepA. Previous studies have determined a prevalence ranging from 0% to 12%.29-31
The current study highlights the emergence of the quinolone resistance plasmid-encoded genes acrA, acrB, qepA, and aac(6’)-Ib-cr in clinical isolates of K. pneumoniae. A high prevalence of all these genes was found, except for qepA. Moreover, the existence of quinolone resistance genes was associated with resistance to aminoglycosides, such as amikacin and gentamicin.
ACKNOWLEDGMENTS
The author would like to thank the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work.
FUNDING
The study was funded by the Deputyship for Research & Innovation, Ministry of Education, Saudi Arabia with project grant number: ISP20-12.
ETHICS STATEMENT
Not Applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health, Proc Natl Acad Sci U S A.2015;112(27):3574-3581.
Crossref - Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, Morgan DJ. Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect Control Hosp Epidemiol. 2014;35(4):426-429.
Crossref - Mammina C, Bonura C, Di Bernardo F, et al. Ongoing spread of colistin resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 2012;17(33):20248.
Crossref - Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597-602.
Crossref - Rodriguez-Martinez JM, Machuca J, Cano ME, Calvo J, Martinez-Martinez L, Pascual A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist Updat. 2016;29:13-29.
Crossref - Seale AC, Obiero CW, Berkley JA. Rational development of guidelines for management of neonatal sepsis in developing countries. Curr Opin Infect Dis. 2015;28(3):225-230.
Crossref - Lubell Y, Ashley EA, Turner C, Turner P, White NJ. Susceptibility of community-acquired pathogens to antibiotics in Africa and Asia in neonates–an alarmingly short review. Trop Med Int Health. 2011;16(2):145-151.
Crossref - Heidary M, Goudarzi H, Hashemi A, et al. Prevalence of Quinolone Resistance Genes in Klebsiella pneumoniae Strains Isolated from Hospitalized Patients During 2013 – 2014. Arch Pediatr Infect Dis. 2017;5(4):e38343.
Crossref - Jacoby GA, Strahilevitz J, Hooper DC, Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014;2(5).
Crossref - Salah FD, Soubeiga ST, Ouattara AK, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lome, Togo. Antimicrob Resist Infect Control. 2019;8:104.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2019.
- Altschul SF, Thomas LM, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25(17):3389-3402.
Crossref - Karah N, Poirel L, Bengtsson S, et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-crin Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagn Microbiol Infect Dis. 2010;66(4):425-431.
Crossref - Briales A, Rodriguez-Martinez JM, Velasco C, et al. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Int J Antimicrob Agents. 2012;39(5):431-434.
Crossref - Bouchakour M, Zerouali K, Gros Claude JD, et al. Plasmid-mediated quinolone resistance in expanded spectrum beta lactamase producing Enterobacteriaceae in Morocco. J Infect Dev Ctries. 2010;4:779-803.
Crossref - Vetting MW, Park CH, Hegde SS, Jacoby GA, Hooper DC, Blanchard JS. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC (6′)-Ib and its bifunctional, fluoroquinolone-active AAC (6′)-Ib-cr variant. Biochemistry. 2008;47(37):9825-9835.
Crossref - Ramirez, Nikolaidis N and Tolmasky ME. Rise and dissemination of aminoglycoside resistance: the aac(6′)-Ib paradigm, Front Microbiol. 2013;17(4):121.
Crossref - Azargun R, Barhaghi MHS, Kafil HS, et al. Frequency of DNA gyrase and topoisomerase IV mutations and plasmid-mediated quinolone resistance genes among Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in Azerbaijan, Iran. J Glob Antimicrob Resist. 2019;17:39-43.
Crossref - Poirel L, Cattoir V, Nordmann P. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin Microbiol Infect. 2008;14(4):295-297.
Crossref - Lavilla S, Gonzalez-Lopez JJ, Sabate M, et al. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2008;61(2):291-295.
Crossref - Pitout JD, Wei Y, Church DL, Gregson DB. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary Health Region, Canada: the emergence of aac(6′)-Ib-cr. J Antimicrob Chemother. 2008;61(5):999-1002.
Crossref - Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49(8):3523-3525.
Crossref - Bialek-Davenet S, Lavigne JP, Guyot K, et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):81-88.
Crossref - Nagasaka Y, Kimura K, Yamada K, et al. Genetic profiles of fluoroquinolone-nonsusceptible Klebsiella pneumonia among cephalosporin-resistant K. pneumoniae. Microb Drug Resist. 2015;21(2):224 -233.
Crossref - Buffet-Bataillon S, Tattevin P, Maillard JY, Bonnaure-Mallet M, Jolivet Gougeon A. Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol. 2016;11(1): 81-92.
Crossref - Huang S, Dai W, Sun S, Zhang X, Zhang L. Prevalence of Plasmid-Mediated Quinolone Resistance and Aminoglycoside Resistance Determinants among Carbapeneme Non-Susceptible Enterobacter cloacae, Plos one 2021;7 (10):e47636.
Crossref - Robicsek A, Strahilevitz J, Jacoby GA, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12(1):83-88.
Crossref - Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50(11):3953-3955.
Crossref - Goudarzi M, Azad M, Seyedjavadi SS. Prevalence of Plasmid-Mediated Quinolone Resistance Determinants and OqxAB Efflux Pumps among Extended-Spectrum β-Lactamase Producing Klebsiella pneumoniae Isolated from Patients with Nosocomial Urinary Tract Infection in Tehran, Iran. Scientifica. 2015;2015:518167.
Crossref - Shams E, Firoozeh F, Moniri R, Zibaei M. Prevalence of Plasmid-Mediated Quinolone Resistance Genes among Extended-Spectrum β -Lactamase-Producing Klebsiella pneumoniae Human Isolates in Iran. J Pathog. 2015;2015:434391.
Crossref - El-Badawy MF, Tawakol WM, El-Far SW, et al. Molecular Identification of Aminoglycoside-Modifying Enzymes and Plasmid-Mediated Quinolone Resistance Genes among Klebsiella pneumoniae Clinical Isolates Recovered from Egyptian Patients. Int J Microbiol. 2017;2017:8050432.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.