ISSN: 0973-7510
E-ISSN: 2581-690X
Enzyme linked Immunosorbent Assay (ELISA) and Immunofluorescence Assay (IFA) are specific serological tests for Scrub Typhus (ST). The etiological agent, Orientia tsutsugamushi DNA is demonstrable by PCR. Different countries apply ST IgM IFA cut-off titres ranging from 1:10 to 1:10240. To avoid confusion, Scrub Typhus Inclusion Criteria (STIC) was proposed. One criterion was a fourfold increase to or single IFA titre of ≥1:10240, later lowered to ≥1:3200. Equally sensitive and specific ELISA has no place in STIC. Modification of STIC is needed for robust laboratory confirmation of ST. One hundred and forty clinically suspected ST patients, including 58 patients who provided paired blood samples, were subjected to ST IgM ELISA, IgM IFA and Real time PCR targeting 56kDa, 47kDa and groEL genes. Among paired samples, all 58 (100%) acute sera were positive for IgM ELISA while 49 were positive in IgM IFA (84.5%). Among convalescent samples, 50 and 43 were positive in ST IgM ELISA and ST IgM IFA (86.2% and 74.1%) respectively. Regarding 82 unpaired samples, IgM ELISA and IgM IFA positivity was observed in 82 (100%) and 72 (87.8%) cases, respectively. PCR positivity in 140 ST cases for all three or any two gene targets, 56kDa, 46kDa and groEL as per STIC was 84, while an additional 40 samples were positive for any one gene. In Indian context, STIC requires modification to include IgM ELISA positivity, lower IgM cutoff IFA titre of ≥1:64 and any one gene positivity in Real time PCR.
Orientia tsutsugamushi, Eschar, ST IgM ELISA, ST IgM IFA, 56kDa, 47kDa, groEL genes
Scrub Typhus (ST) or Mite borne typhus is an emerging/re-emerging infectious zoonotic disease transmitted by the bite of infected larval stage (chiggers) of Trombiculid mites, Leptotrombidium deliense which are found in the areas of heavy scrub vegetation. ST is known for its endemicity in the so called “Tsutsugamushi Triangle” comprising many areas of Northern India and Australia, China, Japan, Indonesia, Malaysia, Thailand, Pakistan, and Korea.1-9 This triangle extended to another two more countries, one from Djibouti region of Africa with three cases10 and another from Kenya.11 Now ST is an endemic disease throughout South, North, East and West India3
Isolation of this pathogen is attempted only in rickettsial research/reference laboratories, since it requires Bio-safety Level III containment facilities. O. tsutsugamushi can be grown in several cell lines like EVC304, L929, ECV304 cells and Vero cells lines.5 Scrub typhus is now being reported throughout India and different parts of the world. For the past four decades, serological diagnosis of ST in India has witnessed the use of ST specific ELISA.12-31 and Lateral flow based Rapid Detection Test (RDT) kits.1,9,12,21 Development of immunofluorescence assay (IFA) for ST has an important milestone in the diagnosis and research of ST.15,18,20,21,24,25,32-39 Demonstration of O. tsutsugamushi DNA in the patients’ blood/eschar tissue by the application of molecular tests like Polymerase Chain Reaction (PCR) is as specific and relevant in the diagnosis of ST and on par with isolation in culture.6,7,18, 22,23,26,40-46
Different countries recorded prevalence of ST based on IFA, but applying cut-off titres ranging from 1:10 to 1:10240.15,20,21,24,25,31-36,38,39,44 To avoid confusion, Scrub Typhus Inclusion Criteria (STIC) was proposed with a cut off titre of ≥1:10240, which is quite high and does not align with findings from India and other countries.33,38 Besides, the equally sensitive and specific serological ELISA did not find a place in STIC. The need for incorporating modifications to STIC is discussed in the light of our experience and published reports from several authors.
This prospective laboratory-based study was carried out in the department of Microbiology, from August, 2016 to December, 2018, after getting approval from our Institutional Human Ethics committee (IHEC). (Approval Project number PG Dissertation/2015/10/01). Informed written consent from the participants was obtained prior to collection of blood. Blood samples from out-patients/in-patients, who attended our tertiary hospital with febrile illness and were clinically suspected as ST and tested positive for ST by Rapid detection test (RDT) kits were archived and anonymised. One hundred and forty clinically suspected ST patients, which included 58 who provided paired blood samples were subjected to ST IgM ELISA, ST IgM IFA and Real time PCR targeting, 47kDa and groEL genes.
Scrub typhus case definitions33
Suspected clinical case
Fever for more than 7 days of illness with one or more of the following clinical symptoms: Eschar, headache, rash, cough, malaise, myalgia, lymphadenopathy, hepatosplenomegaly.
Probable case
Suspected patient with the clinical symptoms and elevated levels of ALT/AST and defervescence of fever within 48-72 hours of antibiotic treatment and positive in the ST screen by Rapid Immunochromatographic test (ICT) like Standard Diagnostics SD Bioline Tsutsugamushi -Assay (IgM, IgG & IgA)/Inbios Rapid test for scrub typhus IgM/ ImmuneMed Scrub Typhus Rapid (IgM & IgG)
Definitive case to be confirmed by any one of the following
Positive for IgM ELISA with an OD value ≥0.5, Positive IgM IFA titre of ≥1:64 and Detection of O. tsutsugamushi DNA in whole blood samples by qPCR. Serological Diagnosis of Scrub typhus:
IgM ELISA
For ST IgM ELISA, Scrub typhus DetectTM IgM ELISA kit (InBios International, Seattle, USA) was used. The ELISA plates were coated with ten recombinant antigens of O. tsutsugamushi, targeting antibodies to the 56kDa antigen. The procedure was performed in strict compliance with the manufacturer’s instructions. Cut-off values were calculated and interpretation of the test results was computed as reported earlier.19 The samples with OD value of ≥0.5 for IgM ELISA,1,3 were considered positive and those below the cut off were taken as negative. Borderline samples were tested in triplicate.
Immunofluorescence assay (IFA)
OTM-120 Orientia tsutsugamushi IFA IgM Antibody kit was purchased from Fuller laboratories, Fullerton, California, USA and these slides was incorporated with four serotypes namely Kato, Karp, Boryong and Gilliam, propagated in L-292 cells in vitro and presented in a linear array of infected cells within each slide well. The procedure was followed according to the technical brochure supplied in the kit. As per the kit, the cut off titre of ≥1:64 for IFA IgM were considered as positive. A small sharply defined fluorescent rod within each antigen spot was taken as positive. Positivity in any one/or more of the four serotypes was taken as positive.
Molecular diagnostic tests
Real time PCR was performed for detection of O. tsutsugamushi DNA in blood samples, targeting three different genes: Detection of the most common 56kDa gene was performed by a commercial Geno-Sen’s Real Time PCR (Genome Diagnostics Pvt. Ltd., India) as described earlier,12 followed by in-house TaqMan Probe PCR for 47kDa and SYBR Green Real Time PCR for groEL gene. Known Scrub typhus positive patient DNA was used as the positive control in these molecular tests. The PCR primers and probes were purchased from Sigma Aldrich, Bangalore.
Commercial real time PCR for 56kDa
ST Real Time PCR was performed to amplify O. tsutsugamushi DNA by the gene targeting 56kDa by a commercial kit Geno-Sen’s Real Time PCR (Genome Diagnostics Pvt. Ltd., Solan, Himachal Pradesh, India).12 This qPCR was carried out in a CFX96 C1000 Touch machine (Bio-Rad, USA). The primers for Commercial qPCR kit were selected from Accession No.: KP334159.1 (www.genomediagnostics.co.in). The amplification protocol was followed as per the kit’s technical brochure. Briefly, the kit targeted 56kDa type specific gene of O. tsutsugamushi DNA. The master mix contains reagents and enzymes for the specific amplification of the target gene through FAM Channel. Amplification Reaction and its cyclic conditions were mentioned as earlier.12
TaqMan probe based real time PCR for 47kDa
TaqMan probe qPCR was performed for 47kDa and the primers and probes were selected from Kim et al.43 The primers and probes were standardized according to the kit brochure of TaqMan master mix. A Ct value ≤40.0 was considered as positive. Briefly, the details of Ingredients and the cyclic conditions are as follows: The reaction mixture contains 25.0 µl which comprises of 12.5 µl of TaqMan probe qPCR (Premix Ex Taq, TakaraBio) followed by 0.5 µl of each 2 µM forward and reverse primers with inclusion of 1.0 µl of probe specific, 8.5 µl of Nuclease free water and 2.0 µl of genomic DNA was added as a template for amplification. The cyclic conditions are as follows: 95 °C for 10 min for Initial denaturation followed by 45 cycles of 95 °C for 10 secs for Denaturation, 58 °C for 30 secs for annealing and 72 °C for 10 secs for Extension and the final extension at 72 °C for 5 mins.
SYBR green real time PCR for groEL gene
An in-house SYBR green qPCR was performed for groEl gene and the primers were selected from the publication of Paris et al.43 In each run positive and negative controls were included. All samples were run in duplicates for confirmation of the result. A Ct value ≤35.0 cycle was considered as positive. The reaction mixture contains 20.0 µl which comprises of 10.0 µl of SYBR Green Master Mix (Thermo Fisher Scientific) followed by 1.0 µl of each 2 µM forward and reverse primers, 6.0 µl of Nuclease free water and 2.0 µl of genomic DNA was added as a template for amplification. The cyclic conditions are as follows: 95 °C for 5 mins for Initial denaturation followed by 35 cycles of 95 °C for 15 secs for Denaturation, 54 °C for 15 secs for annealing and 72 °C for 5 secs for Extension and the final extension at 72 °C for 30 secs. Positive and negative controls were included in each run. Human Beta actin (ACTB) was used as the housekeeping gene to check the quality of the genomic DNA. The ACTB primers were as suggested by Mediannikov et al.47
Statistical analysis
Percentages were calculated for categorical variables. Mean and standard deviations (SD) were calculated with 95% confidence interval for numerical variables. Chi-square test was applied for numerical variables and Fisher’s exact test for small sample sizes. Statistical tests were analysed by QuickCalcs, GraphPad Prism. P values ≤0.05 were considered statistically significant.
All 58 (100%) acute sera were positive for IgM ELISA while 49 were IgM IFA positive (84.5%) with titres ranging from 1:64 to 1:512. Among convalescent samples, 50 and 43 were positive in ST IgM ELISA and ST IgM IFA with a percentage positivity of 86.2% and 74.1% respectively. In the second category of 82 patients from whom only one sample was collected, ST IgM ELISA was positive for all samples (100%), while only 87.8% positivity was observed in ST IgM IFA. Concordance between IgM ELISA and IgM IFA was observed for 49 acute (84.5%) and 37 convalescent samples (63.8%). IgM IFA titres ranged from ≥1:64 to
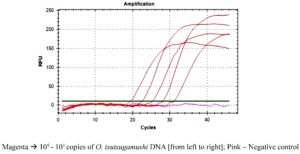
≥1:1024. Serum samples had antibodies against any one or more of the four different serotypes, Kato, Karp, Boryong and Gilliam. Figure 1 shows the limit of detection of O. tsutsugamushi DNA copies for the detection of positivity in patient’s samples.
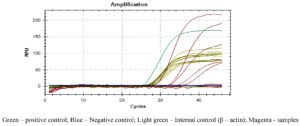
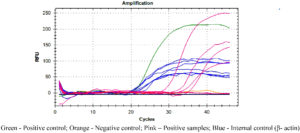
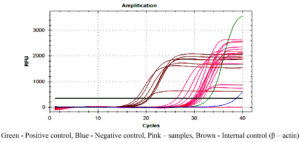
Figures 2, 3 and 4 represent the results of Real Time PCR with positivity of 31, 29 and 38 respectively for 56kDa, 47kDa and groEL gene targets. All three genes were detected in 11 patients, two genes detected in 25 patients and only one gene detected in 15 patients. Thus PCR positivity in any one/two/three genes was 51 (87.9%). Among 15 cases positive for single gene in qPCR, all were ST IgM ELISA positive and 14 were ST IgM IFA positive as well. Table 1 Compares the results of ST IgM ELISA, ST IgM IFA and qPCR in viz., 58 patients from whom paired blood samples were collected and 82 patients from whom only the acute samples could be collected. Regarding 82 unpaired samples, nine were positive for all three genes, 39 for any two genes and 25 had only single gene positivity, with an overall PCR positivity of 73 out of 82 samples (89.0%). Among the 25 single gene positive cases, all were positive for ST IgM ELISA while 23 were positive for IgM IFA (92.0%). Table 2 compares the results of ST IgM ELISA and ST IgM IFA in both acute and convalescent sera. Statistical significance was observed between ST IgM ELISA and IFA among acute samples (p = 0.0028) and unpaired samples (p = 0.0007). Significance was not observed in convalescent samples (p = 0.1030). Table 3 shows the comparison between acute and unpaired samples positivity of Real Time PCR. The results have shown that there is no statistical difference between the positivity of the three gene targets employed.
Table (1):
Comparison of the results of ST IgM ELISA, ST IgM IFA and qPCR (n = 140)
| Category-A | PCR positivity for gene targets in Acute Samples Only of Paired sera (n = 58) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All three seen 56kDa + 47kDa + groEL | Two genes detected | Only one gene detected | PCR Positive | PCR Negative | Total | |||||
| 56kDa + 47kDa | 56KDa + groEL | 47kDa + groEL | 56 kDa | 47 kDa | groEL | |||||
| IgM ELISA & IFA Positive | 11 | 4 | 6 | 9 | 3 | 3 | 8 | 44 | 5 | 49 |
| Only IgM ELISA positive | 0 | 2 | 4 | 0 | 1 | 0 | 0 | 7 | 2 | 9 |
| Sub-Total | 11 | 6 | 10 | 9 | 4 | 3 | 8 | 51 | 7 | 58 |
| Category-B | PCR positivity for gene targets in the Unpaired Single Samples (n = 82) | |||||||||
| IgM ELISA & IFA Positive | 7 | 10 | 12 | 16 | 4 | 9 | 7 | 65 | 6 | 71 |
| Only IgM ELISA positive | 2 | 0 | 0 | 1 | 2 | 0 | 3 | 8 | 3 | 11 |
| Sub-total | 9 | 10 | 12 | 17 | 6 | 9 | 10 | 73 | 9 | 82 |
| Total | 20 | 16 | 22 | 26 | 10 | 12 | 18 | 124 | 16 | 140 |
Table (2):
Comparison of Serological test results of ST IgM ELISA and ST IgM IFA in all samples tested (n = 198) (58 + 58 + 82)
| Category | Test | Positive (%) | Negative (%) | Total | P value |
|---|---|---|---|---|---|
| Acute sera (n = 58) | ST IgM ELISA | 58 (100%) | 0 | 58 | 0.0028 |
| ST IgM IFA | 49 (84.5%) | 9 (15.5%) | 58 | ||
| convalescent sera(n = 58) | ST IgM ELISA | 50 (86.2%) | 8 (13.8%) | 58 | 0.1030 |
| ST IgM IFA | 43 (75.9%) | 15 (24.1%) | 58 | ||
| Unpaired samples (n = 82) | ST IgM ELISA | 82 (100%) | 0 | 82 | 0.0007 |
| ST IgM IFA | 71 (86.6%) | 11 (13.4%) | 82 | ||
| Total ST IgM ELISA positivity: 190 (95.96%) (n = 198) Total ST IgM IFA positivity: 163 (85.78%)(n = 198) |
|||||
Table (3):
Real Time PCR results for Acute and Unpaired single samples tested (n = 140)
| Acute samples (n = 58) | Positivity | Unpaired single samples Positivity (n = 82) | Total | P value |
|---|---|---|---|---|
| All three genes | 11 | 9 | 20 | 0.2230 |
| Two genes | 25 | 39 | 64 | 0.7268 |
| Only one gene | 15 | 25 | 40 | 0.6841 |
| Total PCR positivity | 51 | 73 | 124 | |
| Positivity as per STIC | 36 | 48 | 84 | 0.8063 |
| Details of gene targets detected in Real time PCR | ||||
| 56kDa | 31 | 37 | 68 | 0.4241 |
| 47kDa | 29 | 45 | 74 | 0.6908 |
| groEL | 38 | 48 | 86 | 0.5095 |
The main purpose of this research is to arrive at a consensus towards laboratory confirmation of ST in India. Scrub Typhus Inclusion criteria (STIC) proposed by Paris et al.33 is yet to gain universal acceptance. There is full agreement with reference to two points, viz., isolation of O. tsutsugamushi from patients’ blood and demonstration of O. tsutsugamushi DNA in the blood or eschar tissue of ST patients. The two other criteria which Rickettsiologists from several countries differ are ST confirmation by serological and molecular diagnosis targeting three genes, 56kDa, 47kDa and groEL.
Serological diagnosis
IgM ELISA
There is no mention of IgM ELISA for the serological confirmation of ST in STIC. This is not withstanding the benefits of ELISA, which is an affordable, commercial kit available in India and other developing countries and results are easy to interpret. However, there has been a lot of confusion regarding the cut off OD values for ELISA, since the values vary from as low as 0.06416 to as high as 1.848 as reviewed by Saraswati et al.,16 Chunduru et al.,47 and Kannan et al.18 from several countries like China, Japan, Korea, Thailand and different states in India. Patricia et al.13 arrived at cut off of 0.406. Vanlalruati et al.,27 Pote et al. from Wardha, Central India,9 Stephen et al. from Puducherry, South India,1 consider ELISA diagnostic cut-off titre 0.5 OD. Kalal et al. from Andhra Pradesh set their ELISA cut off OD at 0.6.28 Manjunathachar et al. recommended cut off value of >0.72 for diagnostic ELISA for Madhya Pradesh in central India.17 Indian Council of Medical Research has recommended ST IgM ELISA OD of ≥0.5.3 for ST Inbios IgM ELISA kit, which is used by most researchers in India and abroad.12,13,15,17,18,21,31 Varghese et al. and Kannan et al. employing ST Detect IgM ELISA kit (InBios International, Seattle, USA) set OD cut off values of ≥0.8.18,37
A recent report from Karnataka by Chunduru et al. revealed an OD value cut-off of 1.309 with a sensitivity and specificity of 98.7% for IgM ELISA.47
Jain et al. screened serum samples from 29 labs across India for anti-IgM antibodies by ELISA. ROC curve-based cut-off at an OD of 0.554 had a sensitivity of 95.2 per cent and specificity of 95.1 per cent, whereas OD of >1 had 100 per cent specificity.29 Gupta et al. recommends ELISA OD of ≥0.87/0.8915, 22 and fix a range of 0.5 to 1.0 which should be geography specific OD values. We endorse this view as our recommendations to modifications in STIC. Several authors are of the opinion that ST IgM ELISA positivity must be considered on par with ST IgM IFA. ELISA may be a better option than IFA, which is an expensive, subjective, technically demanding test and can have the interpreter bias.24 In resource poor countries and remote locations, the Point of care Rapid detection Test (RDT) kits for ST would be equally useful like ST IgM ELISA and some times better than IFA, a view shared by reports from Nepal25 and Vellore, India18 and based on our experience as well.1,12,19,21,23,26 Recently Zhang et al. developed a chimeric 56-21kDa antigen-based ELISA, which has no cross reaction with other rickettsioses.30
IgM IFA
IFA is considered ‘the serological gold standard’ for ST diagnosis. STIC by Paris et al.,33 has set a very high cut off of ≥1:10,240 for ST IgM IFA. This was later reduced to ≥1:3,200 by the same group of researchers.38 Among two commercial ST IgM IFA kits manufactured from USA, viz., Focus Diagnostics and Fuller Laboratories, the later has been used by majority of Indian researchers. Cut off titres recommended by the kit’s brochure is ≥1:64, although Koraluru et al. recommends a cut off of ≥1:128.20 Gautam et al. from Nepal using their in-house IFA followed cut off titre of ≥1:128.25 We are of the opinion that in India the cut off ST IgM IFA titre could be fixed at ≥1:64, which is also in line with the guidelines given in the technical brochure of Fuller Laboratories kit and also supported by other authors from Central India (Maharashtra, Madya Pradesh), Delhi and surrounding areas, Vellore (Tamil Nadu)Andhra Pradesh, Puducherry and different states across India.2,9,15,18,19,22,24,29 Lim and associates recommended IFA cut off of ≥1:512 due to the imperfect quality of this test.32 Korea Centers for Disease Control and Prevention (KCDC) recommends ST IgM IFA cut off titre of ≥1:16 or ≥4-fold increase in paired sera. KCDC ST IgM IFA cut of ≥1:1039 is considered too low according to Kim et al., since it gives more false positivity.34 Demonstrating a fourfold increase in IgM IFA titres of paired sera as set by STIC could not be achieved by majority of rickettsiologists. The highest ST IgM IFA titre among our 58 ST patients was only 1:512 and seroconversion was observed in four patients only. Of the 82 unpaired samples the highest IFA titre of 1:1024 was seen in five samples. It is to be remembered that ST IgM antibodies can persist for longer periods from one8 to 13 years34 due to false positivity and cross reactions with other diseases,4,14,36 thus causing diagnostic difficulty in acute ST. Recently, systematic review and meta-analysis was conducted in India, and recorded the burden of scrub typhus in acute febrile illness and in association with liver diseases.48,49
Molecular diagnosis
Several authors have independently confirmed ST in molecular tests employing 56 kDa/47kDa/groEL/16S rRNA. Anitha et al. from Puducherry have targeted three genes, 56kDa, 47kDa and groEL,23 whereas Patricia et al. have identified 56kDa, groEL and 16S rRNA genes in their study on ST in Puducherry.13 Two genes viz., 56kDa and 47kDa were included in their PCR by Kannan et al.,18 47kDa and groEL together were targeted by researchers in India and abroad.13,40 According to Tantibhedhyangkul et al. the, sensitivity and specificity of the multiplex PCR with these two targets is 86% and 100% respectively.40 An analysis of our molecular study, positivity in the two genes could be grouped as follows: 26 were positive for 47kDa + groEL followed by 22 and 16 for 56kDa + groEL and 56kDa + 47kDa gene respectively. Only 56kDa was targeted by some authors.22,24,26,41 According to STIC, PCR confirmation is permissible only with the detection of any two of three genes. Since each of the three genes play an independent role in laboratory confirmation of ST, our view is that real time PCR positivity for any one gene should be recognised. Among our first group of 58 ST patients, single gene positivity in qPCR was corroborated by ST IgM ELISA positivity in 15 patients and 14 of them were also positive in ST IgM IFA with titres of ≥1: 64. In the second category of 82 patients, single gene positivity was observed in 25 patients. While all of these 25 were positive for IgM ELISA, 20 were positive for IgM IFA. Thus, single gene positivity cannot be altogether ignored.
In view of the complexity, need for technical expertise, an expensive fluorescent microscope and further validation of IFA kits in India and its cut-off value, ELISA may be considered as a viable alternative to the reference test IFA at present. Scrub Typhus Inclusion Criteria (STIC) should be aligned to Indian research findings to include IgM ELISA positivity (OD value between 0.5 to 1.0), IgM cutoff titre of ≥1:64 and any one gene positivity in Real time PCR.
O. tsutsugamushi DNA detection by real time PCR targeting all three genes will result in identifying additional ST cases.
Limitations of this study
Sample size is small (n = 140) representing clinically confirmed cases of Scrub typhus patients. ST IgM IFA needs further evaluation from different parts of India. False positivity in IgM ELISA/IFA due to cross reactions with other diseases and persistence of ST IgM positivity in some patients for one year to several years makes it difficult to confirm the acute stage of ST infection based on serological evidence alone.
ACKNOWLEDGMENTS
The authors are grateful to the Honourable Chancellor, Vice-Chancellor, Vice-president (R.I.D.), Dean-Research, Sri Balaji Vidyapeeth (Deemed-to-be University), and Dean, Mahatma Gandhi Medical College and Research Institute, Puducherry, for providing the research facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
VA, JP and SS conceptualized and designed the study. VA and JP carried out the molecular work and collected the dataset for the study. VA, JP and SS drafted the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by Seed money through Intramural Research (IMR), vide IMR Reference No.: SBV/IRC/Seed Money/07/2021.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Mahatma Gandhi Medical College and Research Institute, Puducherry, India (IEC: PG DISSERTATION/2015/10/01).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Stephen S, Sangeetha B, Ambroise S, et al. Outbreak of scrub typhus in Puducherry & Tamil Nadu during cooler months. Indian J Med Res. 2015;142(5):591-597.
Crossref - Mahajan SK, Rolain JM, Kashyap R, et al. Scrub typhus in Himalayas. Emerg Infect Dis. 2006;12(10):1590-1592.
Crossref - Rahi M, Gupte MD, Bhargava A, Varghese GM, Arora R. DHR-ICMR guidelines for diagnosis & management of Rickettsial diseases in India. Indian J Med Res. 2015;141(4):417-422.
Crossref - Paris DH, Shelite TR, Day NP, Walker DH. Unresolved Problems Related to Scrub Typhus: A Seriously Neglected Life-Threatening Disease. Am J Trop Med Hyg. 2013;89(2):301-307.
Crossref - Ko Y, Choi JH, Ha NY, Kim IS, Cho NH, Choi MS. Active escape of Orientia tsutsugamushi from cellular autophagy. Infect Immun. 2013;81(2):552-559.
Crossref - Biswal M, Zaman K, Suri V, et al. Use of eschar for the molecular diagnosis and genotypic characterisation of Orientia tsutsugamushi causing scrub typhus. Indian J Med Microbiol. 2018;36(3):422-425.
Crossref - Varghese GM, Janardhanan J, Mahajan SK, et al. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with scrub typhus in 3 regions of India. Emerg Infect Dis. 2015;21(1):64-69.
Crossref - Trowbridge P, Divya P, Premkumar PS, Varghese GM. Prevalence and risk factors for scrub typhus in South India. Trop Med Int Health. 2017;22(5):576-582.
Crossref - Pote K, Narang R, Deshmukh P. Diagnostic performance of serological tests to detect antibodies against acute scrub typhus infection in central India. Indian J Med Microbiol. 2018;36(1):108-112.
Crossref - Horton KC, Jiang J, Maina A, et al. Evidence of Rickettsia and Orientia infections among abattoir workers in Djibouti. Am J Trop Med Hyg. 2016;95(2):462-465.
Crossref - Thiga JW, Mutai BK, Eyako WK, Ng’ang’a Z, Jiang J, Richards Al, Waitumbi JN. High seroprevalence of antibodies against spotted fever and scrub typhus bacteria in patients with febrile Illness, Kenya. Emerg Infect Dis. 2015;21(4):688-691.
Crossref - Anitharaj V, Stephen S, Pradeep J, Pooja P, Preethi S. Validation of Geno-Sen’s Scrub Typhus Real Time Polymerase Chain Reaction Kit by its Comparison with a Serological ELISA Test. J Global Infect Dis. 2017;9(3):108-112.
Crossref - Patricia KA, Hoti SL, Kanungo R, Jambulingam P, Shashikala N, Naik AC. Improving the diagnosis of scrub typhus by combining groEL based polymerase chain reaction and IgM ELISA. J Clin Diagn Res. 2017;11(8):DC27-DC31.
Crossref - Prakash JAJ, Abraham OC, Mathai E. Evaluation of tests for serological diagnosis of scrub typhus. Trop Doct. 2006;36(4):212-213.
Crossref - Gupta N, Chaudhry R, Thakur CK. Determination of cutoff of ELISA and immunofuorescence assay for scrub typhus. J Glob Infect Dis. 2016;8(3):97-99.
Crossref - Saraswati K, Phanichkrivalkosil M, Day NPJ, Blacksell SD. The validity of diagnostic cut-offs for commercial and in-house scrub typhus IgM and IgG ELISAs: A review of the evidence. PLoS Negl Trop Dis. 2019;13(2):e0007158.
Crossref - Manjunathachar HV, Barde PV, Raut CG, et al. Determination of cut-off of diagnostic ELISA for Scrub typhus in endemic setup: Central India. J Vector Borne Dis. 2021;58(1):90-93.
Crossref - Kannan K, John R, Kundu D, et al. Performance of molecular and serologic tests for the diagnosis of scrub typhus. PLoS Negl Trop Dis. 2020;4(11):e0008747.
Crosssref - Anitharaj V, Stephen S, Pradeep J, et al. Serological diagnosis of acute scrub typhus in Southern India: Evaluation of InBios Scrub Typhus Detect IgM Rapid Test and comparison with other serological tests. J Clin Diagn Res. 2016;10(11):DC07-DC10.
Crossref - Koraluru M, Bairy I, Varma M, Vidyasagar S. Diagnostic validation of selected serological tests for detecting scrub typhus. Microbiol Immunol. 2015;59(7):371-374.
Crossref - Stephen S, Kim S, Pradeep J,et al. Evaluation of ImmuneMed scrub typhus rapid test kit, for diagnosis of scrub typhus. J Vector Borne Dis. 2016;53(3):283-287.
Crossref - Gupta N, Chaudhry R, Kabra SK, et al. Comparative evaluation of serological and molecular methods for the diagnosis of scrub typhus in Indian settings. Jpn J Infect Dis. 2017;70(2):221-222.
Crossref - Anitharaj V, Stephen S, Pooja P. Scrub typhus in Puducherry, India: Application of nested PCR targeting three different genes – 56kDa, 47kDa and groEL of Orientia tsutsugamushi and comparison with ST IgM ELISA. J Vector Borne Dis. 2020;57(2):147-152.
Crossref - Gupta A, Singh D, Verma S, Kanga A, Mahajan S. Comparative evaluation of ELISA, PCR and Mico-IFA in diagnosis of Scrub typhus. Int. J. Cur. Adv. Res. 2017;06(07):4534-4538
- Gautam R, Parajuli K, Tshokey T, Stenos J, Sherchand JB. Diagnostic evaluation of IgM ELISA and IgM Immunofluorescence assay for the diagnosis of Acute Scrub Typhus in central Nepal. BMC Infect Dis. 2020;20(1):138.
Crossref - Anitharaj V, Pradeep J, Amsaveni S, Stephen S, Pooja P. Application of Nested PCR and Loop Mediated Isothermal Amplification to target 56kDA gene in Scrub Typhus patients and Phylogenetic analysis to identify Orientia tsutsugamushi strains circulating in and around Puducherry. J Pure Appl Microbiol. 2023;17(4):2131-2139.
Crossref - Vanlalruati RSC, Hmingmawii L,Ngurchamliana SR, et al. Serological evidence of scrub typhus in Mizoram, North Eastern Region of India. Infect Dis Clin Microbiol. 2022;4(1):55-61.
Crossref - Kalal BS, Puranik P, Nagaraj S, Rego S, Shet A. Scrub typhus and spotted fever among hospitalized children in South India: Clinical profile and serological epidemiology. Indian J Med Microbiol. 2016;34(3):293-298.
Crossref - Jain A, Jain P, Rebello SC, et al. Determination of a cut-off value for the serological diagnosis of scrub typhus by detecting anti-Orientia tsutsugamushi immunoglobulin M. Indian J Med Res. 2023;157(6):519-523.
Crossref - Zhang X, Teng Z, Mo T, et al. Development of a chimeric 56-21kDa antigen-based ELISA for serodiagnosis of Orientia tsutsugamushi infection. Microbiol Infect Dis. 2025;112(4):116841.
Crossref - Blacksell SD, Tanganuchitcharnchai A, Nawtaisong P, et al. Diagnostic accuracy of the InBios scrub typhus detect enzyme-linked immunoassay for the detection of IgM antibodies in Northern Thailand. Clin Vaccine Immunol. 2015;23(2):148-154.
Crossref - Lim C, Blacksell SD, Laongnualpanich A, Kantipong P, Day NPJ, Paris DH, Limmathurotsakul D. Optimal cutof titers for indirect immunofuorescence assay for diagnosis of scrub typhus. J Clin Microbiol. 2015;53:3663-3666.
Crossref - Paris DH, Blacksell SD, Nawtaisong P, et al. Diagnostic Accuracy of a Loop-Mediated Isothermal PCR Assay for Detection of Orientia tsutsugamushi during Acute Scrub Typhus Infection. PLoS Negl Trop Dis. 2011;5(9):e1307.
Crossref - Kim CM, Kim DM, Yun NR. Follow-up investigation of antibody titers and diagnostic antibody cut-off values in patients with scrub typhus in South Korea. BMC Infect Dis. 2021;21(1):69.
Crossref - Blacksell SD, Lim C, Tanganuchitcharnchai A, et al. Optimal cut-off and accuracy of an IgM enzyme-linked immunosorbent assay for diagnosis of acute scrub typhus in northern Thailand: an alternative reference method to the IgM immunofluorescence assay. J Clin Microbiol. 2016;54(6):1472-1478.
Crossref - Blacksell SD, Bryant NJ, Paris DH, Doust JA, Sakoda Y, Day NP. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: a lack of consensus leads to a lot of confusion. Clin Infect Dis. 2007;44(3):391-401.
Crossref - Varghese GM, Rajagopal VM, Trowbridge P, Purushothaman D, Martin SJ. Kinetics of IgM and IgG antibodies after scrub typhus infection and the clinical implications. Int J Infect Dis. 2018;71:53-55.
Crossref - Lim C, Paris DH, Blacksell SD, et al. How to determine the accuracy of an alternative diagnostic test when it is actually better than the reference tests: a re-evaluation of diagnostic tests for scrub typhus using Bayesian LCMs. PLoS One. 2015a;10(5):e0114930.
Crossref - Kim C-M, Kim D-M, Yun NR. Evaluation of the diagnostic accuracy of antibody assays for patients with scrub typhus. J Clin Microbiol. 2021;59(7):e02942-20.
Crossref - Tantibhedhyangkul W, Wongsawat E, Silpasakorn S,et al. Use of multiplex real-time PCR to diagnose scrub typhus. J Clin Microbiol. 2017;55(5):1377-1387.
Crossref - Usha K, Kumar E, Kalawat U, Kumar BS, Chaudhury A, Gopal DVRS. Molecular characterization of Orientia tsutsugamushi serotypes causing scrub typhus outbreak in southern region of Andhra Pradesh, India. Indian J Med Res. 2016;144(4):597-603.
Crossref - Paris DH, Aukkanit N, Jenjaroen K, Blacksell SD, Day NPJ. A highly sensitive quantitative real-time PCR assay based on the groEL gene of contemporary Thai strains of Orientia tsutsugamushi. Clin Microbiol Infect. 2009;15(5):488-495.
Crossref - Kim DM, Park G, Kim HS, et al. Comparison of Conventional, Nested, and Real-Time Quantitative PCR for Diagnosis of Scrub typhus. J Clin Microbiol. 2011;49(2):607-612.
Crossref - Anitharaj V, Pradeep J, Stephen S. Diagnostic dilemma in laboratory confirmation of acute Scrub Typhus infection: Relevance of ST IgM, IgG antibodies and molecular markers. J Vector Borne Dis. 2025.
Crossref - Dwivedi PP, Singh AK, Murthy R, Dwivedi S, Verma AR. Evaluation of the Performance of Various Diagnostic Modalities Available for the Detection of Scrub Typhus in Acute Undifferentiated Febrile Illness (AUFI) Cases at a Teaching Hospital in North Chhattisgarh, India. Cureus. 2025;17(2):e78977.
Crossref - Mediannikov O, Fenollar F, Socolovschi C, et al. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 2010;4(4):e654.
Crossref - Chunduru K, Manoj ARM, Poornima S, et al. Persistence of scrub typhus IgM and IgG antibodies among patients from Karnataka, India. Ann Med. 2025;57(1):2468258.
Crossref - Sondhiya G, Manjunathachar HV, Singh P, Kumar R. Unveiling the burden of scrub typhus in acute febrile illness cases across India: A systematic review & meta-analysis. Indian J Med Res. 2024;159(6):601-618.
Crossref - Balamurugan S, Mohan R, Mary JF, et al. A systematic review and meta-analysis of scrub typhus and its association with liver disease. J Global Infect Dis. 2025;17(2):71-76.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.