ISSN: 0973-7510
E-ISSN: 2581-690X
Brucellosis is a zoonotic disease with veterinary, public health, and economic implications. The study aimed to estimate the seroprevalence of Brucella spp. among ruminants in Rafha, Saudi Arabia during January to October 2022 and to identify camel strains based on a glycosyltransferase gene sequence. Sera (n=1012) were collected from non-vaccinated sheep, goats, camels and cattle of different sex, age and breed randomly from the abattoirs to investigate the circulating brucella antibodies using RBPT. One hundred and eighteen sera (9.7%) were reactive for Brucella spp. IgG immunoglobulins, with higher percentages detected in sheep (11.4%), females (13.3%), adults (10.7%), and naieme breed (13.9%). Significant correlation between Brucella spp. antibodies and animal species (0.095), age (0.077) was found, while strong correlation between antibodies and sex was observed. Glycosyltransferase gene was amplified and sequenced from camel reactive sera (n=6). Camel strains displayed multiple nucleotide substitutions and deletions, nucleotide identity among local strains is 96.2-100%. Phylogenetic analysis showed that Brucella spp. strains clustered in two groups, Rafha strains clustered in one group together with other strains. Further investigation is needed to determine the prevalence of the bacteria among farm animals and to identify the strains involved to improve the preventive measures and strategies adopted for control.
Brucella spp., Seroprevalence, Glycosyltransferase Gene, Ruminants, Rafha, Saudi Arabia
Brucellosis is a zoonotic disease causing serious consequences for animal production and human health.1-3 Brucella spp. is the causal agent of the disease with B. abortus and B. melitensis being mainly important in ruminant and human infections. It infects almost all domestic species and cross transmission can occur between cattle, sheep, goat, camel and human.1 Human illness spread via contaminated dairy products and intimate contact with infected animals.4,5 Families bred camel, along with goats and sheep for a variety of purposes. In addition to providing physical labor, a camel’s wool can be woven into cloth, milk can be drunk, and for meat and leather. The disease was reported in different parts of the Kingdom of Saudi Arabia.6-8 It is regarded as a main public health and agricultural problem in the country.9-15 The RBT is an affordable, quick, simple and efficient screening and a diagnostic test for individual animals and herds.7,16 To adopt effective control measures, large-scale epidemiological studies are required. The glycosyltransferase gene coding for O-antigen production is a crucial virulence protein is conserved among Brucella species.17-20 Studies have shown that it is conserved in all Brucella species. Furthermore, characterization of the gene can clarify the relationship between genotype and their use in differential diagnosis. Analysis of this gene can identify the circulating strains and elucidate the similarity between genetic constitution of Brucella spp.16,20
No published research on the prevalence of brucellosis in ruminants or on genotyping of Brucella species based on the glycosyltransferase gene in Rafha, Saudi Arabia. The objective of the current study aimed to assess the prevalence of brucellosis among ruminants in Rafha, Saudi Arabia and to identify camel Brucella strains based on a glycosyltransferase gene sequence.
Ethical Approval
Animals were treated in accordance with ethical considerations and the research was authorized by the Local Committee of Bioethics (HAP-09-A-043) at Northern Border University, KSA, issued the decision no. (3/44/H/2022).
Sample Collection
Sera (n = 1212) were collected randomly from the slaughterhouse in Rafha, Saudi Arabia during January to October 2022 from sheep, goats, camels, and cattle of various sex, age, and breed (Table 1).
Rose Bengal Plate Test (RBPT)
The presence of anti-Brucella IgG antibodies were examined in serum samples. The antigen was obtained from Lillidale Diagnostics, BH21 4HU, United Kingdom. Test sera and RBPT antigen were mixed in an equal volume (30µl), shaken for 5 minutes, and then read.
Statistical analysis
The correlation between seropositivity for IgG and species, sex, age, and animal breed were assessed utilizing Spearman’s rank correlation coefficient (P-value = 0.01). Analysis was done using SPSS25 (Statistical Package for Social Sciences 25) (Table 2).
DNA extraction
Following the manufacturer’s protocol, DNA were extracted from camel sera (n=10) that were reactive by RBPT using the DNeasy Blood Kit (QIAGEN).
Amplification of a glycosyltransferase gene
The glycosyltransferase gene was amplified using the procedure and the primer sequences (F:5-GAGTAGACACGGGAAATC-3 and R:5- GATAAACACGCCGAGCTT-3) published by Etemadi et al. (2008). Conventional PCR was carried out using QIAGEN kits as follows; denaturation at 94°C for 5 min followed by 30 cycles at 94°C for 30s, 55°C for 30s, 72°C for 90s, and a final extension at 72°C for 8 min.
Purification of amplicons
Amplicons (5µl) were combined with 25µl of ExoASP-IT® (usb) for a total reaction volume of 75µl. ExoSAP-IT was incubated at 37°C for 15 minutes before being deactivated by heating at 80°C for 15 minutes. Purified PCR products were Sanger-sequenced using a 3730xl automated sequencer and the BigDye terminator v3.1 Cycle Sequencing Kit (ABI PRISM 3730XL Analyzer). The Macrogen sequencing facility sequenced both strands (Macrogen Inc., Seoul, Korea).
Sequence analysis
Lasergene 7.1.0 was used to edit and assemble nucleotides using EditSeq and SeqMan (DNASTAR, Inc, Madison, WI, USA) (DNASTAR, Inc, Madison, WI, USA). The BLASTn program (https://blast.ncbi.nlm.nih.gov4/Blast) was used to align local sequences with those from GenBank (Table 3), and a phylogenetic tree was constructed using the neighbor-joining approach.
RBPT
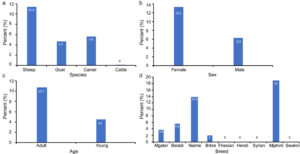
Out of the tested sera one hundred and eighteen sera (9.7%) were reactive for Brucella spp. IgG immunoglobulins, 11.4% among sheep with higher incidence among naieme breed (13.9%), 13.3% among females and 10.7% among adults across all species (Figure 1).
Figure 1. Seroprevalence rate of Brucella spp. antibodies as tested by RBPT according to species (a), sex (b), age (c) and breed (d) in Rafha, Saudi Arabia during January to October 2022
Statistical analysis
Seropositivity significantly correlated with animal species (0.095), age (-0.077) and sex (-0.118) (Table 2).
Table (1):
Seroprevalence of brucellosis as detected by RBPT among species, sex, age and breed in Rafha, Saudi Arabia during January to October 2022
| Species | Sheep | Goat | Camel | Cattle | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | N | 102 | 6 | 10 | 0 | 118 | |||||||
| % within Species | 11.4 | 4.7 | 5.6 | 0.0 | 9.7 | ||||||||
| Negative | N | 789 | 123 | 170 | 12 | 1094 | |||||||
| % within Species | 88.6 | 95.3 | 94.4 | 100.0 | 90.3% | ||||||||
| Total | N | 891 | 129 | 180 | 12 | 1212 | |||||||
| Sex | Male | Female | |||||||||||
| Positive | N | 39 | 79 | 118 | |||||||||
| % within Sex | 6.3 | 13.3 | 9.7 | ||||||||||
| Negative | N | 579 | 515 | 1094 | |||||||||
| % within Sex | 52.9 | 47.1 | 100.0 | ||||||||||
| Total | N | 618 | 594 | 1212 | |||||||||
| Age | Young | Adult | |||||||||||
| Positive | N | 9 | 109 | 118 | |||||||||
| % within Age | 4.5 | 10.7 | 9.7 | ||||||||||
| Negative | N | 189 | 905 | 1094 | |||||||||
| % within Age | 95.5 | 89.3 | 90.3 | ||||||||||
| Total | N | 198 | 1014 | 1212 | |||||||||
| Breed | Mgater (Camels) | Baladi (Goats) | Naime (Sheep) | Brbre (Sheep) | Friesian (Cattle) | Hendi (Cattle) | Syrian (Goats) | Mjahim (Camels) | Swakni (Sheep) | ||||
| Positive | N | 6 | 6 | 99 | 3 | 0 | 0 | 0 | 4 | 0 | 118 | ||
| % within Breed | 3.8 | 5.6 | 13.9 | 2.0 | 0.0 | 0.0 | 0.0 | 19.0 | 0.0 | 9.7 | |||
| Negative | N | 153 | 102 | 615 | 144 | 6 | 6 | 21 | 17 | 30 | 1094 | ||
| % within Breed | 96.2 | 94.4 | 86.1 | 98.0 | 100.0 | 100.0 | 100.0 | 81.0 | 100.0 | 90.3 | |||
| Total | N | 159 | 108 | 714 | 147 | 6 | 6 | 21 | 21 | 30 | 1212 | ||
Table (2):
Spearman correlation of seroprevalence of brucellosis as detected by RBPT with species sex, age and animal breed in Rafha, Saudi Arabia during January to October 2022
| Seropositivity | Species | Sex | Age | Breed | |||
|---|---|---|---|---|---|---|---|
| Seropositivity | Correlation Coefficient | 1.000 | .095** | -.118** | -.077** | .001 | |
| Sig. (2-tailed) | . | .001 | .000 | .007 | .968 | ||
| N | 1212 | 1212 | 1212 | 1212 | 1212 | ||
| Species | Correlation Coefficient | .095** | 1.000 | -.327** | -.655** | -.576** | |
| Sig. (2-tailed) | .001 | . | .000 | .000 | .000 | ||
| N | 1212 | 1212 | 1212 | 1212 | 1212 | ||
| Sex | Correlation Coefficient | -.118** | -.327** | 1.000 | .366** | .014 | |
| Sig. (2-tailed) | .000 | .000 | . | .000 | .620 | ||
| N | 1212 | 1212 | 1212 | 1212 | 1212 | ||
| Age | Correlation Coefficient | -.077** | -.655** | .366** | 1.000 | .450** | |
| Sig. (2-tailed) | .007 | .000 | .000 | . | .000 | ||
| N | 1212 | 1212 | 1212 | 1212 | 1212 | ||
| Breed | Correlation Coefficient | .001 | -.576** | .014 | .450** | 1.000 | |
| Sig. (2-tailed) | .968 | .000 | .620 | .000 | . | ||
| N | 1212 | 1212 | 1212 | 1212 | 1212 | ||
**Correlation is significant at the 0.01 level (2-tailed)
Table (3):
Annotation of sequences retrieved from GenBank using BLASTn tool for phylogenetic analysis
NO |
GenBank |
Host |
Species |
strain |
Country |
Reference |
|---|---|---|---|---|---|---|
1 |
CP025821 |
Homo sapiens |
Brucella melitensis |
CIT31 |
China |
Unpublished |
2 |
LT962945 |
Homo sapiens |
Brucella melitensis |
1 |
Norway |
Unpublished |
3 |
CP018506 |
Homo sapiens |
Brucella melitensis |
BwIM_SOM_36a |
Somalia |
21 |
4 |
CP007717 |
Sus scrofa |
Brucella suis |
513UK |
United Kingdom |
22 |
5 |
CP001578 |
Vole |
Brucella microti |
CCM 4915 |
Czech Republic |
23 |
6 |
AY065979 |
Unknown |
Brucella melitensis |
16M |
USA |
Unpublished |
7 |
CP033079 |
Elk |
Brucella abortus |
BJ1 |
China |
Unpublished |
8 |
CP027643 |
Dog |
Brucella canis |
GB1 |
China |
Unpublished |
9 |
LT671512 |
Bos taurus |
Brucella abortus |
Wisconsin |
USA |
Unpublished |
10 |
LT963350 |
Homo sapiens |
Brucella melitensis |
1 |
Norway |
Unpublished |
11 |
LT962916 |
Homo sapiens |
Brucella melitensis |
1 |
Norway |
Unpublished |
12 |
CP023308 |
Bubalus bubalis |
Brucella abortus |
9510 |
Italy |
Unpublished |
13 |
CP023223 |
Bubalus bubalis |
Brucella abortus |
67761 |
Italy |
Unpublished |
14 |
CP022875 |
Bos taurus |
Brucella melitensis |
BL |
China |
Unpublished |
15 |
CP018554 |
Homo sapiens |
Brucella melitensis |
BwIM_TUR_39 |
Turkey |
21 |
16 |
CP018532.1 |
Homo sapiens |
Brucella melitensis |
BwIM_SYR_41 |
Syria |
21 |
17 |
CP066175 |
Sheep |
Brucella abortus |
68 |
Ukraine |
Unpublished |
18 |
CP061816 |
Cystophora cristata |
Brucella pinnipedialis |
23a-1 |
Svalbard |
Unpublished |
19 |
CP054955 |
Sus scrofa |
Brucella suis |
CVI_72 |
Slovenia |
Unpublished |
Glycosyltransferase gene sequencing
Glycosyltransferase gene was amplified and sequenced from camel sera (n=6). Sequences were deposited in the GenBank (Accession numbers MN934944, MN934945, MN934946, MN934947, MN934948 and MN934949).
Sequence analysis
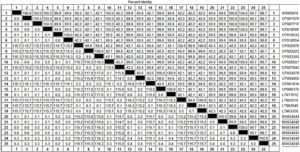
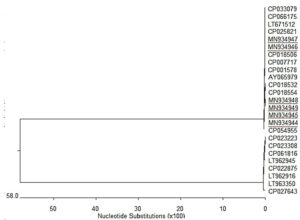
Local strains exhibited multiple nucleotide substitutions and deletions (Figure 2), identity among sequences was 96.2-100% (Figure 3), while identity with strains retrieved from GenBank was 42.1-99.9% (Figure 2). Phylogenetic analysis displayed Brucella spp. in two branches, Rafha strains clustered in one group together with other strains (Figure 4).
Figure 2. Sequence alignment of a glycosyltransferase gene identified from brucella spp. strains from camel in Rafha, Saudi Arabia during January to October 2022
Figure 3. Percentage of identity of a glycosyl transferase gene sequences from Brucella spp. strains identified from camel in Rafha, Saudi Arabia and sequences retrieved form GenBank
Brucellosis is a zoonotic bacterial disease caused by various Brucella species, which mainly infect cattle, swine, goats, sheep and dogs.24 In the current study, prevalence of brucellosis in Rafha. Saudi Arabia in sheep, goats and camels was determined by RBPT, as well as, camel strains were identified based on a glycosyltransferase gene analysis. Sero-positivity was assumed to be attributable to infection of brucellosis since immunization has never been practiced in the area.
The findings showed that the disease spread among ruminants and it occurred at a higher rate in sheep. The detected prevalence of brucellosis in sheep (11.4%), is similar to that published in Saudi Arabia14 as well as in India.25 It was slightly lower than that reported in other parts of the country, including western region (15.6%) and Makkah (12.3-14.2%).7,8,26 Much higher seroprevalence (31.7%) was reported at Duhok in northern Iraq.27 However, it was slightly higher than the reported one (7.3%) in Aljouf region, Saudi Arabia,15 and in Aseer and Jazan (5.1%) in southern Saudi Arabia,28 also that found (8.3%) in India.29 Nevertheless, none of tested sheep sera in Farasan Islands in the Red Sea in southwestern Saudi Arabia were found to be positive.30 Discrepancy may be due to the breed involved, herd size, management, and seasonality of the disease.
The detected percent of caprine brucellosis (4.7%) is alike to previous records in Bangladesh.31,32 However, Abdellatif et al. 14 detected much higher seropositivity (12.1%) in Hail, Saudi Arabia. Meanwhile it was 8.8%, in Medina.33 A very low seroprevalence (0.6%) was detected in goats in Farasan Islands in Saudi Arabia 30 which is expected due to the isolated nature of the island. Brucellosis is endemic in many countries, variable seroprevalence of the disease had been reported. A very high prevalence (34%) in goats have been detected in Iraq 27 and (27.7%) in Jordan.34 In Dhofar Province at Southern Oman, goat sera showed 13% seropositivity.35 In India, 5.8% seropositivity was determined.29 It was found to be 14.8% in North West Libya,36 which was higher than our present study. In contrast, Rahman & Ahasan37 declared that the rate of brucellosis was 1.98% in Bangladesh. Despite contradictions in the literature, reports showed that sheep was more likely to be reactive than goats,38-40 which may be influenced by sampling, circulating strain, immunity of the species, management (animal, herd, farm), and/or owner’s awareness about the disease.41-43 The occurrence of camel brucellosis (5.6%) was slightly comparable to the obtained results in Hail (6.2%)14 and Alzulfi, Saudi Arabia (6.5%) Salih et al, higher than that reported (3.5%.) at Aseer and Jazan in southern Saudi Arabia28 and disagree with the previous data (1.9%) reported in Riyadh by Alshaikh et al. Variable seroprevalence were found in other Gulf countries, in Dhofar, Southern Oman, 3.4% of camel sera tested positive.35 A far higher seroprevalence of brucellosis in camel sera (20.6%) was reported in Qatar.35 The rate also contrast with that reported in Ethiopia (2.2%),44 4.1%,45 and 3.37%.46 Higher rate was recorded in Sudan ((37.5%) 47 and Somalia (7%).48 The exposure may be increased as a consequence of the intense animal rearing.49 Furthermore, the infections can also spread due to the absence of control measures, mixed grazing of camel herds with other herds and animals in the pasture, and drinking points. All cattle sera in the current study were seronegative, it may be related to the number of animals tested because cattle were not bred in the region. Previous research reported bovine brucellosis as 18.1% in Jordan,12 1.9% in China,50 6.3% in Pakistan.51 Seroprevalence was found to be variable depending on sampling size, species, sex and size of herd.3
Sex wise, seroprevalence was comparable to the previous reports.15,52 But it differs with other literature.34,53,54 Difference may be due to the immune response or the interaction of other risk factors. Age-level showed that was higher in adult animals, it’s supported by previous research.15,40 It may be owing to sex hormones, which may stimulate the growth of the bacteria, and tend to increase in concentration with age and sexual maturity. This might be true since older animals keep on in the flock for a long time, and they had a longer duration of contact. The higher percentages in naimee breed may be due the number of animals tested.
Analysis of data revealed significant correlation between seroprevalence of Brucella spp. and species, sex and age. In contrary, there was no association between the prevalence and animal breed. Results were in accordance to that observed by Rahman et al. However, it opposed with Akhter et al.55 who found that none of these factors was linked with brucellosis. Inconsistency may be attributed to variance in the risk factors involved at both animal and herd level.56
Traditional identification of brucellosis depends on the isolation of the bacteria.57 Owing to various restrictions in the isolation of the bacteria including requirement for biosafety facilities, workers expertise, and hazard of contamination, numerous molecular procedures to identify and discriminate Brucella species have been developed.58 In the present investigation, camel strains were identified as B. melitensis based on glycosyltransferase gene amplification and sequencing. Comparative analysis of the nucleotide sequence among camel strains exhibited 96.2-100% similarity with multiple nucleotide substitutions and deletions. Phylogenetic analysis based on Glycosyltransferase gene sequence to clarify the genetic relation between local strains and other sequences deposited in GenBank. Unfortunately, there were no sequences of the Glycosyltransferase gene of the Brucella from Saudi Arabia found in GenBank to be added in the tree. Analysis display Brucella strains in two branches, Rafha strains clustered in one group together with other strains. The results agreed with Etemady et al., who report considerable genetic diversity among B. melitensis and conservation of B. abortus strains.
The prevalence of brucellosis was 11.4% in sheep, 4.7% in goats and 5.6% in camels. The seropositivity was higher in females (13.3%), adults (10.7%), and naieme breed (13.9%). Circulating strains were identified from camel sera as B. melitensis based on a glycosyltransferase gene. Analysis revealed multiple nucleotide substitutions and deletions, displaying variable identities. Further investigation to identify the circulating strains and to understand factors implicated in the epidemiology is needed to improve the preventative measures and control policy adopted.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the approval and the support of this research study by the grant no. SCAR-2022-11-1610 from the Deanship of Scientific Research at Northern Border University, Arar, Saudi Arabia.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by Deanship of Scientific Research at Northern Border University, Arar, Saudi Arabia, with grant no. SCAR-2022-11-1610.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Local Committee of Bioethics (HAP-09-A-043) at Northern Border University, Arar, Saudi Arabia, wide letter number 3/44/H/2022.
- Mantur B, Amarnath S, Shinde R. Review of clinical and laboratory features of human brucellosis. Indian J Med Microbiol. 2007;25(3):188-202.
Crossref - Christopher S, Umapathy BL, Ravikumar KL. Brucellosis: review on the recent trends in pathogenicity and laboratory diagnosis. J Lab Physicians. 2010; 2(2):55-60.
Crossref - Khurana SK, Sehrawat A, Tiwari R, et al. Bovine brucellosis-a comprehensive review. Vet Q. 2021;41(1):61-88.
Crossref - Christopher S, Umapathy BL, Ravikumar KL. Brucellosis: review on the recent trends in pathogenicity and laboratory diagnosis. J Lab Physicians. 2010;2(2):55-60.
Crossref - Franc K, Krecek R, Hasler B, Arenas-Gamboa A. Brucellosis remains a neglected disease in the developing world: a call for interdisciplinary action. BMC Public Health. 2018;18(1):1-9.
Crossref - Al Anazi M, AlFayyad I, AlOtaibi R, Abu-Shaheen A. Epidemiology of brucellosis in Saudi Arabia. Saudi Medical Journal. 2019, 40(10):981.
- EL-Rahim A, Asghar AH. Brucellosis in ruminant animals and their close contact humans in Western Region of Saudi Arabia in 2012. Assiut Vet Med J. 2014;60(140):1-6.
Crossref - Radwan AI, Asmar JA, Frerichs WM, Bekairi SI, Al-Mukayel AA. Incidence of brucellosis in domestic livestock in Saudi Arabia. Trop Anim Health Prod. 1983;15(3):139-143.
Crossref - Human W. animal brucellosis epidemiological surveillance in the MZCP Countries. Report of a WHO. 1998.
- Memish Z, Venkatesh S. Brucellar epididymo orchitis in Saudi Arabia: a retrospective study of 26 cases and review of the literature. BJU International. 2001;88(1):72-76.
Crossref - Asaad AM, Alqahtani JM. Serological and molecular diagnosis of human brucellosis in Najran, Southwestern Saudi Arabia. J Infect Public Health. 2012;5(2):189-194.
Crossref - Musallam I, Abo-Shehada M, Hegazy Y, Holt H, Guitian F. Systematic review of brucellosis in the Middle East: disease frequency in ruminants and humans and risk factors for human infection. Epidemiol Infect. 2016;144(4):671-685.
Crossref - Alyousef M, Aldoghaither R. First case of cervical epidural abscess caused by brucellosis in Saudi Arabia: A case report and literature review. IDCases. 2018;12:107-111.
Crossref - Abdellatif M, Osman YHA, Arafat HH, Mahmoud AZE. Seroprevalence and 16S rRNA gene sequence analysis of Brucella spp. among domestic ruminants in Northern Border, Saudi Arabia Medical Science. Med Sci. 2019;24(101):165-173.
- Alsharari AA, Altuwaijri S. Seroprevalence Rate of Brucellosis in Sheep at Aljouf Region, Saudi Arabia. Journal of Animal, Poultry & Fish Production. 2021;10(1):71-81.
Crossref - Teng Y-H, Teng J-J, Chao S, Chao H, Waghela SD. Comparison of the rose bengal plate and the complement fixation tests with the tube agglutination test for diagnosis of human brucellosis. Open Journal of Clinical Diagnostics. 2017;7(3):73-82.
Crossref - Vemulapalli R, He Y, Buccolo LS, Boyle SM, Sriranganathan N, Schurig GG. Complementation of Brucella abortus RB51 with a functional wboA gene results in O-antigen synthesis and enhanced vaccine efficacy but no change in rough phenotype and attenuation. Infect Immun. 2000;68(7):3927-3932.
Crossref - Wang Z, Niu J, Wang S, Lv Y, Wu Q. In vivo differences in the virulence, pathogenicity, and induced protective immunity of wboA mutants from genetically different parent Brucella spp. Clin Vaccine Immunol. 2013;20(2):174-180.
Crossref - Zygmunt MS, Blasco JM, Letesson J-J, Cloeckaert A, Moriyon I. DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. BMC Microbiol. 2009;9(1):1-13.
Crossref - Etemady A, Mohammdi M, Esmaelizad M, et al. Genetic characterization of the wboA gene from the predominant biovars of Brucella isolates in Iran. Electron Physician. 2015;7(6):1381-1386.
- Georgi E, Walter MC, Pfalzgraf M-T, et al. Whole genome sequencing of Brucella melitensis isolated from 57 patients in Germany reveals high diversity in strains from Middle East. PLoS one. 2017;12(4):e0175425.
Crossref - Minogue T, Daligault H, Davenport K, et al. Whole-genome sequences of 24 Brucella strains. Genome Announc. 2014;2(5):e00915-14.
Crossref - Audic S, Lescot M, Claverie J-M, Scholz HC. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics. 2009;10(1):1-18.
Crossref - Erdenebaatar J, Bayarsaikhan B, Yondondorj A, et al. Epidemiological and serological survey of brucellosis in Mongolia by ELISA using sarcosine extracts. Microbiol Immunol. 2004;48(8):571-577.
Crossref - Suryawanshi S, Tembhurne P, Gohain S, Ingle V. Prevalence of Brucella antibodies in sheep and goats in Maharashtra. Indian Research Journal of Extension Education. 2016;14(4):75-77.
- Bilal N-E, Jamjoom G, Bobo R, ALY OM, El-Nashar N. Brucellosis in the Asir region of Saudi Arabia. Saudi Med J. 1991;12(1):37-41.
- Alhamada AG, Habib I, Barnes A, Robertson I. Risk factors associated with Brucella seropositivity in sheep and goats in Duhok Province, Iraq. Vet Sci. 2017;4(4):65.
Crossref - Al-Hakami AM, Alqahtani AJ, Moosa RA, et al. Seroprevalence of brucellosis among exposed agro-pastoral communities in southern Saudi Arabia. Asian Pac J Trop Med. 2019;12(12):454-551.
Crossref - Natesan K, Kalleshamurthy T, Nookala M, et al. Seroprevalence and risk factors for brucellosis in small ruminant flocks in Karnataka in the Southern Province of India. Vet World. 2021;14(11):2855-2862.
Crossref - Soares J, Wronski T. Preliminary disease survey of domestic ruminants on Farasan Islands, Saudi Arabia. Approaches in Poultry, Dairy & Veterinary Sciences. 2021;8(4).
Crossref - Munsi MN, Akther S, Rahman MH, Hassan MZ, Ali MZ, Ershaduzzaman M. Seroprevalence of Brucellosis in goats in some selected areas of Bangladesh. J Adv Vet Anim Res. 2021;8(1):123-128.
Crossref - Rahman M, Faruk M, Her M, Kim J, Kang S, Jung S. Prevalence of brucellosis in ruminants in Bangladesh. Vet Med. 2011;56(8):379-385.
Crossref - Shabana II, Krimly RA. Seroprevalence of some viral and bacterial zoonoses in domestic ruminants in Medina. J Adv Vet Anim Res. 2019;7(1):42-50.
Crossref - Al-Majali AM. Seroepidemiology of caprine brucellosis in Jordan. Small Rumin Res. 2005;58(1):13-18.
Crossref - Al-Marzooqi W, Elshafie EI, Al-Toobi A, et al. Seroprevalence and Risk Factors of Brucellosis in Ruminants in Dhofar Province in Southern Oman. Vet Med Int. 2022;176147.
Crossref - Al-Griw HH, Kraim ES, Farhat ME, Perrett LL, Whatmore AM. Evidence of ongoing brucellosis in livestock animals in North West Libya. J Epidemiol Glob Health. 2017;7(4):285-288.
Crossref - Ahasan M, Rahman M, Song H-J. A sero-surveillance of Brucella spp. antibodies and individual risk factors of infection in cattle in Bangladesh. Korean J Vet Serv. 2010;33(2):121-128.
- Hegazy YM, Moawad A, Osman S, Ridler A, Guitian J. Ruminant brucellosis in the Kafr El Sheikh Governorate of the Nile Delta, Egypt: prevalence of a neglected zoonosis. PLoS Negl Trop Dis. 2011;5(1):e944.
Crossref - Kandeel A, Gamal T, Sediek A, Salauddin H, Fadlelmoula A. Seroprevalence of Brucellosis within sheep and goat flocks in Alkamil province in Saudi Arabia. Bothalia J. 2014;44(5):131-138.
- Rajala EL, Grahn C, Ljung I, Sattorov N, Boqvist S, Magnusson U. Prevalence and risk factors for Brucella seropositivity among sheep and goats in a peri-urban region of Tajikistan. Trop Anim Health and Prod. 2016;48(3):553-558.
Crossref - Burridge MJ. Epidemiological approaches to disease control. Diseases of Cattle in the Tropics. Springer. 1981:53-64.
Crossref - Kadohira M, McDermott J, Shoukri M, Kyule M. Variations in the prevalence of antibody to Brucella infection in cattle by farm, area and district in Kenya. Epidemiol Infect. 1997;118(1):35-41.
Crossref - Cowie CE, Marreos N, Gortazar C, Jaroso R, White PC, Balseiro A. Shared risk factors for multiple livestock diseases: A case study of bovine tuberculosis and brucellosis. Res Vet Sci. 2014;97(3):491-497.
Crossref - Alshaikh M, Al-Haidary A, Aljumaah R, et al. First detection of Brucella abortus in camel serum in Saudi Arabia using the polymerase chain reaction. J Appl Anim Res. 2007;31(2):149-152.
Crossref - Angesom H, Mahendra P, Tesfu K, Fikre Z. Sero-epidemiology of camel brucellosis in the Afar region of Northeast Ethiopia. J Vet Med Anim Health. 2013;5(9):269-275.
- Habtamu T, Richard B, Dana H, Kassaw A. Camel brucellosis: its public health and economic impact in pastoralists, Mehoni district, Southeastern Tigray, Ethiopia. J Microbiol Res. 2015;5(5):149-156.
- Omer M, Musa M, Bakhiet M, Perrett L. Brucellosis in camels, cattle and humans: associations and evaluation of serological tests used for diagnosis of the disease in certain nomadic localities in Sudan. Rev Scie Tech. 2010;29(3):663-669.
Crossref - Mohamud AS, Kothowa JP, Mfune RL, Mubanga M, Godfroid J, Muma JB. Seroprevalence and Risk Factors Associated with Brucella Infection in Camels in the Puntland State of Somalia. Vet Sci. 2021;8(7):137.
Crossref - Gwida M, El-Gohary A, Melzer F, Khan I, Rosler U, Neubauer H. Brucellosis in camels. Res Vet Sci. 2012;92(3):351-355.
Crossref - Ran X, Cheng J, Wang M, et al. Brucellosis seroprevalence in dairy cattle in China during 2008-2018: A systematic review and meta-analysis. Acta Tropica. 2019;189:117-123.
Crossref - Ali S, Saleem S, Imran M, et al. Detection of Brucella antibodies in selected wild animals and avian species in Pakistan. Indian J Anim Res. 2020;54(4):478-481.
- Ebid M, El Mola A, Salib F. Seroprevalence of brucellosis in sheep and goats in the Arabian Gulf region. Vet World. 2020;13(8):1495-1509.
Crossref - Kabagambe E, Elzer P, Geaghan J, Opuda-Asibo J, Scholl D, Miller J. Risk factors for Brucella seropositivity in goat herds in eastern and western Uganda. Prev Vet Med. 2001;52(2):91-108.
Crossref - Samadi A, Ababneh M, Giadinis N, Lafi S. Ovine and caprine brucellosis (Brucella melitensis) in aborted animals in Jordanian sheep and goat flocks. Vet Med Int. 2010.
Crossref - Akhter L, Islam MA, Das S, Khatun MM. Seroprevalence of brucellosis and its associated risk factors in sheep and goat in the farms and slaughter house in Mymensingh, Bangladesh. Microbes and Health. 2014;3(1):25-28.
Crossref - Gebremedhin EZ. Seroepidemiology of ovine brucellosis in East and West Shewa Zones of Oromia Regional State, Central Ethiopia. J Veterinar Sci Technol. 2015;6(6):1000265.
Crossref - Yagupsky P. Detection of Brucella melitensis by BACTEC NR660 blood culture system. J Clin Microbiol. 1994;32(8):1899-1901.
Crossref - Nagalingam M, Shome R, Balamurugan V, et al. Molecular typing of Brucella species isolates from livestock and human. Trop Anim Health Prod. 2012;44(1):5-9.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.