ISSN: 0973-7510
E-ISSN: 2581-690X
We aimed to develop a multiplex PCR (mPCR) assay utilizing the afaC gene for the detection of DAEC pathotypes and the blaTEM gene for the identification of ESBL production in clinical E. coli isolates along with an Internal Amplification Control (IAC) to rule out false negative test results. Monoplex PCR assays were first established for the afaC gene and blaTEM gene using 60 characterized E. coli isolates from various clinical samples. Subsequently, an mPCR assay was designed to detect both the genes simultaneously along with an IAC to rule out false negative reactions. The effectiveness of this assay was validated using 80 additional clinical isolates. The overall occurrence of DAEC in the study was found to be 0.7% (1/140). ESBL production was detected in 40.7% of the tested isolates, indicating a concerning prevalence of drug-resistant strains. This study emphasizes the value of an in-house mPCR assay as a crucial tool for simultaneously identifying DAEC and ESBL E. coli strains. The inclusion of an IAC in the PCR protocol bolstered the assay’s reliability. This innovation offers a vital resource for effective infection management and contributes to the comprehension of pathogenicity and resistance mechanisms in clinical E. coli isolates.
Diffusely Adherent Escherichia coli (DAEC), Extended Spectrum β-lactamase (ESBL), Internal Amplification Control (IAC), multiplex PCR (mPCR), afaC gene, blaTEM gene
Diarrheal diseases are major cause of illness and death, especially among infants and young children in developing countries.1,2 Among the various causes of diarrhoea, pathogenic strains of E. coli are frequently associated with both epidemic and endemic outbreaks.3,4 E. coli belongs to the “Enterobacterales” order and forms part of the harmless normal gut flora in humans and animals. However, certain strains of E. coli, known as diarrheagenic E. coli (DEC), are responsible for enteric infections. The major pathotypes of DEC include enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enterohemorrhagic E. coli (EHEC), and enteroinvasive E. coli (EIEC). 5 Each pathotype possesses different virulence attributes, causing diarrhoea through various mechanisms and leading to different clinical symptoms. The virulence genes can be transferred horizontally from these pathotypes and are therefore, potential source for the emergence of novel hybrid and hetero-pathogenic E. coli strains.6
Diffusely Adherent Escherichia coli (DAEC) is another pathotype of diarrheagenic E. coli that is characterized by its distinct adherence pattern to intestinal epithelial cells. Compared to the other pathotypes, DAEC exhibits a specific mechanism of adhesion to the intestinal mucosa. DAEC strains adhere diffusely to the entire surface of the intestinal epithelial cells, forming a fuzzy, homogenous adherence pattern.7 This adherence is mediated by fimbriae or adhesins, which are surface structures that facilitate attachment to host cells. The adhesins involved in DAEC adherence include Afa/Dr (afimbrial and Dr family of adhesins) and F1845 fimbriae.8,9 The pathogenesis of DAEC is not fully understood, and its association with diarrhoea remains a subject of ongoing research. However, studies have suggested that DAEC strains can cause intestinal inflammation and disrupt the normal function of the intestinal epithelium, leading to diarrhoea. DAEC has been implicated in not only acute and persistent diarrhoea in children10 and adults but also in inflammatory bowel disease and colon cancer.11 The afa-1 DAEC have the ability to adhere to, and invade epithelial cells and possibly play a role in epithelial-to-mesenchymal transition.11-13 The afaC gene is an integral part of the afa1 operon encoding for components of these adhesins. These afimbrial adhesins enable DAEC strains to adhere diffusely to intestinal epithelial cells in a “stacked-brick” manner.8,14 Our current research project was initiated in continuation of our previous work, where we had successfully devised a multiplex PCR (mPCR) assay for the identification of various E. coli pathotypes.4 However, this reported mPCR assay did not include the relevant genes to identify DAEC. We also wanted to study the drug resistance among the E. coli strains isolated from our population. Multi Drug Resistant (MDR) strains of DEC pose a significant clinical challenge. The primary mode of resistance to β-lactam antibiotics in DEC is attributed to the production of extended-spectrum β-lactamases (ESBLs) encoded by plasmids.15 ESBL-producing organisms have become widespread worldwide and genes responsible for ESBL production such as blaCTX-M, blaSHV, blaOXA, and blaTEM show varying prevalence across the regions. These genes also exhibit different frequencies among the strains and hence play a significant role in the management of infections.1,10 Hence in this study, we chose to design a multiplex PCR assay (mPCR) for detection of the afaC gene with blaTEM gene for the rapid identification of DAEC among the clinical isolates of E.coli and the drug resistance mechanism among these strains. Reported mPCR assays often lack an Internal Amplification Control (IAC), which is necessary to prevent false negative results. Hence, we also incorporated an IAC in this mPCR assay.
Identification and sensitivity test of E. coli isolates
Total 60 E. coli isolates from clinical samples comprising of stool specimens from diarrheic patients (n= 35), suspected urinary tract infections (n= 14), blood stream infection (n= 4), exudates including pus (n= 4) and endotracheal samples (n= 3) were collected. The isolates were phenotypically identified using standard biochemical and physiological tests including amino acid assimilation and sugar fermentation tests. The antimicrobial sensitivity assay was performed by standard disk diffusion test as per CLSI 2019 guidelines. The sensitivity test panel included CAZ (ceftazidime) and CAC (ceftazidime/clavulanic acid) disks for determination of extended spectrum β-lactamase (ESBL) production. Isolates showing ≥ 5 mm increase in the zone size around the CAC disk compared to CAZ disk were considered as ESBL producers.16
Bacterial DNA extraction
Phenotypically confirmed, purified isolates of E. coli from various clinical samples viz. blood, urine, exudates and stool, were sub-cultured in Luria Bertani broth (LB broth) which was incubated at 37°C for 18 to 24 hours. The LB broth was centrifuged at 4000 rpm for 5 minutes to get a pellet of bacterial cells. The pellet was subjected to DNA extraction by phenol-chloroform method. 17
Primer designing for afaC gene
Designing the specific primers is a critical step to establish a PCR assay. Utmost care was taken while designing the primers to avoid the formation of primer dimers and non specific amplification. A set of primers targeting the afaC gene was designed for detection of DAEC pathotype. The coding DNA sequence (CDS) for the selected gene was chosen from NCBI GenBank Database. The selected gene sequence was verified for its conservancy using NCBI BLAST. Primers were designed for the most conserved region using Gene Runner version 3.01 software (Hastings Software Inc., Hastings on Hudson, NY, USA) with reference to GenBank sequences afaC gene. The designed primers were ordered and obtained from Sigma Aldrich, India (Table 1).
Standardizing the monoplex PCR for detection of DAEC
Monoplex PCR to determine the ideal conditions for amplification of the individual selected target gene was standardized as described by Aradhya et al. 4 The PCR reaction mixture of 20µl contained 2.0µl PCR Buffer (10X), 1.6µl MgCl2 (25mM), 1.0µl dNTP mix (2 mM), 0.1µl Taq polymerase, 1.2µl of 10 pmol of afaC gene Primers (F & R), 1.0µl extracted DNA and 13.1µl PCR grade water. The thermal cycling was carried out in Quanta Biotech Thermal Cycler QB-96 Satellite thermal cycler with following conditions: initial denaturation at 94°C for 4 minutes, followed by 30 cycles of denaturation at 94°C for 1 minute, annealing at 53°C for 1 minute and extension at 72°C for 1 minute followed by final elongation at 72°C for 10 min.
The PCR product was resolved on 1% agarose gel containing ethidium bromide in Tris acetate ethylenediaminetetraacetic acid buffer at 100 V for 1 h followed by documentation using gel doc system (Zenith Research, Mumbai) and screened for 208 bp amplicon with reference to 100 bp DNA ladder. We tested a total of 60 phenotypically identified E. coli isolates from stool (n=35), urine (n=14), pus (n=4), endotracheal (n=3) and blood (n=4) samples. Only one isolate obtained from urine sample tested positive for afaC gene. The amplicon of this isolate was further sent for sequencing (Chromous Biotech, Bangalore) and analyzed by Basic Local Alignment Search Tool and was confirmed it to belong to the DAEC family. The sequence was submitted to BankIt (accession no. MT444431). This isolate, U-6065, was used as the reference strain in the study.
Standardizing the monoplex pcr for detection of blaTEM gene
Previously reported primers were used for standardising this PCR. 18 (Table 1). Similar methods as described above were used to establish this blaTEM ESBL monoplex PCR. The PCR conditions were found to be optimum using the following protocol – Initial denaturation at 94°C for 4 minutes, followed by 30 cycles of denaturation at 94°C for 1 minute, annealing at 53°C for 1 minute, extension at 72°C for 1 minute 30 seconds and final extension at 72°C for 12 minutes. The 12 minutes of final elongation time was found to be ideal for the amplification of blaTEM gene with 1080 bp. All the 60 E. coli isolates were subjected to this blaTEM ESBL monoplex PCR.
Table (1):
List of primers used in multiplex PCR study
| No. | Pathotypes | Gene | Primer | Size (bp) | Source |
|---|---|---|---|---|---|
| 1 | DAEC | afaC | F-CTGAAAGGTGGCGGTAAG | 208 | This study |
| R-CAATCCTGACCCCGTAAC | |||||
| 2 | ESBL | blaTEM | F – AAAATTCTTGAAGACG | 1080 | 18 |
| R – TTACCAATGCTTAATCA | |||||
| 3 | pUC 19 | IAC | F- CAATTCCACACAACATACGA | 660 | 29 |
| R- CGGATAAGGCGCAGCG |
Standardization of multiplex PCR
A multiplex PCR targeting 2 pathotype specific genes viz., afaC (208 bp), blaTEM(1080 bp) was developed along with an IAC (660 bp) (Table 1). E. coli isolates U-6065 and E-5455 which were confirmed by monoplex PCR for the presence of afaC gene and blaTEM genes respectively were employed as the reference strains for establishing this multiplex PCR. Once successful amplification of the target genes was achieved, the optimization of multiplex PCR was conducted, determining the optimal concentrations of PCR reagents such as MgCl2, primers, dNTPs, and Taq polymerase. To standardize the multiplex PCR, a gradient range of annealing temperatures spanning from 50°C to 58°C was set. The optimum annealing temperature was concluded to be 53°C for multiplex PCR.
The reaction was finally optimized using the following components: 2.0µl PCR Buffer (10X), 2.0µl MgCl2 (25 mM), 1.2µl dNTP mix (2 mM), 0.2µl Taq polymerase, 0.4µl of 10 pmol of afaC gene Primers (F & R), 0.8µl of 10 pmol of blaTEM gene Primers (F & R), 50-100 ng of template DNA from the reference strains. Additionally, a minimum quantity of 0.3 pmol of the IAC primer was included in the reaction along with IAC DNA. The final PCR reaction volume was adjusted to 20µl using PCR-grade water. The PCR conditions for amplification, included initial denaturation at 94°C for 4 min, 30 cycles of denaturation at 94°C for 1 min, 53°C of annealing for 1 min, 72°C of extension for 1 min 30 seconds followed by final extension at 72°C for 12 minutes. The multiplex PCR products were visualized in 1% agarose (Hi-media, Mumbai, India) gel stained with ethidium bromide after electrophoresis in a 1X Tris-acetate EDTA buffer at 100 V for 1 hour 30 minutes. The amplicons were examined and documented using GelDoc system.
A total 80 freshly isolated E. coli from diarrheal stool samples were tested using this multiplex PCR.
Initially 60 E. coli phenotypically identified E. coli isolates were subjected to monoplex PCR. Of these 60 isolates, 33 were MDR strains and were shown to be ESBL producers. However, among these strains only 12 isolates tested positive by monoplex blaTEM gene PCR (Table 2). Twenty-seven phenotypically confirmed Non-ESBL E. coli isolates tested negative for this target gene. Only one strain isolated from urine tested positive for DAEC PCR (Table 2).
Table (2):
Results of Monoplex PCR (n=60)
No. |
Pathotype |
Gene |
Size |
Positives |
|---|---|---|---|---|
1 |
ESBL |
blaTEM |
1080 bp |
20% (12/60) |
2 |
DAEC |
afaC |
208 bp |
1.6% (1/60) |
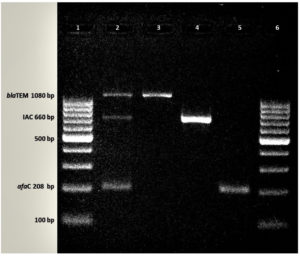
Multiplex PCR to detect DAEC and ESBL strains among phenotypically identified E. coli isolates was successfully established (Figure). All three genes, i.e., afaC, blaTEM and pUC 19 (as Internal amplification control) could be detected in this multiplex PCR assay. This standardized multiplex assay was used to test a fresh set of 80 E. coli isolates. Of these 80 isolates 24 were positive for ESBL production by phenotypic method. Of these 24 isolates, 16 were positive for blaTEM gene (Table 3). The IAC had amplified in all the reactions successfully. We did not detect any afaC gene among these strains.
Table (3):
Results of multiplex PCR assay (n=80)
No. |
Pathotype |
Gene |
Size |
Positives |
|---|---|---|---|---|
1 |
ESBL |
blaTEM |
1080 bp |
20% (16/80) |
2 |
DAEC |
afaC |
208 bp |
Nil |
Figure. Amplification of DAEC, ESBL and IAC genes
Footnote: Agarose gel electrophoresis showing multiplex and monoplex PCR pattern
Lane 1: 100 bp DNA Ladder, Lane 2: multiplex PCR, Lane 3: blaTEM, Lane 4: IAC, Lane 5: afaC, Lane 6:100 bp DNA Ladder
The overall incidence of DAEC strain tested using the molecular assay was 0.7% (1/140). The incidence of ESBL when tested phenotypically was 40.7% (57/140) and the incidence of ESBL based on the detection of the blaTEM gene was 20% (28/140). The overall frequency of blaTEM gene among ESBL producers was 49.1% (28/57).
DAEC has not only been linked to diarrhoea but also to several chronic disease conditions related to colon, e.g. Crohn’s disease and even to colorectal cancer.2,13,10,19,20 The afaC gene is one of the molecular marker to identify DAEC and helps to differentiate DAEC from the other diarrheagenic E. coli pathotypes.19,14 The Afa-1 operon is responsible for expression of afimbrial adhesin. This gene is also present in uropathogenic E. coli (UPEC) strains giving them the ability to adhere to epithelial cells.8,11 The only afaC + E. coli in this study was a urinary isolate. The possibility of this strain being a UPEC cannot be ruled out. However, further molecular characterization is essential for its confirmation. This isolate was subjected to pap gene PCR for identification of UPEC; however, it turned out to be negative. Finding an afaC gene in other E. coli strains is not uncommon as intra and interspecies horizontal gene transfers are well known.6 E. coli strains not only from diarrheic samples but also from other infectious samples need to be screened for the presence of this virulence gene.6,11,21 The isolates we included in the study were from different clinical samples like blood, stool, urine, pus, exudates, endotracheal tube and aspirations. The PCR amplicon from afaC positive isolate was confirmed by sequencing and BLAST analysis. This novel sequence is submitted to GenBank (accession no. MT444431).
Initially, we used 60 phenotypically characterized E. coli isolates recovered from various clinical samples and established monoplex PCR assays for both the targets. After having satisfactorily established the monoplex PCR assays, we designed a multiplex PCR assay for their simultaneous detection. A fresh set of 80 E. coli isolates from clinical samples was tested by this mPCR assay.
The overall occurrence/frequency of DAEC in our study was 0.7% (1/140). Such low prevalence has been reported by other investigators as well. These studies have employed the detection of daaE gene for identification of DAEC among diarrheic samples.2,10,22 Low rates of DAEC could be attributed to various factors, such as the specific patient population or the limited number of samples analysed. Studies with larger sample size and diverse patient populations are needed to establish a dependable estimation of DAEC prevalence. The detection rates of DAEC are higher when investigators have used the afaC gene.23,24 Interestingly, the presence of afaC gene of E. coli strains has been more frequently linked to inflammatory bowel disease and colorectal cancer. The DAEC have the ability to adhere to and invade epithelial cells and are likely play a role in epithelial to mesenchymal transition.11,13,20
Most studies focusing on the detection of extended spectrum β-lactamase (ESBL) producing strains and diffusely adherent E. coli (DAEC) pathotypes have worked on isolates from stool samples. In this work we have included isolates from various clinical samples as the pathogenicity of E. coli is extensive. Identification of E. coli pathotypes is essential as they display different virulence and resistance mechanisms. Precise pathotype identification helps to understand the mechanisms involved in the pathogenesis of the infection and directs the management decision for the infection.14,21 Drug resistance is a frequent problem in E. coli infections and isolation of MDR strains in common. Both genotypic and phenotypic tests were used in this work to determine the prevalence of ESBL producers. Of the total 140 isolates 57 (40.7%) were found to be positive for ESBL production by phenotypic detection while 28 showed presence of blaTEM gene. All the 28 isolates positive for blaTEM PCR were also positive by phenotypic method for ESBL production. The prevalence of 40.7% ESBL-producing strains among the tested clinical isolates is alarming. With blaTEM gene only 20% isolates were positive for ESBL activity. In this work we have targeted only one gene from among the various other targets available for ESBL production. Prevalence rates of ESBL producers and distribution of various drug resistance mechanisms vary among different hospitals in the country and worldwide.1,10,25 The blaCTX-M in particular has gained much prominence due to its global prevalence. The prevalence of ESBLs reported from various other studies from India ranges from 37% to as high as 91%.1,10,25 It is, therefore, important to continue monitoring and understanding the significance of ESBL production by both molecular and phenotypic methods. Inclusion of other targets to determine ESBL production like blaCTX-M, blaSHV, blaOXA in an mPCR for this purpose is essential and we hence intend to establish such an assay in future. Molecular methods for detection of resistance mechanisms are ever evolving as gene alterations keep occurring in Gram negative pathogens. Phenotypic methods for identifying ESBL production may not consistently align with existing molecular assays because of incessant appearance of newer drug resistance mechanisms. Therefore, updating or development of novel molecular methods is imperative to accurately identify ESBL production in such cases.26 Mandal et al. reported 37.6% of DEC isolates to be ESBL producers of which 51.8% were blaTEM positive. 1Govindaswamy et al., found 59.53% of their E. coli isolates producing ESBL when tested by disk potentiation test of which 67.3% isolates tested positive for blaTEM.25 Chellapandi K et al., reported 91.6% of E. coli isolates were ESBL producers by phenotypic method. They did not test for blaTEM gene in their isolates.10 El Aila et al., evaluated various gram-negative isolates phenotypically and found 51.6% of the isolates produced ESBL of which 57.6% were blaTEM positive. 27 It is imperative to know the local prevalence of drug resistance and the drug resistance mechanisms in commonly isolated pathogens.
A major challenge with mPCR is the potential for false negative results. The absence of a positive signal may not truly indicate the absence of the target DNA. False negatives may occur due to various other factors like PCR inhibition, sample degradation or technical issues. To address this issue, The European Standardization Committee (CEN) and International Organization for Standardization (ISO) have suggested the inclusion of an Internal Amplification Control (IAC) in PCR testing protocols for detecting pathogens.28,29
The use of mPCR assay in our study played a crucial role in evaluating DAEC strains and detecting the presence of the blaTEM for ESBL production. The importance of mPCR in identifying these pathogens and resistance genes lies in its ability to simultaneously detect multiple targets in a single assay. The detection of DAEC strains using mPCR allows for efficient identification of this pathotype, which has been linked not only to diarrhoea but also to chronic conditions such as Crohn’s disease and colorectal cancer. This is valuable in understanding the role of DAEC in various diseases and their potential impact on human health. Furthermore, the detection of the blaTEM gene, known for conferring resistance to β-lactam antibiotics, is of great significance in monitoring and combating antimicrobial resistance.
The implementation of mPCR in routine diagnostic laboratories can aid in timely and accurate identification of pathotypes and resistance profiles, enabling appropriate treatment strategies and effective management of infectious diseases. Although our mPCR assay currently focuses on a limited number of gene targets, it is flexible to incorporate additional targets without compromising the accuracy and specificity. In future, we desire to expand the assay by including more genes of interest to this assay. The flexibility of mPCR allows it to be used for specific diagnostic needs and has great applicability in a rapidly evolving field of biomedical sciences.
ACKNOWLEDGMENTS
The authors would like to thank Defence Food Research Laboratory (DFRL), Mysore and Shri Dharmasthala Manjunatheshwara University, Dharwad, for providing the necessary facilities for this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Mandal A, Sengupta A, Kumar A, et al. Molecular Epidemiology of Extended-Spectrum b-Lactamase-Producing Escherichia coli Pathotypes in Diarrheal Children from Low Socioeconomic Status Communities in Bihar, India: Emergence of the CTX-M Type. Infect Dis Res Treat. 2017;10:117863361773901.
Crossref - Rajendrana P, Ajjampura SSR, Chidambarama D, et al. Pathotypes of diarrheagenic Escherichia coli in children attending a tertiary care hospital in South India. Diagn Microbiol Infect Dis. 2010;68(2):117-122.
Crossref - Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123-140.
Crossref - Aradhya S, Ramlal S, Madalgi R, et al. Simultaneous Identification of Diarrheagenic Escherichia coli Using Thermostabilized Multiplex PCR Formulation Shruthi. Int J Sci Res Biol Sci. 2020;7(5):38-46.
Crossref - Gomes TAT, Elias WP, Scaletsky ICA, et al. Diarrheagenic Escherichia coli. Braz J Microbiol. 2016;47(Suppl 1):3-30.
Crossref - Santos AC de M, Santos FF, Silva RM, Gomes TAT. Diversity of Hybrid- and Hetero-Pathogenic Escherichia coli and Their Potential Implication in More Severe Diseases. Front Cell Infect Microbiol. 2020;10:339.
Crossref - Benevides-Matos N, Pieri FA, Penatti M, Orlandi PP. Adherence and virulence genes of Escherichia coli from children diarrhoea in the Brazilian Amazon. Braz J Microbiol. 2015;46(1):131-137.
Crossref - Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev. 2005;18(2):264-292.
Crossref - Pokharel P, Dhakal S, Dozois CM. The Diversity of Escherichia coli Pathotypes and Vaccination Strategies against This Versatile Bacterial Pathogen. Microorganisms. 2023;11(2):344.
Crossref - Karuppasamy C, Chellapandi K, Ralte L, et al. Diffusely Adherent E. coli Burden in Low Socio-Economic Pediatric Population. J Med Bacteriol. 2019;8(5):44-55. https://jmb.tums.ac.ir/index.php/jmb/article/view/418
- Prorok-Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63(5):761-770.
Crossref - Rombaux P, Huart C, Levie P, Cingi C, Hummel T. Olfaction in Chronic Rhinosinusitis. Curr Allergy Asthma Rep. 2016;16(5):41.
Crossref - Eklof V, Lofgren-Burstrom A, Zingmark C, et al. Cancer associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528-2536.
Crossref - Garcia MI, Labigne A, Le Bouguenec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol. 1994;176(24):7601-7613.
Crossref - Yandag M, Tsend-ayush A, Gunregjav N, Erdenebayar O. Detection and antibiotic resistance of diarrheagenic Escherichia coli from patients with diarrhea in Ulaanbaatar , Mongolia. J Infect Dev Ctries. 2023;17(2):202-209.
Crossref - CLSI. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 33rd ed.; 2019. www.clsi.org.
- Sambrook J, Russell DW. Molecular Cloning A Laboratory Manual, Fourth Edition. Cold Spring Harbor Laboratory Press. New York. 1989.
- Sharma J, Sharma M, Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332-336.PMID: 20847381.
- Fujihara S, Arikawa K, Aota T, et al. Prevalence and properties of diarrheagenic Escherichia coli among healthy individuals in Osaka City, Japan. Jpn J Infect Dis. 2009;62(4):318-323.
Crossref - Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. 2017;18(6):1310.
Crossref - Meza-Segura M, Zaidi MB, Vera-Ponce de Leon A, et al. New Insights Into DAEC and EAEC Pathogenesis and Phylogeny. Front Cell Infect Microbiol. 2020;10.
Crossref - Walczuk U, Sobieszczanska B, Turniak M, Rzeszutko M, Duda-Madej A, Iwanczak B. The prevalence of mucosa-associated diffusely adherent Escherichia coli in children with inflammatory bowel disease. Adv Clin Exp Med. 2019;28(7):899-905.
Crossref - Javadi K, Mohebi S, Motamedifar M, Hadi N. Characterization and antibiotic resistance pattern of diffusely adherent Escherichia coli (DAEC), isolated from paediatric diarrhoea in Shiraz, southern Iran. New Microbes New Infect. 2020;38:100780.
Crossref - Abbasi P, Kargar M, Doosti A, Mardaneh J, Ghorbani-Dalini S, Dehyadegari MA. Molecular detection of diffusely adherent Escherichia coli strains associated with diarrhea in Shiraz, Iran. Arch Pediatr Infect Dis. 2017;5(2):e37629.
Crossref - Govindaswamy A, Bajpai V, Khurana S, et al. Prevalence and characterization of beta-lactamase-producing Escherichia coli isolates from a tertiary care hospital in India. J Lab Physicians. 2019;11(2):123-127.
Crossref - Jubeh B, Breijyeh Z, Karaman R. Resistance of gram-positive bacteria to current antibacterial agents and overcoming approaches. Molecules. 2020;25(12):2888.
Crossref - El Aila NA, Al Laham NA, Ayesh BM. Prevalence of extended spectrum beta lactamase and molecular detection of blaTEM, blaSHV and blaCTX-M genotypes among Gram negative bacilli isolates from pediatric patient population in Gaza strip. BMC Infect Dis. 2023;23(1):99.
Crossref - Hoorfar J, Malorny B, Abdulmawjood A, Cook N, Wagner M, Fach P. Practical Considerations in Design of Internal Amplification Controls for Diagnostic PCR Assays. J Clin Microbiol. 2004;42(5):1863-1868.
Crossref - Nagaraj S, Ramlal S, Kingston J, Batra HV. Thermostabilization of indigenous multiplex polymerase chain reaction reagents for detection of enterotoxigenic Staphylococcus aureus. J Microbiol Immunol Infect. 2018;51(2):191-198.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.