ISSN: 0973-7510
E-ISSN: 2581-690X
Marine bacteria (n = 1,928) were isolated using Actinomycetes Isolation Agar (AIA) media and screened against bacteria causing nosocomial lower respiratory tract infections via agar overlay method. Nine Actinomycetes displayed significant antimicrobial efficacy against target pathogens, viz. Streptococcus pyogenes, Staphylococcus aureus, Klebsiella pneumoniae and Streptococcus pneumoniae. Based on the inhibitory efficiency, SZ33 was chosen for further studies and identified as a new strain Streptomyces rochei SZ33 (OQ726229). Production of antimicrobial secondary metabolites was optimized and best results were achieved at pH 6, 20 °C, with 2% salt concentration. FTIR analysis revealed the presence of alcohol and amide carbonyl compounds. Analysis of diethyl ether extract using GC-MS confirmed the presence of antimicrobial compounds, viz. β-carotene-3,3′-diol, (3R, 3’R)-all-trans; 2H-pyran, 2-(7-heptadecynyloxy) tetrahydro; astaxanthin and aurin etc. Furthermore, docking studies demonstrated that natural compounds derived from SZ33, exhibited superior binding affinities, with larger allosteric binding pocket sites compared to conventional synthetic antibiotics.
Secondary Metabolites, GC-MS, Optimization, Molecular Docking, Binding Pockets, Antibiotics, Crude Extract
Despite groundbreaking advancements, medical science continues to face substantial challenges, particularly in the management of post-surgical patients who are susceptible to various infections.1,2 Amongst these, nosocomial infections are one of the most concerning. Patients with severe illnesses, age, or compromised immune systems are particularly vulnerable, with hospital-acquired infections exacerbating their conditions. Among the various types of nosocomial infections, those affecting the respiratory tract, both upper and lower, are amongst the most severe. Lower respiratory tract infections (LRTIs), caused by both gram-positive and gram-negative bacteria, is a significant risk of morbidity if not promptly identified and treated with appropriate antibiotics.3,4 During 2019, LRTIs accounted for approximately 2.4 million deaths globally, with 12,000 of these occurring in India.5 Broad-spectrum antibiotics like penicillin derivatives and carbapenems are typically employed to combat these infections6; however, the rise of resistant strains and the difficulty of attaining therapeutic concentrations in the lung epithelium pose significant barriers to successful therapy. Consequently, research focus has significantly increased on identifying novel antibiotics with broad-spectrum activity.
Marine Actinomycetes, an underexplored source of antimicrobial agents, have garnered attention in recent years. These microorganisms are known to produce some of the most effective antibiotics, such as amphotericin, neomycin, novobiocin, and tetracycline.7 However, the rise of antibiotic resistance has limited the efficacy of many current treatments. Therefore, the identification and successful in vitro cultivation of antibiotic-producing marine Actinomycetes, alongside the production of antimicrobial secondary metabolites targeting multidrug-resistant pathogens, is pivotal.8 This study aims to isolate and identify marine Actinomycetes effective against bacteria, viz. Streptococcus pyogenes, Staphylococcus aureus, Streptococcus pneumoniae and Klebsiella pneumoniae, responsible for nosocomial lower respiratory tract infections.9
In the present study, isolation of marine actinomycetes and screening for antimicrobial secondary metabolites was carried out under in vitro conditions against the target pathogens. Optimization for the higher production of antimicrobial secondary metabolites was carried out, and Gas Chromatography-Mass Spectrometry (GC-MS) analysis was performed to identify the secondary metabolites. Molecular docking and visualization analyses investigated the binding affinities and binding pocket sizes of the identified compounds towards the active sites of the target pathogens. We have also, in this study, compared the aforesaid docking results with commercially available antibiotics for the aforementioned pathogens to demonstrate the heightened antibacterial potency of these compounds. Therefore, in the attempts to find novel sources of antimicrobial compounds, this study has successfully generated a reservoir of potent natural antibacterials to combat nosocomial LRTIs.
Sample collection
Marine sediments from 12 different sites, viz. Dhanushkodi [L1A – L1G; L2A – L2E] in Ramanathapuram District and Pichawaram [L3A – L3D], in Cuddalore District, Tamil Nadu, India. Sampling was carried out between midday and afternoon during low tide conditions. The sediment samples were obtained from a depth of 2-3 feet using sterilized glass bottles. A total of twenty-two samples were collected, transported in ice boxes, and subsequently stored under refrigeration until further use.
Isolation of Actinomycetes
One gram of each sediment sample was serially diluted up to 10-6 using a 1:1 mixture of sterile distilled water and seawater. From the 10-3, 10-4, and 10-5 dilutions, 0.3 mL aliquots were inoculated onto Actinomycetes Isolation Agar (AIA) using the spread plate technique. The plates were incubated at 28 °C for 4 to 5 days. Marine Actinobacteria exhibiting diverse morphologies were randomly selected and sub-cultured. Gram’s staining was performed for initial confirmation of the Actinomycetes.10-12
Pathogen cultures
Pathogens known to cause lower respiratory tract infections, including Streptococcus pyogenes (MTCC1927), Staphylococcus aureus (MTCC7443), Klebsiella pneumoniae (MTCC9509), and Streptococcus pneumoniae (MTCC2672), were selected based on previous studies and obtained from the Microbial Type Culture Collection (MTCC), Chandigarh, India. The pathogens were subcultured on nutrient agar media and stored under refrigeration until further application.
Screening for antimicrobial activity against target pathogens
Primary screening
The isolated Actinomycetes strains were screened against target pathogens by the standard method on AIA media.13 The plates were spread with the respective bacterial pathogen cultures using sterile cotton swabs and allowed to remain for 1-2 minutes. Each labelled plate was then streaked at the center with Actinomycetes isolates and incubated at 37 °C for 7 days. The plates were observed for development of zone of inhibition.12-15
Optimization of growth parameters and production parameters for antibacterial secondary metabolites
Effect of different parameters, viz. pH (4, 5, 6, 7, 8, 9), temperature (20 °C, 25 °C, 30 °C, 35 °C, 40 °C, 45 °C), carbon sources (glucose, dextrose, starch, fructose, lactose), nitrogen sources (asparagine, peptone, tryptone, urea, yeast extract), and salt concentrations (0%, 1%, 2%, 3%, 5%, 7%, 9%, 11%) on both growth and secondary metabolite production was assessed. Actinomycetes strains were inoculated into AI broth with the varying parameters, and growth was monitored by measuring the optical density at 570 nm for up to 10 days. After 14 days of incubation, crude extracts of secondary metabolites were obtained through centrifugation and filtration. Well diffusion assay was carried out to evaluate the antimicrobial activity of these extracts against the target pathogens.16,17
Secondary screening
Potential Actinomycetes strain was grown under optimised conditions. The culture was centrifuged at 12,000 rpm for 10 minutes. The supernatant was filtered using a 0.45 micron membrane filter (Sartorius) to obtain cell-free crude extract of secondary metabolites. Pathogens were inoculated separately onto AIA plates using the spread plate method. On each plate, three equidistant wells (6 mm diameter) were made using a sterile cork borer. 75 µL of crude extract was added in each well, subsequently the plates were incubated at 37 °C for 24 hours. Streptomycin (30 units) served as positive control. All the tests were conducted in triplicates. The inhibition zones were measured in millimeters (mm). Based on these results, a potential actinobacterial strain was selected for further studies.18
Molecular characterization
Selected Actinomycete strain was identified through 16S rRNA sequencing. DNA was amplified using the forward primer (5′-TCACGGAGAGTTTGATCCTG-3′) and the reverse primer (5′-7GCGGCTGCTGGCACGTAGTT-3′). The resulting sequence was analyzed using BLAST (Basic Local Alignment Search Tool). A phylogenetic tree was constructed with the selected sequences from GenBank database (NCBI) using the maximum likelihood method, followed by alignment using Clustal W algorithm implemented in MEGA11 (Molecular Evolutionary Genetics Analysis version 11) software. A neighbor-joining tree was then constructed based on evolutionary distances calculated with the Kimura-2-parameter substitution model. The robustness of the tree was assessed by bootstrap analysis with 1000 replications.
Solvent extraction of secondary metabolites
The selected Actinomycete strain was inoculated into 100 mL of AI broth and incubated at 37 °C for 7 days on a rotary shaker set at 200 rpm. The culture broth was centrifuged at 12,000 rpm for 10 minutes to separate the biomass. The supernatant was extracted using diethyl ether in a 1:1 (v/v) ratio. The mixture was agitated continuously for 24 hours to ensure thorough extraction. After the extraction period, the immiscible phases were separated using a separating funnel, and the organic phase (diethyl ether layer) was collected. The organic extract was then assessed for its antibacterial activity using well diffusion assay, following the procedure described previously.19
Detection and identification of bioactive antimicrobial metabolites
Gas chromatography-mass spectrometry (GC-MS) analysis
The lyophilized diethyl ether extract was reconstituted in separate solvents, acetonitrile and methanol for GC-MS analysis. The compounds were identified by comparing their retention times, peak areas, and molecular weights with known standards from the GC-MS database.20,21
Fourier transforms infrared (FTIR) spectral analysis
FTIR spectrum of the active extract was detected using Shimadzu IR-470 plus. The spectra were also scanned in the 400 to 4000 cm-1 range and plotted as intensity versus wave number.22
Retrieval of the structure of the receptors and the compounds
The chemical structures of the compounds (obtained from GC-MS) were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The receptor protein structures were obtained from the Protein Data Bank (https://www.rcsb.org/). The specific targets selected for docking were Penicillin-Binding Protein (PBP) 3 from S. aureus (3VSK), S. pneumoniae (1XP4), K. pneumoniae (8GPW), and PBP 2 from P. aeruginosa (7KIS). The specific receptor targets were chosen based on the ability of both compounds under study and commercial antibiotics to bind the receptors.
Docking
The receptor protein structures were cleaned and prepared for docking using Discovery Studio software (Dassault Systemes BIOVIA. Discovery Studio Visualizer (v24.1.0.23298) [Software]. Available from https://www.3ds.com/products-services/biovia/. The protein structures were optimized by the omission of water molecules and the inclusion of hydrogen atoms. The compounds were prepared for docking using PyRx software, a virtual drug discovery screening tool. Molecular docking was then performed using Discovery Studio, where each compound was docked into the binding sites of the respective receptor proteins. Discovery Studio was employed to perform the docking calculations and generate docking scores for evaluating the binding affinities between the compounds and the receptors. The binding affinities were evaluated based on the docking scores, with more negative scores indicating stronger binding interaction.
Pocket visualization
The binding pockets of the docked complexes were visualized using PyMOL software Schrodinger, LLC. The PyMOL Molecular Graphics System (version 2024) [Software]. Available from https://pymol.org/. This permits the optimal assessment of the interaction between the query compound and the target receptor proteins, including identifying key binding sites and specific interactions that may contribute to the binding affinity.
Statistical analysis
All assays were performed in triplicates, and the results are depicted as the mean ± standard deviation (SD). Statistical analysis was conducted using one-way analysis of variance (ANOVA), followed by Tukey’s post-hoc test to determine significant differences between groups. Statistical significance was assigned to a p-value of <0.05. All analyses were performed using IBM SPSS Statistics version 25.
Isolation and Primary Screening of Actinomycetes
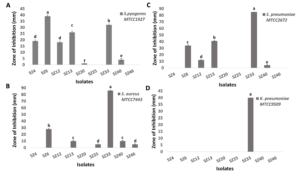
The study yielded 1,928 isolates in total from marine sediment and mangrove soil samples, of which 78 were formerly identified as Actinomycetes owing to colony morphology and Gram staining characteristics. Out of these 78 isolates, only 9 isolates showed significant antimicrobial activity (P <0.01) against target bacterial pathogens. Most of the isolates demonstrated strong antagonism against S. pyogenes and S. pneumoniae, with moderate activity against S. aureus. Notably, only one isolate, SZ33, displayed inhibitory activity against K. pneumoniae too. Hence, isolate SZ33 was selected for further studies for exhibiting broad-spectrum inhibitory activity, effectively inhibiting the growth of all four target pathogens (Figures 1 and 2). Table 1 shows the comparative antimicrobial activity of purified secondary metabolites from SZ33 with standard antibiotic.
Table (1):
Comparative antimicrobial activity of purified secondary metabolites from SZ33 with standard antibiotic
| Pathogens | Inhibition of pathogens Zone of inhibition (mm in diameter) Purified Antibiotic (Positive control) |
Relative inhibition (%) | |
|---|---|---|---|
| S. pyogenes MTCC1927 | 22.73 + 0.05 | 11.00 + 0.10 | 106.667 |
| S. aureus MTCC7443 | 13.43 + 0.05 | 10.1 + 0.10 | 33.003 |
| S. pneumoniae MTCC2672 | 16.53 + 0.05 | 16.57 + 0.05 | -0.201 |
| K. pneumoniae MTCC9509 | 29.50 + 0.10 | 19.33 + 0.05 | 52.586 |
Note: The values are the mean values of three replication
Figure 1. Zone of inhibition in primary screening of 9 selected Actinomycetes against selected respiratory pathogens: (A) S. pyogenes, (B) S. aureus, (C) S. pneumoniae, (D) K. pneumoniae
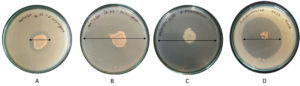
Figure 2. Zone of inhibition in primary screening of SZ33 against pathogens. (A) S. pyogenes, (B) S. aureus, (C) S. pneumoniae, (D) K. pneumoniae
Optimization of growth parameters
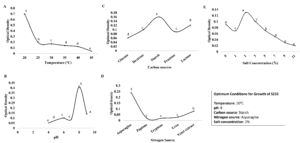
Maximum growth of isolate SZ33 was observed on the seventh day post-inoculation. Optimal growth conditions were achieved at pH 8 and 20 °C, with starch, asparagine and a salt concentration of 2% (Figure 3).
Figure 3. Optimization of growth parameters; (A) pH, (B) Temperature, (C) Carbon source, (D) Nitrogen source, (E) Salt concentration
Optimization of production parameters for antibacterial secondary metabolites
Optimization studies for the generation of antibacterial secondary metabolites by isolate SZ33 revealed that pH 6 was optimal for antimicrobial activity against all target bacterial pathogens. The cell-free extract produced at 20 °C exhibited maximum inhibition against all pathogens, except for S. pyogenes, where the maximum inhibition was observed with the extract obtained at 25 °C. The medium containing starch as the nitrogen and carbon source, yeast extract, and 2% salt concentration was found to be the most optimal for the production of antibacterial metabolites against all tested pathogens (Supplementary Figure S1).
Secondary screening
Purified secondary metabolites (75 µl) showed significant inhibition of tested pathogens in well diffusion assay. Maximum inhibition was observed against K. pneumoniae (29.50 mm) followed by S. pyogenes (27.33 mm), S. aureus (13.43 mm) and S. pneumoniae (16.53 mm) (Figure 4). In comparison, the standard antibiotic Streptomycin produced inhibition zones of 11.00 mm, 10.10 mm, and 19.33 mm against S. pyogenes, S. aureus, and K. pneumoniae, respectively. Thus, the purified metabolites of SZ33 exhibited substantially greater inhibition against these pathogens. Additionally, SZ33 showed comparable inhibitory activity to Streptomycin against S. pneumoniae (16.57 mm).23
Figure 4. Zone of inhibition in purified secondary metabolites from SZ33 against pathogens (A) S. pyogenes, (B) S. aureus, (C) S. pneumonia, (D) K. pneumoniae
Note: Antimicrobial activity by secondary metabolites obtained from SZ33 extracts against S. pyogenes, S. aureus, S. pneumoniae, and K. pneumoniae
Characterization of SZ33
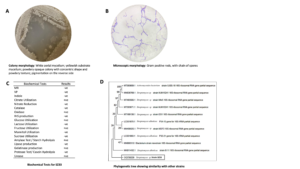
Strain SZ33 is aerobic, Gram-positive bacillus. The colonies appeared yellowish in colour and exhibited a powdery texture on AIA media. Based on biochemical and molecular characterization, SZ33 was identified as Streptomyces rochei SZ33 (NCBI Accession No. OQ726229) (Figure 5).
Figure 5. (A, B) Morphological, (C) physiological, and biochemical characteristics of SZ33.19,60 (D) Phylogenetic tree displaying the relationship of Streptomyces species (NCBI Accession No. OQ726229) and its nearest relatives based on the 16S rRNA sequences
Detection and identification of bioactive antimicrobial metabolites by GC-MS analysis
GC-MS analysis of the diethyl ether extract from SZ33 identified 26 compounds (Figure 6). Table 2 using methanol and acetonitrile. The majority of the identified compounds exhibited antimicrobial, antifungal, and antibacterial activities, targeting a variety of receptors and employing diverse mechanisms of action. Notably, compounds such as β-carotene-3,3′-diol,(3R,3’R)-all-trans, 2H-pyran, 2-(7-heptadecynyloxy) tetrahydro, astaxanthin, and aurin, etc., which are known for their antimicrobial properties, are detailed in Table 3.
Table (2):
List of distinct compounds identified from the GC-MS analysis of the crude extract
| Retention Time (min) | Identified Compound | Molecular Formula (MF) | Molecular weight (MW) | Peak Area % |
|---|---|---|---|---|
| GC-MS of crude sample using methanol | ||||
| 18.825 | 10,13-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 4.684 | cis-5,8,11,14,17-Eicosapentaenoic acid | C20H3002 | 302 | 18.94 |
| 4.684 | 10-Heptadecen-8-ynoic acid, methyl ester, (E)- | C18H3002 | 278 | 18.94 |
| 18.825 | 12,15-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 18.825 | 8,11-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 19.116 | Rhodopin | C40H580 | 554 | 32.79 |
| 4.864 | Doconexent | C22H3202 | 328 | 18.94 |
| 18.825 | Methyl 10,12-pentacosadiynoate | C26H4402 | 388 | 42.21 |
| 18.825 | Cholesta-8,24-dien-3-ol, 4-methyl-, (3β,4α)- | C28H460 | 398 | 42.21 |
| GC-MS of crude sample using acetonitrile | ||||
| 3.681 | .psi.,.psi.-Carotene, 1,1′,2,2′-tetrahydro-1, 1′-dimethoxy- | C40H6402 | 600 | 100 |
| 3.681 | Astaxanthin | C40H5204 | 596 | |
| 3.681 | Rhodopin | C40H580 | 554 | |
| 3.681 | Carda-16,20(22)-dienolide, 3-[(6-deoxy-3, 4-O-methylenehexopyranos-2-ulos-1-yl)oxy] -7,8-epoxy-11,14-dihydroxy-12-oxo-, (3β,5β,7β,11α) | C30H36011 | 572 | |
| 3.681 | β-Carotene-3,3′-diol, (3R,3’R)-all-trans- | C40H5602 | 568 | |
| GC-MS of extract using methanol | ||||
| 3.584 | 8,11-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 10.78 |
| 19.116 | Rhodopin | C40H580 | 554 | 32.72 |
| 19.116 | .psi.,.psi.-Carotene, 1,1′,2,2′-tetrahydro-1, 1′-dimethoxy- | C40H6402 | 600 | 2.21 |
| 19.116 | 10-Heptadecen-8-ynoic acid, methyl ester, (E)- | C18H3002 | 278 | 2.21 |
| 19.116 | 9-Octadecenoic acid, (2-phenyl-1,3-dioxolan -4-yl)methyl ester, trans- | C28H44O4 | 444 | 2.21 |
| 18.857 | 9-Octadecenoic acid, (2-phenyl-1,3 -dioxolan-4-yl)methyl ester, cis- | 444 | 4.61 | |
| 19.116 | 10,13-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 18.857 | cis-5,8,11,14,17-Eicosapentaenoic acid | C20H3002 | 302 | 32.79 |
| 4.669 | Cholesta-8,24-dien-3-ol, 4-methyl-, (3β,4α)- | C28H460 | 398 | 1.35 |
| 18.825 | 12,15-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 11.682 | 2H-Pyran, 2-(7-heptadecynyloxy)tetrahydro- | C22H40O2 | 336 | 43.70 |
| 3.584 | Methyl 10,12-pentacosadiynoate | C26H4402 | 388 | 10.78 |
| 11.682 | Methyl 9,11-octadecadiynoate | C19H30O2 | 290 | 43.70 |
| 4.863 | N-(5-Hydroxy-2-oxo-5-phenyl-1-aza-bicyclo [4.2.0]oct-3-yl)carbamic acid, benzyl ester | C21H22N2O4 | 366 | 2.63 |
| 4.863 | Morphinan-4,5-epoxy-3,6-di-ol, 6-[7-nitrobenzofurazan-4-yl]amino | C26H27N5O6 | 505 | 2.63 |
| 11.682 | Z,Z,Z-1,4,6,9-Nonadecatetraene | C19H32 | 260 | 43.70 |
| GC-MS of extract using Acetonitrile | ||||
| 3.568 | Phenyl-β-D-glucoside | C12H1606 | 256 | 34.72 |
| 3.568 | Aurin | C19H14O3 | 290 | 34.72 |
| 3.568 | 1,4,4a,5,8,8a-Hexahydronaphthalene, 4a, 8a-dicyano-1,8:4,5-diethano-1,4:5,8-diepoxy | C16H12N2O2 | 264 | 34.72 |
| 3.568 | Benzene, [(1-methyl-2-propenyl)oxy]- | C10H120 | 148 | 34.72 |
| 18.825 | 8,11-Octadecadiynoic acid, methyl ester | C19H3002 | 290 | 42.21 |
| 19.116 | .psi.,.psi.-Carotene, 1,1′,2,2′-tetrahydro-1, 1′-dimethoxy- | C40H6402 | 600 | 32.79 |
| 11.909 | Astaxanthin | C40H5204 | 596 | 37.59 |
| 11.909 | Carda-16,20(22)-dienolide, 3-[(6-deoxy-3, 4-O-methylenehexopyranos-2-ulos-1-yl)oxy]-7, 8-epoxy-11,14-dihydroxy-12-oxo-, (3β,5β,7β,11α) | C30H36011 | 572 | 37.59 |
| 11.909 | β-Carotene-3,3′-diol, (3R,3’R)-all-trans- | C40H5602 | 568 | 37.59 |
Table (3):
Antimicrobial activity Profile of the compounds identified in the Crude Extract
No. |
GC compounds |
Property |
Classification |
Reference |
|---|---|---|---|---|
1 |
2-(7-Heptadecynyloxy)tetrahydro-2H-pyran |
– |
– |
50 |
2 |
Z, Z, Z-1,4,6,9-Nonadecatetraene |
antibacterial, antifungal |
Hydrocarbon |
51 |
3 |
Rhodopin |
antioxidant |
antioxidant |
9 |
4 |
Phenyl beta-D-glucopyranoside |
antibacterial, antifungal |
Glucoside |
52 |
5 |
N-(5-Hydroxy-2-oxo-5-phenyl-1- aza-bicyclo [4.2.0]oct-3-yl)carbamic acid, benzyl ester |
antibacterial, antifungal |
Carbamic acid derivative |
53 |
6 |
Methyl 9,11-octadecadiynoate |
antibacterial |
Ester |
54 |
7 |
cis-5,8,11,14,17-Eicosapentaenoic acid (2) |
antibacterial |
Fatty acid |
55 |
8 |
beta-Carotene-3,3′-diol, (3R,3’R)-all-trans (Zeaxanthin) |
antioxidant |
Carotenoid |
56 |
9 |
beta, epsilon-Carotene-3,3′-diol, (3R,3’R,6’R) (Lutein) |
antimicrobial, antioxidant |
Carotenoid |
57 |
10 |
Benzene, [(1-methyl-2-propenyl) oxy]- |
– |
– |
– |
11 |
Aurin |
antibacterial |
Phenol derivative |
– |
12 |
Astaxanthin |
antimicrobial, antioxidant |
Carotenoid |
58 |
13 |
10,13-Octadecadiynoic acid, methyl ester |
antimicrobial |
Ester |
59 |
14 |
8,11-Octadecadiynoic acid, methyl ester |
antibacterial |
Ester |
60 |
15 |
1,4,4a,5,8,8a-Hexahydronaphthalene, 4a,8a-dicyano-1,8:4,5-diethano-1,4:5,8-diepoxy |
antibacterial |
Naphthalene derivative |
61 |
16 |
Benzene, [(1-methyl-2-propenyl) oxy |
antibacterial, antifungal |
Benzene derivative |
– |
17 |
Carda-16,20(22)-dienolide, 3-[(6-deoxy-3,4-O- methylenehexopyranos-2-ulos-1-yl) oxy]-7,8-epoxy-11,14-dihydrox |
antimicrobial |
Cardanol derivative |
– |
18 |
Methyl 10,12-pentacosadiynoate |
antibacterial, antifungal |
Ester |
62 |
19 |
Morphinan-4,5-epoxy-3,6-di-ol,6-[7-nitrobenzofurazan-4-yl] amino |
antibacterial |
Morphinan derivative |
63 |
20 |
2H-Pyran, 2-(7-heptadecynyloxy) tetrahydro |
antibacterial |
Pyran derivative |
64 |
21 |
9-Octadecenoic acid, (2-phenyl-1,3-dioxolan-4-yl) methyl ester, trans |
antibacterial |
Ester |
65 |
22 |
psi.,psi.-Carotene, 1,1′,2,2′-tetrahydro-1,1′-dimethoxy |
antioxidant |
Carotenoid |
66 |
23 |
Cholesta-8,24-dien-3-ol, 4-methyl-, (3β,4α)- |
antifungal |
Alcohol |
66 |
FTIR spectral analysis
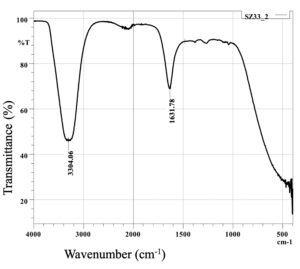
The assay of SZ33 (Figure 7) displayed intense absorption bands at 3304.06 and 1631.78 cm-1, suggesting normal “polymeric” OH stretch being alcohol compound and amide carbonyl compound respectively.24
Docking analysis
The molecular docking analysis of 14 natural compounds (identified by GC-MS analysis) against the selected PBP targets is summarized in Table 4. The reported binding affinity scores, measured in kcal/mol, demonstrated substantial interactions between numerous natural compounds of S. rochei SZ33 and the PBPs. From this set, the natural compounds exhibiting the maximum binding affinities were shortlisted for detailed investigation, alongside a commercially available antibiotic for comparative purposes (Table 5).
Table (4):
Binding affinities of GC compounds with PBPs
GC compounds |
Binding Affinity (PBP3 S. aureus) |
Binding Affinity (PBP2 P. aeruginosa) |
Binding Affinity (PBP3 K. pneumoniae) |
Binding Affinity (PBP3 S. pneumoniae) |
|---|---|---|---|---|
2-(7-Heptadecynyloxy) tetrahydro-2H-pyran |
-4.6 |
-4.6 |
-5.8 |
-4.9 |
Z,Z,Z-1,4,6,9-Nonade-catetraene |
-4.4 |
-4.6 |
-6 |
-5.6 |
Rhodopin |
-8.2 |
-8 |
-8.3 |
-7.7 |
Phenyl beta-D-gluco-pyranoside |
-8.3 |
-7 |
-7 |
-7.4 |
Table (5):
Binding affinities of commercial antibiotics with PBPs
Organism |
Receptor |
Commercially available drug |
Binding Affinity |
|---|---|---|---|
S. aureus |
PBP3 |
Oxacillin |
-8.9 |
P. aeruginosa |
PBP2 |
Doripenem |
-7.4 |
S. pneumoniae |
PBP3 |
Amoxicillin |
-7.5 |
K. pneumoniae |
PBP3 |
Imipenem |
-6.7 |
S. aureus
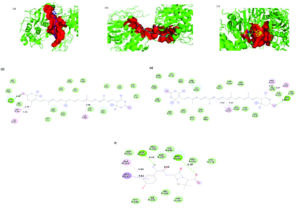
Zeaxanthin, a carotenoid also known as beta-carotene-3,3′-diol, (3R,3’R)-all-trans, exhibited the highest binding affinity for S. aureus PBP3, with a docking score of -9.9 kcal/mol. The interaction was primarily stabilized by alkyl bonds with TYR275B, TYR278B, ILE381B, PRO500B, LYS494A and VAL493A, as well as a hydrogen bond at GLN548A.
Lutein, or beta, epsilon-carotene-3,3′-diol, (3R,3’R,6’R), displayed a binding affinity of -9.7 kcal/mol, forming alkyl and π-alkyl interactions with LEU256B, TYR275B, TYR278B, ILE381B, and ILE507A. Lutein exhibited similar binding behaviour, sharing key interactions with zeaxanthin.
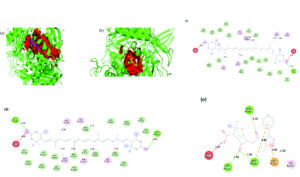
The reference antibiotic, oxacillin, demonstrated a lower binding affinity of -8.9 kcal/mol. Its interaction network included π-anion bonds at GLU255A and hydrogen bonds with TYR275A, ASN487B, and ARG504B. A π-alkyl bond at LYS273A was also observed, although an unfavorable interaction with ARG483B was present. Structural analysis revealed distinct binding pockets for these compounds were different from oxacillin, with zeaxanthin and lutein sharing a 288-atom pocket, in contrast to oxacillin’s smaller pocket of 192 atoms. 3D representation of the ligand-binding pocket of Staphylococcus aureus PBP3 with zeaxanthin (blue) and lutein (pink) and oxacillin (yellow). 2D interaction diagram of Staphylococcus aureus PBP3 with zeaxanthin, lutein, and oxacillin (Figure 8).
Figure 8. 3D representation of the ligand-binding pocket of Staphylococcus aureus PBP3 with a) Zeaxanthin (blue) and lutein (pink) and b) Oxacillin (yellow). 2D interaction diagram of Staphylococcus aureus PBP3 with c) Zeaxanthin d) Lutein e) Oxacillin
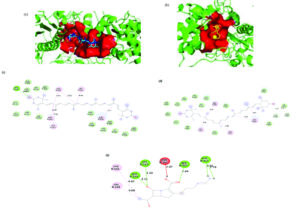
P. aeruginosa
Astaxanthin displayed the highest binding affinity of -9.4 kcal/mol for P. aeruginosa PBP2, with the interaction stabilized by a 264-atom binding pocket. Key stabilizing interactions included alkyl and π-alkyl bonds with residues VAL248B, ILE476B, PRO488B, and LYS482B, as well as hydrogen bonds with ARG74A and ASN75A. Zeaxanthin also exhibited a strong binding affinity of -9.2 kcal/mol for P. aeruginosa PBP2, occupying a larger 310-atom binding pocket. Both compounds had alternate binding sites in contrast to doripenem. The core interactions were alkyl and π-alkyl bonds with TYR364A and ILE447A, along with a π-sigma bond with TYR441A. Conventional hydrogen bonds with SER592A and TYR360A, as well as extensive van der Waals forces, further stabilized this complex.
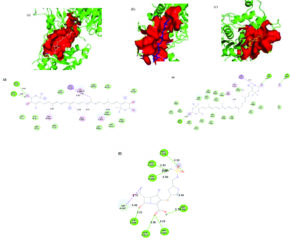
In contrast, the reference antibiotic doripenem exhibited a substantially lower binding affinity of -7.4 kcal/mol and engaged a smaller 180-atom binding pocket, with key interactions involving hydrogen bonds at ARG365A, ARG369A, THR541A, GLN452A, SER384A, ASP386A, and TYR390A. Both compounds bound to different pockets than doripenem. 3D representation of the ligand-binding pocket of Pseudomonas aeruginosa PBP2 with Astaxanthin (wheat), zeaxanthin (blue), and doripenem (yellow). 2D interaction diagram of Pseudomonas aeruginosa PBP2 with astaxanthin, zeaxanthin, and doripenem (Figure 9).
Figure 9. 3D representation of the ligand-binding pocket of Pseudomonas aeruginosa PBP2 with a) Astaxanthin (wheat) b) Zeaxanthin (blue) c) Doripenem (yellow). 2D interaction diagram of Pseudomonas aeruginosa PBP2 with d) Astaxanthin e) Zeaxanthin f) Doripenem
S. pneumoniae
Lutein demonstrated the strongest binding affinity of -8.3 kcal/mol for S. pneumoniae PBP, with the 265-atom binding pocket stabilized by alkyl and π-alkyl interactions at HIS270A, PRO275A, and TYR276A, as well as a hydrogen bond at THR221A. Zeaxanthin followed closely with a binding score of -8.1 kcal/mol and a 242-atom pocket, forming alkyl bonds at TYR305A, PHE344A, and LYS369A, along with hydrogen bonding at GLN306A. The reference antibiotic amoxicillin had a lower binding affinity of -7.5 kcal/mol, engaging a smaller 167-atom binding pocket. The interactions formed by amoxicillin were relatively weaker, including a π-alkyl bond at ALA154D, a π-sigma bond at LEU147D, and hydrogen bonds at GLN152D and ARG143D. 3D representation of the ligand-binding pocket of Streptococcus pneumoniae PBP3 with zeaxanthin (blue), lutein (cyan) and amoxicillin (yellow). 2D interaction diagram of Streptococcus pneumoniae PBP3 with zeaxanthin, lutein and amoxicillin (Figure 10).
Figure 10. 3D representation of the ligand-binding pocket of Streptococcus pneumoniae PBP3 with (a) Zeaxanthin (blue), (b) Lutein (cyan) and (c) Amoxicillin (yellow). 2D interaction diagram of Streptococcus pneumoniae PBP3 with (d) Zeaxanthin (e) Lutein (f) Amoxicillin
K. pneumoniae
For the K. pneumoniae PBP3 target, zeaxanthin showed the highest binding affinity at -9.5 kcal/mol, occupying a sizeable 300-atom binding pocket. The complex was stabilized by a network of alkyl interactions with ALA242A, ALA242B, ARG246B, LEU243B, and ARG559B, as well as a hydrogen bond with ASN249A. The carotenoid lutein showed a similar binding affinity of -9.3 kcal/mol and shared the same binding pocket and interaction profile. In contrast, the reference antibiotic imipenem demonstrated a substantially lower binding affinity of -6.7 kcal/mol, with a smaller 152-atom binding pocket. Its binding interactions, including alkyl and hydrogen bonds with residues such as SER307B and LYS310B, PHE417B, SER359B. 3D representation of the ligand-binding pocket of Klebsiella pneumoniae PBP3 with zeaxanthin (blue), lutein (cyan), and imipenem (yellow). 2D interaction diagram of Klebsiella pneumoniae PBP3 with zeaxanthin, lutein, amoxicillin (Figure 11).
Hospital-acquired infections (HAIs) are rapidly increasing on a global scale, driven by factors such as inadequate facility maintenance, ineffective infection control practices, resistance development in pathogens, increasing cross and co-infections, etc. The magnitude of this problem is substantial and warrants immediate attention. Our study targets the nosocomial lower respiratory tract bacterial pathogens which are frequently reported to cause severe infections. K. pneumoniae is a common opportunistic pathogen that causes nosocomial infections, particularly in immunocompromised patients, leading to fatal disease manifestations such as meningitis, chronic pneumonia, necrotizing fasciitis, etc.25,26 S. pneumoniae-induced pneumococcal community-acquired pneumonia (pCAP) is a major cause of LRTIs in the elderly, often leading to disorientation, confusion, and exacerbation of pre-existing conditions; and morbidity and mortality among children. Although, S. pyogenes rarely causes LRTIs, it can lead to severe and fatal pneumonias when infects. S. aureus is the most common Gram-positive bacterium in health care systems. Therefore, the present study was focused around four key bacterial pathogens, viz. S. pyogenes, S. aureus, S. pneumoniae, and K. pneumoniae, which are commonly implicated in nosocomial lower respiratory tract infections.27
A major risk factor associated with these pathogens is, the rise in MDRs which undermines various strategies aimed at mitigating nosocomial infections.28,29 For instance, S. aureus, shows a resistance rate of nearly 83% to Penicillin G. Of the several reasons attributed to the inefficacy or resistance seen in commercially used beta-lactam antibiotics, a suboptimal binding affinity (between -6 to -9 kcal/mol) along with a small binding pocket size has been amongst the most prevalent. Beta-lactam antibiotics, such as penicillin, target PBPs, which are required for bacterial cell wall formation. However, when the binding affinity of these antibiotics falls between -6 and -9 kcal/mol, the interaction may be insufficient to effectively block the PBPs, resulting in diminished effectiveness. Furthermore, the size restrictions of the PBP binding pocket can prohibit bigger or structurally modified antibiotics from binding well, lowering the drug’s efficacy.30 Among macrolides, erythromycin has been documented with the highest resistance rate (51.9%) since 2019, with a notable increase in levofloxacin resistance, ranging from 28.1% to 54.9%. Additionally, K. pneumoniae, a Gram-negative bacterium demonstrated an average resistance rate of 70% to cephalosporin and penicillin. The increasing prevalence of MDR pathogens accentuates necessity for the discovery of novel antimicrobial compounds with improved antimicrobial structural capabilities.28
Actinomycetes have demonstrated as a powerful source of antibiotics in the last few decades. However, much of the Actinomycetes studied have been soil-borne, the marine counterparts are underexplored. Actinomycetes found in marine environments are abundant in bioactive compounds exhibiting unique properties. Unlike their terrestrial kin, marine Actinomycetes have developed unique metabolic pathways to adapt to the marine environment. This in turn leads to the production of many unique compounds with distinct structures and functions. These microorganisms thrive in diverse marine habitats, including sediments, seawater, and marine invertebrates, contributing significantly to the biodiversity of marine ecosystems. Marine Actinomycetes derived compounds are highly used in biotechnology and drug discovery.31-33 Many novel promising bioactive metabolites such as, streptopyrrolidine, cyclo-(l-Pro-l-Met), and thiocoraline were discovered from marine Actinomycetes in a recent study.7,34,35 In this regard, our study successfully isolated many marine Actinomycetes and also identified a potential Actinomycete strain, S. rochei SZ33. This strain, originating from a relatively underexplored marine habitat, demonstrated significant antimicrobial activity, effectively inhibiting the target pathogens by the production of antimicrobial metabolites. Utilising S. rochei SZ33 as a source of bioactive compounds warrants further investigation for exploring all possible therapeutic applications. A recent study highlighted the antimicrobial activity of Streptomyces rochei SUN35, against MRSA, Fusarium solani, Aspergillus fumigatus, Pseudomonas aeruginosa, and Penicillium chrysogenum under in vitro conditions.36 This emphasizes the potential of S. rochei species in combating a wide range of pathogenic microorganisms.
To improve the production of the secondary metabolites, and the consequential heightened inhibition of target pathogens, media composition and physical conditions were optimized.37 Optimization studies indicated that the highest production of antimicrobial secondary metabolites occurred at 25 °C and pH 6, with a medium containing starch as the carbon source, yeast extract as the nitrogen source, and a salt concentration of 2%. These optimized conditions are easily achievable, and beneficial for the large-scale production of antimicrobials employing S. rochei SZ33. Post optimization, the inhibition was increased by two-fold against the tested pathogens. Metabolite analysis using GC-MS identified several bioactive compounds,38 including β-carotene-3,3′-diol, (3R,3’R)-all-trans, 2H-pyran,39 2-(7-heptadecynyloxy) tetrahydro, and astaxanthin,4,17 all of which demonstrated antimicrobial properties, while aurin compounds were identified as LsrK inhibitors.40 The observed inhibition of the target pathogens was significantly higher than that of the standard antibiotic, might well be a consequence of inherent synergism present between the metabolites of the crude extract.
The antimicrobial activity of astaxanthin-alpha tocopherol has been shown to be due to its ability to disrupt bacterial cell membranes.41 Previous studies have demonstrated that secondary metabolites produced by marine Actinomycetes exhibit superior antimicrobial activity against respiratory pathogens, including S. pneumoniae, Mycobacterium tuberculosis, K. pneumoniae, and S. aureus, which are commonly associated with pneumonia and other respiratory infections. Marine ecosystems are generally isolated and diversified, with harsh circumstances such as high salinity, fluctuating temperatures, and tremendous pressure. To survive this, the said microorganisms have developed the ability to synthesize new chemical substances. The chemical diversity of secondary metabolites in marine Actinomycetes is primarily driven by the evolutionary pressure to compete with other microorganisms in the competitive environment. This explains why metabolites derived from marine actinomycetes possess distinct inhibitory modes of action, circumventing the resistance mechanisms that many infections have acquired against standard antibiotics.31
Several bioactive compounds derived from marine Actinomycetes have shown significant potential as therapeutic agents for treating respiratory infections. For instance, Streptomyces sp. 6921, isolated from marine sediments in Mauritius, secretes a range of antibacterial compounds, including C-glycosides himalomycins, anthraquinones, fridamycin E, and chromophore.42 Additionally, a marine Streptomyces sp. 182SMLY was shown to produce streptophenazines, which inhibited (MRSA). Another strain, S. psammoticus, produced a novel polyketide tetracycline analog, SBR-22, which also demonstrated potent activity against MRSA.43
The proliferation of multidrug-resistant organisms is severely undermining the treatment using existing drugs.44,45 This accentuates the urgent need for the development of novel drugs and therapeutic strategies that are effective against antibiotic-resistant pathogens.46-49 Crude extracts, which contain a diverse array of active metabolites, offer a promising alternative to traditional single or combination drug therapies, as their complex composition may provide enhanced efficacy through synergistic interactions among the bioactive compounds.
The compounds might be new antimicrobial agents that mitigate antibiotic resistance. Several of these compounds bind strongly to bacterial enzymes, disrupting processes such as cell wall synthesis, DNA replication, and enzyme activity. In addition to hydrophobic interactions, hydrogen bonds, and a double-stacked structure, these molecules are able to bind to microbial targets with exceptional stability. By addressing both traditional pathways and new strategies to combat antibiotic-resistant pathogens, this study reveals a promising mechanism of action. Integrating these molecular insights with marine actinomycetes underscores their potential as a source of innovative antimicrobial therapies.
In this regard, the compounds identified by GC-MS were further analyzed by docking them with PBPs as molecular targets for all the pathogens associated with nosocomial LRTIs. PBPs, the essential enzymes in bacterial cell wall synthesis, are targeted by antibiotics such as Penicillin and Carbapenems, which inhibit their function and lead to bacterial cell death. However, the extensive use of these antibiotics has driven the evolution of drug resistance and multidrug-resistant pathogens, reducing the effectiveness of these drugs. This in turn has led to PBPs being frequently employed in docking studies for nosocomial (hospital-acquired) infections as it is the key target of beta-lactam antibiotics. PBPs being elemental for bacterial cell wall formation, and their blockage causes cell death. Nosocomial bacteria predominantly develop resistance to beta-lactam antibiotics by altering PBPs, thereby reducing the antibiotic’s affinity for these enzymes. This makes PBPs an ideal target for research into finding new therapeutic targets.
This study provides compelling evidence for the potential of naturally derived compounds as potent inhibitors of PBPs across a range of pathogenic bacteria. The findings demonstrate that these natural compounds exhibit superior binding affinities compared to conventional synthetic antibiotics. The GC-MS analysis identified 28 distinct compounds in the crude extract, many of which displayed notable antimicrobial capabilities. Compounds such as zeaxanthin, lutein, and astaxanthin exhibited exceptionally strong interactions with PBPs, compared to commercial antibiotics, suggesting their promise as alternatives or adjuncts in combating antibiotic resistance.
Zeaxanthin and lutein demonstrated strong binding affinities to the PBP3 enzyme of S. aureus, with binding energies of -9.9 kcal/mol and -9.7 kcal/mol, respectively. The antibacterial mode of action, in carotenoids ranges from dissolution of the cell membrane, disruption of cell wall synthesis and alterations in the immune response. Their interactions with key amino acid residues, such as TYR275B and GLN548A, suggest a capacity for competitive inhibition, which could effectively mitigate resistance mechanisms in this pathogen. Furthermore, these carotenoids also exhibited robust binding to the PBP3 enzymes of P. aeruginosa and K. pneumoniae, with binding affinities of -9.5 kcal/mol and -9.3 kcal/mol, respectively. The antimicrobial actions of these compounds include enhancing oxidative stress in bacteria and inhibiting biofilm formation.
Astaxanthin, another naturally derived compound, exhibited a binding affinity of -9.4 kcal/mol for the PBP2 enzyme of P. aeruginosa, suggesting its potential as an alternative to antibiotics like doripenem. Astaxanthin is known for its potent antioxidant properties, which can reduce inflammation and enhance the immune response against bacterial infections. Furthermore, this compound has the capacity to disrupt bacterial cell membranes, subsequently resulting in cell death. The analysis also revealed that the carotenoids lutein and zeaxanthin maintained significant binding affinities of -8.3 kcal/mol and -8.1 kcal/mol, respectively, against the PBP targets of S. pneumoniae. These naturally occurring compounds have demonstrated the ability to inhibit cell wall synthesis and interfere with bacterial metabolic pathways, offering an effective alternative strategy to control S. pneumoniae infections compared to traditional antibiotics like amoxicillin. In addition to substantial differences in binding affinities from commercial antibiotics against PBPs, the naturally derived compounds identified by GC-MS also displayed an alternate binding pocket along with a significant difference in binding pocket sizes. Larger or more open binding pockets may allow for easier entry of larger molecules or those with bulky groups. If the size of the binding pocket is too small, steric hindrance can prevent effective binding of larger compounds, reducing their antibacterial activity. A bigger pocket may also provide more structural flexibility, allowing for the binding of a wider spectrum of substrates or inhibitors. Larger pockets frequently offer more possibilities for interaction between the antimicrobial drug and the target protein. Binding of antimicrobial agents to alternate sites other than the active sites could lead to inhibition of the enzyme without changing the active site.
In addition to the carotenoids, zeaxanthin, lutein, and astaxanthin, several other natural compounds, including carbamic acid derivatives, rhodopin, phenyl-beta-D-glucoside, and aurin, have demonstrated the potential to inhibit synthesis of cell wall by binding to PBPs across various pathogenic strains, as evidenced by molecular docking analyses. Furthermore, certain other naturally derived compounds, such as fatty acid methyl esters and polyunsaturated fatty acids, may exert antimicrobial effects through alternative mechanisms, which may involve increasing membrane fluidity and permeability, promoting lipid peroxidation, or disrupting membrane integrity, ultimately leading to bacterial cell death. There is limited direct evidence for antibacterial activity from compounds such as lutein, zeaxanthin, astaxanthin, etc. However, studies have looked at their ability to work synergistically either with other similar compounds or with synthetic antibiotics.42
The results from the study perhaps favor the argument of using crude extracts as superior antimicrobials. Combining chemicals with varied binding affinities, and allosteric binding pockets could improve antibacterial activity, particularly against resistant species. This method reduces the possibility of resistance building.43
This study not only addresses traditional antimicrobial pathways but also explores novel strategies for combating antibiotic-resistant pathogens, revealing a promising mechanism of action. Integrating these molecular insights underscores the potential of marine Actinomycetes as a valuable reservoir of novel antimicrobial therapies.
Further investigations are necessary to have a more comprehensive understanding the potential of S. rochei SZ33 for the large-scale production of antimicrobial metabolites suitable for industrial applications. The results of this work have far reaching impact in terms of developing effective treatment alternatives for drug resistant pathogens in general. Marine Actinomycetes are earning their place as potent source of novel therapeutic agents, offering a treasure trove of possibilities in the search for potential antimicrobials.
The robust binding capabilities and multifaceted antimicrobial mechanisms of these natural compounds underscore their promising potential as novel therapeutic agents. Additional studies will be essential to evaluate their in vivo efficacy, pharmacokinetic characteristics, safety profiles, and potential synergistic interactions with existing antibiotics. Furthermore, optimizing delivery methods will be crucial in translating these encouraging findings into viable clinical applications to address the growing challenge of antibiotic resistance.
Additional file: Additional Figure S1.
ACKNOWLEDGMENTS
The authors would like to thank Dayananda Sagar University for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SZ, ASR, MPP and BA collected the resources and perofmed investigation. AF, SZ, ASR, MPP and BA designed the experiments. SZ, ASR, MPP and BA performed formal analysis. MAS, MG, SZ, ASR, MPP, AN and BA performed software work. SZ, ASR, MPP, AN and BA performed visualization and data validation. SSM performed supervision. SZ, ASR, MPP, AN, AF and BA wrote, reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available in the GenBank repository, Accession No: OQ726229.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Bucataru A, Balasoiu M, Ghenea AE, et al. Factors Contributing to Surgical Site Infections: A Comprehensive Systematic Review of Etiology and Risk Factors. Clin Pract. 2024;14(1):52-68.
Crossref - Hrynyshyn A, Simões M, Borges A. Biofilms in Surgical Site Infections: Recent Advances and Novel Prevention and Eradication Strategies. Antibiotics. 2022;11(1):69.
Crossref - Elias C, Nunes MC, Saadatian-Elahi M. Epidemiology of community-acquired pneumonia caused by Streptococcus pneumoniae in older adults: a narrative review. Curr Opin Infect Dis. 2024;37(2):144-153.
Crossref - Seray Topçu, Mine Gül Şeker. In Vitro Antimicrobial Effects and Inactivation Mechanisms of 5,8-Dihydroxy-1,4-Napthoquinone. Antibiotics. 2022;11(11):1537-1537.
Crossref - Santella B, Serretiello E, De Filippis A, et al. Lower Respiratory Tract Pathogens and Their Antimicrobial Susceptibility Pattern: A 5-Year Study. Antibiotics. 2021;10(7):851.
Crossref - Antimicrobial Resistance Collaborators. Global Burden of Bacterial Antimicrobial Resistance in 2019: a Systematic Analysis. The Lancet. 2022;399(10325):629-655.
Crossref - Krishna PS, Sudha S, Reddy KA, et al. Studies on wound healing potential of red pigment isolated from marine Bacterium Vibrio sp. Saudi J Biol Sci. 2017;26(4):723-729.
Crossref - Jagannathan SV, Manemann EM, Rowe SE, Callender MC, Soto W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar Drugs. 2021;19(7):365.
Crossref - Youssef AMM, Maaty DAM, Al-Saraireh YM. Phytochemical Analysis and Profiling of Antioxidants and Anticancer Compounds from Tephrosia purpurea (L.) subsp. apollinea Family Fabaceae. Molecules. 2023;28(9):3939.
Crossref - Abdelfattah MS, Elmallah MIY, Hawas UW, Abou El-Kassema LT, Eid MAG. Isolation and characterization of marine-derived actinomycetes with cytotoxic activity from the Red Sea coast. Asian Pac J Trop Biomed. 2016;6(8):651-657.
Crossref - Al-Ansari M, Kalaiyarasi M, Almalki MA, Vijayaraghavan P. Optimization of medium components for the production of antimicrobial and anticancer secondary metabolites from Streptomyces sp. AS11 isolated from the marine environment. J King Saud Univ Sci. 2020;32(3):1993-1998.
Crossref - Mohseni M, Hamed Norouzi, Javad Hamedi, Roohi A. Screening of Antibacterial Producing Actinomycetes from Sediments of the Caspian Sea. Int J Mol Cell Med. 2013;2(2):64-71.
- Dar MS, Ahmad I. Screening and evaluation of antibacterial active strains of Actinomycetes isolated from Northern Indian soil for biofilm inhibition against selected ESKAPE pathogens. J Umm Al-Qura Univ Appll Sci. 2025;11:340-355
Crossref - Jorgensen JH, Turnidge JD. Susceptibility Test Methods: Dilution and Disk Diffusion Methods. In: Jorgensen JH, Carroll KC, Funke G, Pfaller MA, Landry ML, Richter SS, Warnock DW, Richter SS, Patel JB, eds., Manual of Clinical Microbiology, 11th Edition. ASM Press eBooks. 2015:1253-1273.
Crossref - Mondal H, Thomas J. Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens. Antibiotics. 2022;11(11):1546.
Crossref - Mohan YSYVJ, Sirisha B, Haritha R, Ramana T. Selective screening, isolation and characterization of antimicrobial agents from marine actinomycetes. Int J Pharm Pharm Sci. 2013;5:443-449.
- Song P, Wang Z, Sun X, et al. Screening, Identification, and Fermentation of a Biocontrol Strain against Peony Southern Blight and Extraction of Secondary Metabolites. Agriculture. 2024;14(6):833.
Crossref - Kalyani BS, Krishna PS, Sreenivasulu K. Screening and identification of novel isolate Streptomyces sp., NLKPB45 from Nellore costal region for its biomedical applications. Saudi J Biol Sci. 2019;26(7):1655-1660.
Crossref - Delgado-Vargas F, Jiménez AR, Paredes-López O. Natural Pigments: Carotenoids, Anthocyanins, and Betalains — Characteristics, Biosynthesis, Processing, and Stability. Crit Rev Food Sci Nutr. 2000;40(3):173-289.
Crossref - Ibraheam IA, Hussein HM, Hameed IH. Cyclamen persicum: Methanolic Extract Using Gas Chromatography-Mass Spectrometry (GC-MS) Technique. International Journal of Pharmaceutical Quality Assurance. 2017;8(4):200-213
Crossref - Kamarudheen N, Rao KVB. Fatty acyl compounds from marine Streptomyces griseoincarnatus strain HK12 against two major bio-film forming nosocomial pathogens; an in vitro and in silico approach. Microb Pathog. 2019;127:121-130.
Crossref - Al-Zoreky NS. Antimicrobial activity of pomegranate (Punica granatum L.) fruit peels. Int J Food Microbiol. 2009;134(3):244-248.
Crossref - Maskey RP, Helmke E, Laatsch H. Himalomycin A and B: isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J Antibiot. 2003;56(11):942-949.
Crossref - Nandiyanto ABD, Oktiani R, Ragadhita R. How to read and interpret FTIR spectroscope of organic material. Indonesian Journal of Science and Technology. 2019;4(1):97-118.
Crossref - Flores-Díaz M, Monturiol-Gross L, Naylor C, Alape-Girón A, Flieger A. Bacterial sphingomyelinases and phospholipases as virulence factors. Microbiol Mol Biol Rev. 2016;80(3):597-628.
Crossref - Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071-1081.
Crossref - Santella B, Serretiello E, De Filippis A, et al. Lower Respiratory Tract Pathogens and Their Antimicrobial Susceptibility Pattern: A 5-Year Study. Antibiotics. 2021;10(7):851.
Crossref - Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global Geographic Trends in Antimicrobial Resistance: The Role of International Travel. J travel med. 2019;26(8):taz036.
Crossref - Sheykhsaran E, Abbasi A, Memar MY, et al. The role of Staphylococcus aureus in Cystic Fibrosis Pathogenesis and Clinico-Microbiological Interactions. Diagn Microbiol Infect Dis. 2024;109(3):116294-116294.
Crossref - Kumar KM, Anbarasu A, Ramaiah S. Molecular docking and molecular dynamics studies on β-lactamases and penicillin binding proteins. Mol BioSyst. 2014;10(4):891-900.
Crossref - Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiological Research. 2014;169(4):262-278.
Crossref - Chananan Ngamcharungchit, Nutsuda Chaimusik, Watanalai Panbangred, Jirayut Euanorasetr, Bungonsiri Intra. Bioactive Metabolites from Terrestrial and Marine Actinomycetes. Molecules. 2023;28(15):5915-5915.
Crossref - Yu J, Zhang L, Liu Q, Qi X, Ji Y, Beom SK. Isolation and characterization of actinobacteria from Yalujiang coastal wetland, North China. Asian Pac J Trop Biomed. 2015;5(7):555-560.
Crossref - Dhandapani R, Thangavelu S, Ragunathan L, Paramasivam R, Velmurugan P, Muthupandian S. Potential Bioactive Compounds from Marine Streptomyces sp. and Their In Vitro Antibiofilm and Antibacterial Activities Against Antimicrobial-Resistant Clinical Pathogens. Applied Biochemistry and Biotechnology. 2022;194(10):4702-4723.
Crossref - Siddharth S, Rai V R. Isolation and characterization of bioactive compounds with antibacterial, antioxidant and enzyme inhibitory activities from marine-derived rare actinobacteria, Nocardiopsis sp. SCA21. Microbial Pathogenesis. 2019;137:103775.
Crossref - Awad NM, Abdel-Hamied MR, Aboseidah AA, Aziz MA, Elshamy AI. Chemical profile, Antimicrobial and Antitumor Activities of the Streptomyces rochei SUN35 Strain. Egypt J Chem. 2023, 66(13), 2211-2218.
Crossref - Alam K, Mazumder A, Sikdar S, et al. Streptomyces: The biofactory of secondary metabolites. Front Microbiol. 2022;13:968053.
Crossref - Santos JD, Vitorino I, Reyes F, Vicente F, Lage OM. From Ocean to Medicine: Pharmaceutical Applications of Metabolites from Marine Bacteria. Antibiotics. 2020;9(8):455.
Crossref - I Sudaryadi, F Oktaweni, Pramono IE, Fatikasary KW, H Widiawati, Sutikno. Bioactive compound profile of propolis product from Beekeeping (Meliponiculture) in Turi Yogyakarta Indonesia. IOP Conf Ser Earth Environ Sci. 2023;(1200)012002.
Crossref - Linciano P, Cavalloro V, Martino E, et al. Tackling Antimicrobial Resistance with Small Molecules Targeting LsrK: Challenges and Opportunities. J Med Chem. 2020;63(24):15243-15257.
Crossref - Shanmugapriya K, Kim H, Saravana PS, Chun BS, Kang HW. Astaxanthin-alpha tocopherol nanoemulsion formulation by emulsification methods: Investigation on anticancer, wound healing, and antibacterial effects. Colloids and Surfaces B: Biointerfaces. 2018;172:170-179.
Crossref - Olszowy-Tomczyk M. Synergistic, antagonistic and additive antioxidant effects in the binary mixtures. Phytochemistry Reviews. 2020;19(1):63-103.
Crossref - György Abrusán, Marsh JA. Ligands and Receptors with Broad Binding Capabilities Have Common Structural Characteristics: An Antibiotic Design Perspective. 2019;62(21):9357-9374.
Crossref - Al-Dhabi NA, Esmail GA, Duraipandiyan V, Arasu MV, Salem-Bekhit MM. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles. 2015;20(1):79-90.
Crossref - Van Boeckel TP, Pires J, Silvester R, et al. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science. 2019;365(6459):eaaw1944.
Crossref - Ganesh M, Mohankumar M. Extraction and identification of bioactive components in Sida cordata (Burm.f.) using gas chromatography–mass spectrometry. J Food Sci Technol. 2017;54(10):3082-3091.
Crossref - Robinson TP, Bu DP, Carrique-Mas J, et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. 2016;110(7):377-380.
Crossref - Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an Emerging Battleground in Microbial Communities. Antimicrob Resist Infect Control. 2019;8(1).
Crossref - Sharma S, Fulke AB, Chaubey A. Bioprospection of marine actinomycetes: recent advances, challenges and future perspectives. Acta Oceanol Sin. 2019;38(6):1-17.
Crossref - Limboo KH, Singh B. Antibiotic potentiating effect of Bauhinia purpurea L. against multidrug resistant Staphylococcus aureus. Frontiers in microbiology. 2024;15.
Crossref - Jasna TK, Khaleel KM. GC-MS analysis of bioactive components of Kandelia candel (L.) druce. J Adv Sci Res. 2021 11(04 Suppl 9), 193-197.
- Hwang SJ, Lee HJ. Phenyl-b-d-Glucopyranoside exhibits anti-inflammatory activity in Lipopolysaccharide-Activated RAW 264.7 cells. Inflammation. 2015;38(3):1071-1079.
Crossref - Liang HJ, Cheng YJ, Wang LX, et al. Exploration of (3-benzyl-5-hydroxyphenyl)carbamates as new antibacterial agents against Gram-positive bacteria. Arch Pharm. 2020;353(3):e1900294.
Crossref - Nabi M, Tabassum N, Ganai BA. Phytochemical screening and antibacterial activity of Skimmia anquetilia N.P. Taylor and Airy Shaw: A first study from Kashmir Himalaya. Front Plant Sci. 2022;13.
Crossref - Desbois AP. Antimicrobial Properties of Eicosapentaenoic Acid (C20:5n−3). In: Se-Kwon K, Marine Microbiology. 2013:
Crossref - Bouyahya A, El Omari N, Hakkur M, et al. Sources, health benefits, and biological properties of zeaxanthin. Trends Food Sci Technol. 2021;118(Part A):519-538.
Crossref - Kusmiati, Endah Budi Ningsih, Indriati Ramadhani, Amir M. Antibacterial and antioxidant activity test of crude lutein extracted from sunflower (Helianthus annuus L.). AIP Conf Proc. 2021;2331(1):050001.
Crossref - Karpiński TM, Ożarowski M, Alam R, Łochyńska M, Stasiewicz M. What Do We Know about Antimicrobial Activity of Astaxanthin and Fucoxanthin? Mar Drugs. 2021;20(1):36.
Crossref - Alqahtani FY, Aleanizy FS, Mahmoud AZ, et al. Chemical composition and antimicrobial, antioxidant, and anti-inflammatory activities of Lepidium sativum seed oil. Saudi J Biol Sci. 2019;26(5):1089-1092.
Crossref - Wahyuni DK, Junairiah J, Rosyanti C, et al. Computational and in vitro analyses of the antibacterial effect of the ethanolic extract of Pluchea indica L. leaves. Biomed rep. 2024;21(4):137.
Crossref - Seray Topçu, Mine Gül Şeker. In Vitro Antimicrobial Effects and Inactivation Mechanisms of 5,8-Dihydroxy-1,4-Napthoquinone. Antibiotics. 2022;11(11):1537-1537.
Crossref - Venkatesh Kumar R, Gosipatala SB, Kumar R, et al. Characterization, Antioxidant, and Antimicrobial Properties of Mulberry Lattices. ACS Omega. 2023;8(50):47758-47772.
Crossref - Shreadah MA, El Moneam NMA, Al-Assar SA, Nabil-Adam A. Phytochemical and pharmacological screening of Sargassium vulgare from Suez Canal, Egypt. Food Sci Biotechnol. 2018;27(4):963-979.
Crossref - Georgiadis MP, Couladouros EA, Delitheos AK. Synthesis and antimicrobial properties of 2H-pyran-3(6H)-one derivatives and related compounds. Journal of pharmaceutical sciences. 1992;81(11):1126-1131.
Crossref - Abdel RSM, Kamel MA, Ali MA, et al. Comparative Study on the Phytochemical Characterization and Biological Activities of Azolla caroliniana and Azolla filiculoides: In Vitro Study. Plants. 2023;12(18):3229-3229.
Crossref - Kamel DG, Mansour AIA, El-diin MAHN, Hammam ARA, Mehta D, Abdel-Rahman AM. Using Rosemary Essential Oil as a Potential Natural Preservative during Stirred-like Yogurt Making. Foods. 2022;11(14):1993.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.