ISSN: 0973-7510
E-ISSN: 2581-690X
Salt pan water collected from Kovalm, Kanyakumari district, Tamil Nadu, used for bacterial isolation using Zobal marine agar. Red pigmented colonies were selected for further studies and labelled as KSB. Microscopic, macroscopic and molecular studies were performed for the identification of isolate and it is confirmed as Delftia. Gen Bank accession number obtained for the same and deposited as Delftia MW172212. The optimization studies concluded that optimum temperature was 40 °C, optimum pH was 8 and optimum lactose concentration was 20%. Acetone precipitation concentrates active compounds, increasing efficacy against Gram-negative bacteria, according to the Delftia sp. acetone-precipitated fraction, which had a maximal inhibitory activity against E. coli MTCC-1671 that was marginally greater than the pure isolate (17 mm). Neutral red assay showed significant inhibition against MCF-7 hence it is considered as anticancer compound.
Salt Pan Water, Zobal Marine Agar, Delftia, Optimization, Antibacterial Study, Anticancer Study
Delftia is a diverse bacterial isolate having industrial and agriculture relevance. This species broadly distributed in all environments and isolated from different sources. The community of Delftia species are notable community of current research due to their less exposure and diverse applications.1-8 They belong to the class of Betaproteobacteria. They are Gram-negative organisms isolated from different environmental niches including polluted water, soil, sea water, etc. Though it is having broad spectrum applications recent potential studies are reported for bioremediation. First time this bacterium reported from Netarland.9 In this study, bacteria are isolated from salt pan water, characterised, and then used for antibacterial and anticancer purposes.
Sample collection
Using sterile bottles, a water sample was taken from Kovalam Saltpan in Kanyakumari, Tamil Nadu, and brought to the lab to be kept at 4 °C until it was needed.
Bacteria isolation
After serially diluting the samples with a 1M NaCl solution, they were plated on a Zobell marine agar plate (one litre contains 5,000 peptones). Ferric citrate: 0.100; NaCl: 19.450; MgCl2: 8.800; Na2SO4: 1.000; yeast extract: 7.6 ± 0.2 pH. After two weeks of incubation at 40 °C, the plates were checked for the development of halophilic bacterial isolates. Pure cultures were separated after morphologically distinct colonies that were seen on the plates were further streaked on halophilic agar plates.10
Characterisation and identification of isolate
Predominant isolate was characterised by microscopic study (Gram staining) and macroscopic studies-Biochemical studies and motility, according to Bergy’s Manual of Determinative Bacteriology and by 16S rRNA gene sequencing.11
Optimisation of parameters on the growth of bacteria
The process parameters such as incubation temperature, pH, salinity, carbon source and nitrogen source were optimized for the growth of bacteria.12
Optimization of temperature
After being inoculated into several tubes with nutritional broths (which included peptone, NaCl and beef extract, the isolated bacteria was cultured for five days at 25, 37, and 40 °C. OD values of bacterial suspensions incubated at different temperatures were determined with colorimeter under 590 nm up to decline phase.
Optimization of pH
1N HCl and 1N NaOH were used to bring the pH of the liquid media down to pH 5, 6, 7, 8, and 9. After being injected into various broths, isolated bacterial cultures were cultured for five days at 40 °C. OD values of bacterial suspensions with different pH were determined with colorimeter under 590 nm.
Optimization of NaCl concentration
Nutrient broths were prepared with 10%, 15%, 20%, 25%, 30% and 35% NaCl, bacterial cultures were introduced in different broths and incubated for 5 days at 40 °C. OD values of bacterial suspensions with varying NaCl concentration were determined with colorimeter under 590 nm up to decline phase.
Effect of different carbon source
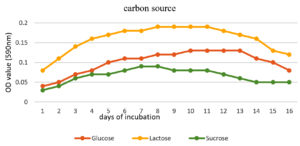
Nutrient broths were prepared with 20% NaCl, 0.3% beef extract and 0.5% different carbon sources (dextrose, lactose and sucrose). After being injected into various broths, isolated bacterial cultures were cultured for five days at 40 °C. OD values of bacterial suspensions with different carbon sources were determined with colorimeter under 590 nm up to its decline phase.
Effect of different nitrogen source
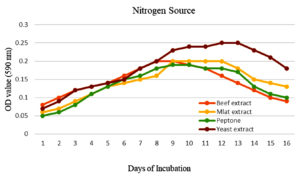
Nutrient broths were prepared with 0.5% peptone, 20% NaCl and 0.5% different N2 sources. Isolated bacterial cultures were injected in different broths then incubated to 72 hours at 40 °C. OD values of bacterial suspensions with different nitrogen sources were determined with colorimeter under 590 nm up to its decline phase.
Mass scale cultivation of bacteria
Isolated culture was seeded in 150 ml of optimised nutrient broth medium in a 250 ml conical flask with optimised culture conditions in a shaker at 150 rpm.
Partial purification of protein
The culture filtrate of the incubated culture was obtained by centrifugation at 1000 rpm for 15 minutes. The protein in the culture filtrate precipitated with ammonium sulphate, and 2.9 g of ammonium sulphate was added to the supernatant while stirring continuously to achieve 100% saturation. The resulted precipitate allowed to centrifuged at 6000 rpm then the precipitate was resuspended in 10 ml of 0.05 M calcium chloride. The mixture was thoroughly stirred, and dialysis was conducted in tubular cellulose membrane against 0.05 M calcium chloride for 24 h at 4 °C. The precipitate then dissolved in 10 ml of 50 mM Tris-HCl (pH 7.5).13
Estimation of protein content
Using bovine serum albumin as the reference, Lowry’s method14 was used to assess the protein content.
Antibacterial activity of partially purified protein
Antimicrobial activity of partially purified Delftin was evaluated against four different human pathogenic strains such as E. coli, Pseudomonas spp., Bacillus spp. and Staphylococcus spp., by agar well diffusion method. 10 µl of sample was added to the well of 6 mm diameter. Incubated at 37 °C for 24 hrs.
In vitro study-antitumor activity-neutral red assay
MCF-7 (Human Breast Adenocarcinoma) cells supplied by National Centre for Cell Sciences (NCCS). The culture medium made with 10% FBS, NaHCO3, L-glutamine, and antibiotics. Cells were maintained in a 25 cm² tissue culture flask and incubated at 37 °C.
96 well plate analysis
Trypsinization was carried out on the cells with confluent monolayer, which were 48 h old and suspended in 10% growth media. Then seeded into 96-well plates (volume of 100 µl with a concentration 5 × 103). Finally, plates were incubated at 37 °C in a humidified 5% carbon dioxide incubator.
Compound stock preparation
1 mg of the compound was solubilized in 1 mL of Dulbecco’s Modified Eagle Medium (DMEM) and the resulted solution was filtered via a 0.22 µm filter. Once the cells reached adequate confluency, they were trypsinized for two minutes using 0.025% (500 µl) trypsin in PBS/0.5 mM EDTA and transferred to T-flasks under sterile conditions. The sample (6.25 µl, 12.5 µl, 25 µl, 50 µl, and 100 µl in 500 µl DMEM) were incubated for 24 hours before adding 10 µl of neutral red. The samples the incubated at 37 °C for 3 h, then the cells were fixed with 200 µl ethanol-acetic acid solution, followed by extraction buffer. Absorbance was measured at 540 nm to calculate cell viability. The growth inhibition percentage was derived as follows.15
% of viability = [Mean Samples OD / OD of control group (Mean)] × 100
Isolation of bacteria
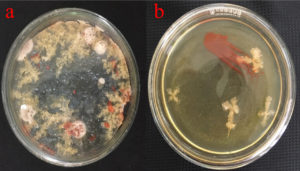
The microbiological investigations were carried out using standard plating methods. Zobal marine agar plate was used for the isolation.16 After two weeks of incubation, orange-red coloured colonies were dominant forming more than 50% noticed on halophilic agar (Figure 1). The red pigmented colonies were further considered for the studies and labelled as KSB (Kovalam Salt Pan Bacteria).
Figure 1. (a) Plate showing colonies of Delftia sp. Growing on agar plates, (b) isolated from salt water
Bacterial isolate identification
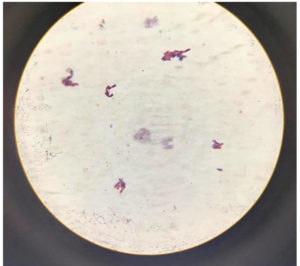
The isolate was identified through 16S rRNA gene sequencing, as well as by analysing its biochemical characteristics and examining its colony and cell morphology. It was discovered that the isolated colony was round, moderately big, orange-red in colour, smooth, elevated, and opaque throughout its whole edge. Macroscopic studies revealed that it is non-motile and biochemical characters are given in Table 1. Organism observed as Gram-negative (Figure 2) and molecular studies (Figure 3) details revealed that isolate belongs to Delftia genus. Gen Bank accession number obtained for the same and deposited as Delftia MW172212.17
Table (1):
Biochemical characteristics of the isolate
Biochemical tests |
Results |
|---|---|
Indole test |
– |
Methyl Red (MR) test |
– |
VP test |
– |
Citrate test |
– |
Urease test |
– |
Catalase test |
– |
Amylase test |
+ |
Gelatinase test |
– |
Nitrate reduction test |
– |
Glucose fermentation |
+ |
Lactose fermentation |
+ |
Sucrose fermentation |
+ |
Maltose fermentation |
– |
Optimization of growth parameters
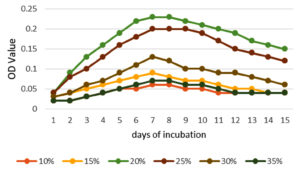
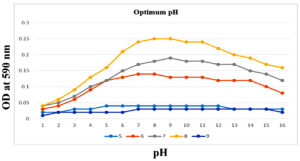
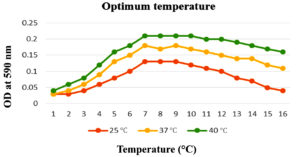
To enhance the growth and for the better production of metabolite external growth factors and media components were optimized. Bacterial isolate, Delftia was grown in wide range of salinity and optimum growth was achieved at 20%. The pH optimum for the growth of the organism was found to be 8. The temperature optimum for the growth was found to be 40 °C for Delftia. Growth was high in the medium containing lactose as the source of carbon and yeast extract as the source of nitrogen.18 Results are given in Figures 4-8.
Figure 6. Optimum temperature
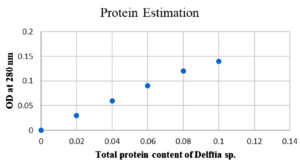
Estimation of protein content
By Lowry’s method, protein content in partially protein was found to be 0.12 mg/ml (Figure 9). The sample was stored and used for further antibacterial and cell line studies.19
Identification of the bacterial isolates
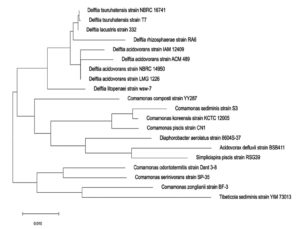
Comprehensive morphological, biochemical, and 16S rRNA examinations established the isolate’s identity.18 The bacterium is a non-sporulating, Gram-negative bacillus that appears as elongated rods, exhibits no catalase activity, hydrolyses starch, and remains negative for gelatin liquefaction. These diagnostic features, together with sequence homology, warrant its placement within the genus/Delftia. The isolate’s 1,458 base pair 16S rRNA gene has been deposited in GenBank, where it is available under accession number/MW172212 (Figure 10).
Phylogenetic tree showing the relationship of Delftia sp. with 16S rRNA
The phylogenetic tree reinforces that the Delftia genus represents a distinct, monophyletic lineage within the family/Comamonadaceae, clearly separated from other related genera such as Comamonas, Diaphorobacter, and Acidovorax.20-24 It delineates two well-supported clades within Delftia-one encompassing D. tsuruhatensis and D. lacustris, and the other comprising various D. acidovorans strains-highlighting significant intra-genus diversity and confirming previous genomic and ecological lineage differentiation. These findings corroborate the taxonomic classification based on 16S rRNA and whole-genome phylogenies, underscoring the evolutionary coherence and distinctness of Delftia in the Betaproteobacteria class.22,24
The strain’s designation as a possible lead for the discovery of new antibiotics is supported by its location within Delftia, which is corroborated by morphological and biochemical profiling as well as 16S rRNA gene sequencing. The isolate may be a potential lead for the discovery of new antibiotics due to its notable suppression of E. coli, increased extract potency, and alignment with previously identified Delftia antimicrobial compounds. The study’s emphasis on in vitro agar diffusion, however, emphasises the necessity of additional research to measure minimum inhibitory concentrations (MICs) and look into antibiotic synergy. Additionally crucial are the structural clarification and chemical characterisation of the acetone and Delftin precipitates. Therapeutic outcomes shall be established by assessing biosafety and in vivo effectiveness in mammalian models.
Antibacterial activity of partially purified protein
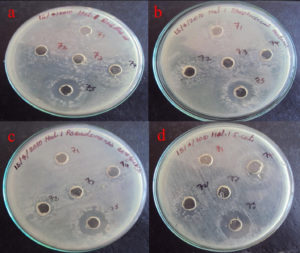
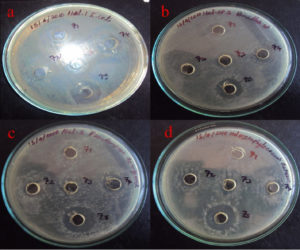
The partially purified protein extract of acetone and ethanol was tested for antibacterial activity against Gram-positive Bacillus sp., and Streptococcus sp., and Gram-negative bacteria, including Pseudomonas, and E. coli, (Figures 11 and 12). It showed significant antibacterial effects against these bacterial strains, leading to the conclusion that the separated protein must exhibit antibacterial activity against these four bacterial isolates.20
Figure 11. Antibacterial activity of Delfitin extracted from Delftia sp. against a). Bacillus sp., b). Streptococcus sp., c). Pseudomonas sp., and d). E. coli sp. at varying concentrations using acetone as solvent
Figure 12. Antibacterial activity of Delfitin extracted from Delftia sp. against a). E. coli sp., b). Bacillus sp., c). Pseudomonas sp., and d). Staphylococcus sp. at varying concentrations using ethanol as solvent
Delftin activity of acetone fractions
The acetone-precipitated fraction from Delftia sp. demonstrated clear antibacterial effects, forming a maximum inhibition zone of 18 mm against Escherichia coli MTCC 1671 and a minimal zone of 10 mm against Pseudomonas aeruginosa MTCC 6538. In parallel, the naked isolate of Delftia sp. achieved an inhibition zone of 17 mm against E. coli MTCC 1671 and just 11 mm against Streptococcus mutans MTCC/896. These findings collectively highlight the notable antibacterial potential of both the acetone-precipitated fraction and the pure isolate, especially in targeting E. coli MTCC/1671 (Figure 11).
Delftin activity of ethanol precipitated samples
The ethanol-precipitated Delftin extract from the Delftia sp. isolate exhibited antimicrobial activity, with inhibition zones ranging from 10 mm against Streptococcus mutans MTCC 896 to a maximum of 16 mm against Escherichia coli MTCC 1671. Notably, the Delftin compound demonstrated its highest inhibitory effect against E. coli MTCC 1671, producing a 17 mm zone of inhibition, while showing the least effect against Bacillus subtilis MTCC 1134, with a 10 mm zone (Figure 12). This study presents novel insights into the antimicrobial potential of Delftia sp., comparing the effects of its crude isolate and solvent-precipitated extracts against a range of bacterial pathogens.
Acetone precipitation concentrates active compounds, increasing efficacy against Gram-negative bacteria, according to the Delftia sp. acetone-precipitated fraction, which had a maximal inhibitory activity against E. coli MTCC-1671 that was marginally greater than the pure isolate (17 mm).20-24 Purified Delftin had the maximum activity (17 mm) against E. coli, and its inhibition zones ranged from 10 mm to 16 mm, reflecting its selective antimicrobial ability.20 The fact that B. subtilis (10 mm) exhibited the lowest activity suggests that Delftin has a predilection for certain Gram-negative bacteria. Baseline generation of antimicrobial metabolites is reflected in the inhibitory zones of the naked isolate.21
In vitro study-antitumor activity
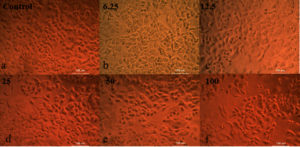
The partially purified extracellular protein demonstrates antitumor activity against breast cancer cells. As the protein concentration increases, the cell viability percentage decreases (Figure 13 and Table 2).25,26 The neutral red cytotoxicity study was used to study the live cells. In this study the dye was absorbed and bound only by live cells, for dead cell nullified activity was observed.27 The results clearly indicate that neutral red accumulation is directly related to number of viable cells. The neutral red solution pH was adjusted to 6.35 by adding KH2PO4 (1M). The LC50 value was determined to be between 137-149 µl, calculated using ED50 PLUS software.
Table (2):
Antitumor activity for partially purified protein
Sample Concen. (µL) |
OD-I |
OD-II |
OD-III |
Mean OD |
% Viability |
|---|---|---|---|---|---|
Control |
0.1572 |
0.1556 |
0.1587 |
0.1572 |
100 |
6.25 |
0.1512 |
0.1508 |
0.1497 |
0.1506 |
94.78 |
12.5 |
0.1408 |
0.1410 |
0.1418 |
0.1412 |
89.82 |
25 |
0.1346 |
0.1346 |
0.1314 |
0.1335 |
84.94 |
50 |
0.1162 |
0.1144 |
0.1161 |
0.1156 |
73.52 |
100 |
0.0959 |
0.1055 |
0.1020 |
0.1011 |
64.33 |
The colony with red pigmentation were selected for further studies and named as KSB. Initial characterization was revealed that it is Gram-negative non-motile organism. IMViC studies negative for the organism, urease negative and gelatinase negative and amylase positive with zone of clearance. Optimization of media components and external factors were identified as 20% NaCl, pH 5, Temperature 40 °C, best carbon source lactose and yeast extract as nitrogen source. Partially purified Delftin had the maximum activity (17 mm) against E. coli, and its inhibition zones ranged from 10 mm to 16 mm, reflecting its selective antimicrobial ability and reported with antitumor activity against MCF-7 by Neutral Red assay.
ACKNOWLEDGMENTS
The authors acknowledge their respective institutions for all the support and guidance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
KP collected resources, performed data curation and supervised the study. APK applied methodology. RG performed formal analysis. Y, KP and KT wrote the mansucript. RG, KP, TM, KK and KT reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Wang SX, Park M, Kim SG. Complete genome sequence of Delftia tsuruhatensis strain HA60 isolated from a commercial hydroxyapatite nano-particle product (nano-hydroxyapatite). Microbiol Resour Announc. 2024;13(6):e00171-24.
Crossref - Sazonova OI, Ivanova AA, Delegan YA, et al. Characterization and genomic analysis of the naphthalene-degrading Delftia tsuruhatensis ULwDis3 isolated from seawater. Microorganisms. 2023;11(4):1092.
Crossref - Scaglione V, Stefanelli LF, Mazzitelli M, et al. Delftia acidovorans Infections in Immunocompetent and Immunocompromised Hosts: A Case Report and Systematic Literature Review. Antibiotics. 2025;14(4):365.
Crossref - Lu TL, Huang C. Retrospective cohort study on Delftia acidovorans infections in patients: a rare and significant infection. Infect Drug Resist. 2024;2024(17):1741-1749.
Crossref - Khan SF, Krishnan T. Delftia acidovorans skin infection occurring in a diabetic with peripheral edema. Journal of Case Reports. 2015;4(1):127-131.
Crossref - Bilgin H, Sarmis A, Tigen E, Soyletir G, Mulazimoglu L. Delftia acidovorans: a rare pathogen in immunocompetent and immunocompromised patients. Can J Infect Dis Med Microbiol. 2015;26(5):277-279.
Crossref - Agarwal N, Jindal A, Bhargava A. Delftia acidovorans: rarely a pathogen: a case report. Pediatr Infect Dis J. 2023;42(4):e130-e131.
Crossref - Ates, G., Vanhaecke, T., Rogiers, V., Rodrigues, R.M.. Assaying Cellular Viability Using the Neutral Red Uptake Assay. In: Gilbert, D., Friedrich, O. (eds) Cell Viability Assays. Methods in Molecular Biology, vol 1601. Humana Press, New York, NY. 2017;160:19-26.
Crossref - Li Y, Guo P, Sun J. Isolation, identification, phylogeny, and growth promoting characteristics of endophytic diazotrophs from tuber and root crops. Chinese Journal of Agricultural Sciences. 2017;50(1):104-122.
Crossref - Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265-275.
Crossref - Morel MA, Ubalde MC, Brana V, Castro-Sowinski S. Delftia sp. JD2: a potential Cr(VI)-reducing agent with plant growth-promoting activity. Arch Microbiol. 2011;193(1):63-68.
Crossref - Stolze Y, Eikmeyer F, Wibberg D, et al. IncP-1β plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet. Microbiology. 2012;158(Pt 8):2060-2072.
Crossref - Vega FE, Emche S, Shao J, et al. Cultivation and genome sequencing of bacteria isolated from the coffee berry borer (Hypothenemus hampei), with emphasis on the role of caffeine degradation. Front Microbiol. 2021;12:644768.
Crossref - Wozniak M, Galazka A, Tyskiewicz R, Jaroszuk-Scisel J. Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their phenotypic/physiological profiles in the Biolog GEN III MicroPlate™ Test. Int J Mol Sci. 2019;20(21):5283.
Crossref - Yan H, Yang X, Chen J, Yin C, Xiao C, Chen H. Synergistic removal of aniline by carbon nanotubes and the enzymes of Delftia sp. XYJ6. J Environ Sci. 2011;23(7):1165-1170.
Crossref - Wang YS, Xu JM, Zheng RC, Zheng YG, Shen YC. Improvement of amidase production by a newly isolated Delftia tsuruhatensis ZJB-05174 through optimization of culture medium. J Microbiol Biotechnol, 2008;18(12):1932-1937
- Stincone P, Brandelli A. Marine bacteria as source of antimicrobial compounds. Crit Rev Biotechnol, 2020;40(3):306-319.
Crossref - Vásquez-Piñeros MA, Martínez-Lavanchy PM, Jehmlich N. et al. Delftia sp. LCW, a strain isolated from a constructed wetland shows novel properties for dimethylphenol isomers degradation. BMC Microbiol. 2018;18:108.
Crossref - Ansari A, Zohra RR, Tarar OM, et al. Screening, purification and characterization of thermostable, protease resistant Bacteriocin active against methicillin resistant Staphylococcus aureus (MRSA). BMC Microbiol. 2018;18:192.
Crossref - Morel MA, Iriarte A, Jara E, Castro Sowinski S. Revealing the biotechnological potential of Delftia sp. JD2 by a genomic approach. J Genom. 2016;3(2):156-175.
Crossref - Ohkubo T, Matsumoto Y, Cho O, Ogasawara Y, Sugita T. Delftia acidovorans secretes substances that inhibit Staphylococcus epidermidis growth through TCA cycle–triggered ROS production. PLOS ONE. 2021;16(7):e0253618.
Crossref - Wen A, Fegan M, Hayward C, Chakraborty S, Li S. Phylogenetic relationships among members of the Comamonadaceae and description of Delftia acidovorans gen. nov. Int J Syst Bacteriol. 1999;49(2):567-576.
Crossref - Bhat SV, Maughan H, Cameron ADS, Yost CK. Phylogenomic analysis of the genus Delftia reveals distinct major lineages with ecological specializations. Microb Genom. 2022;8(9):e000864.
Crossref - Yin Z, Liu S, Qian C, et al. Pan genome analysis of Delftia tsuruhatensis reveals important traits concerning the genetic diversity, pathogenicity, and biotechnological properties of the species. Microbiol Spectr. 2021;9(2):e0207221.
Crossref - Samimi M, Shahriari-Moghadam M. Isolation and identification of Delftia lacustris Strain-MS3 as a novel and efficient adsorbent for lead biosorption: Kinetics and thermodynamic studies, optimization of operating variables. Biochemical Engineering Journal. 2021;173: 108091.
Crossref - Shakibaie M, Amiri-Moghadam P, Ghazanfari M, Adeli-Sardou M, Jafari M, Forootanfar H. Cytotoxic and antioxidant activity of the biogenic bismuth nanoparticles produced by Delftia sp. SFG. Materials Research Bulletin. 2018;104: 155-163.
Crossref - Nikolova MP, Joshi PB, Chavali MS. Updates on biogenic metallic and metal oxide nanoparticles: therapy, drug delivery and cytotoxicity. Pharmaceutics, 2023;15(6): 1650.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.