ISSN: 0973-7510
E-ISSN: 2581-690X
Among the acquired mechanisms of resistance to antibiotics of microorganisms, the production of beta-lactamases, enzymes that inactivate penicillins, cephalosporins, carbapenems, and monobactams, is widespread. Most often, such beta-lactamases, in particular ESBL (extended-spectrum beta-lactamases), are capable of destroying III and IV generations of cephalosporins. One of the important ESBL producers is Escherichia coli and, to a lesser extent, Salmonella enteritidis, which are clinically significant in animals and humans. The purpose of the study was to screen ESBL DDM using cephalosporin markers and screening of mobile extrachromosomal factors of bacterial heredity – plasmids (potentially dangerous factors of genetic transport) in isolates of E. coli and S. enteritidis, polyresistant to aminoderms, from environmental objects, patho- and biological material, raw materials and products of animal origin. Results of our studies have shown the level of their distribution among animals, poultry, since from 13 field isolates of E. coli isolated from the milk of cows with mastitis and pathological material from pigs, ESBL production was found in 3 strains (23.1%) and from 18 field isolates of S. enteritidis isolated from pathological material from poultry, ESBL production was found in 2 strains (11.1%). Based on the results of molecular genetics studies, the presence of resistance plasmids (R-plasmids) in 9 field E. coli isolates was confirmed, 4 of which produced acquired beta-lactamases, incl. ESBL and 8 field isolates of S. enteritidis, 7 of which confirmed the presence of acquired carbapenemases.
Antibiotic resistance, antibiotics, beta-lactamases of extended-spectrum (ESBL), carbapenemases, E. coli, plasmids, S. enteritidis
Currently, there is a worldwide trend of increasing resistance to antibiotics in pathogens of zoonotic diseases to threatening levels1. New mechanisms of resistance in pathogenic bacteria are emerging and spreading everywhere, with great risks associated with a reduction in the effectiveness of human and animal treatment and an increase in its cost. Antibiotic resistance is gaining momentum due to their misuse, low level of prevention and control of bacterial infections, due to the ingress of antibiotic-resistant bacteria into the food chain due to contamination of raw materials and food2.
Measures to reduce the resistance of bacteria and limit their spread have to be taken at all levels of society. At the session of the United Nations General Assembly in New York in September 2016, the Heads of State pledged to deploy broad and coordinated action to address the root causes of antibiotic resistance, especially in the human and animal health sectors and agriculture with further development of national action plans to address antibiotic resistance. Ukraine is also involved in these.
One of the initiatives proposed by the WHO to improve the situation is particularly relevant and concerns the Global Action Plan on antimicrobial resistance (GLASS). This system is based on standardized approaches to the collection, analysis and exchange of data on antibiotic resistance on a global scale using the data obtained for decision-making at the local, national and regional levels3-6.
Thus, a detailed study and analysis of the resistance of zoonotic pathogens obtained by passive monitoring after microbiological studies of patho- and biomaterial, samples of raw materials and products of animal origin, poultry and environmental facilities for Ukraine is an urgent problem today.
Leading experts of the World Health Organization (WHO) in the field of antibiotic resistance recommend the development and implementation of effective approaches to overcome this dangerous global phenomenon by conducting comprehensive monitoring studies of the resistance of microorganisms to antibiotics. Antibiotic-resistant microflora enters the body in different ways. One of these is through raw materials and food products of animal origin contaminated with antibiotic-resistant microorganisms7-10. Clinically significant beta-lactam antibiotics (beta-lactams) are now widely used in foods of animal origin. This is a large group of antibiotics containing a beta-lactam ring. The group of beta-lactams includes large subgroups of cephalosporins, carbapenems, monobactams and penicillins. The mechanism of action of beta-lactam antibiotics is based on the violation of the microorganisms’ cell wall synthesis. The target of their action is trans- and carboxypeptidases, which promote the synthesis of the main component of the outer membrane of gram-positive and gram-negative bacteria – peptidoglycan11,12.
Such antibiotics are inactivated by the action of special enzymes – beta-lactamases, and the detection of mechanisms of resistance to beta-lactams is of great epidemiological importance. Beta-lactamases include enzymes: carbapenemases (beta-lactamases, hydrolyzing penicillins, some cephalosporins, carbapenems and monobactams), extended-spectrum beta-lactamases (ESBL, hydrolyzing cephalosporins of the third and fourth generation, and aztreonam, in addition to cephamycin and carbapenems) and acquired AmpC beta-lactamases (hydrolyze penicillins, third-generation cephalosporins and monobactams and do not hydrolyze fourth-generation cephalosporins)13,14.
In most countries of the world and in Europe, in particular, experts of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) for national reference centers of health and veterinary authorities recommend the detection of ESBL-producing bacteria and their characteristics5,15.
ESBL beta-lactamases are resistant to cephalosporins and, as studies show, have a high level of polyresistance to antibiotics16.
Numerous studies have shown that the genes that encode the synthesis of the vast majority of already identified beta-lactamases are contained on extrachromosomal elements of bacterial heredity – plasmids17.
The problem today is the anthropogenic pressure of beta-lactam antibiotics and the increased risk of the spread of plasmid associated antibiotic resistance genes of zoonotic Enterobacteriaceae. Therefore, the monitoring of plasmid-associated genes in R-plasmims will be able to provide information on the possibility of developing alternative ways to combat antibiotic resistance of zoonotic pathogens18-20.
Thus, the study of the prevalence of enterobacteriaceae circulating in Ukraine with acquired mechanisms of antibiotic resistance will promote a standardized approach to data collection, analysis and exchange on a global scale and ensure the implementation of the tasks of the National Action Plan for Combating Antibiotic Resistance to Antimicrobials.
The purpose of the study was to screen ESBL DDM using cephalosporin markers and screening of mobile extrachromosomal factors of bacterial heredity – plasmids (potentially dangerous factors of genetic transport) in isolates of E. coli and S. enteritidis, polyresistant to aminoderms, from environmental objects, patho- and biological material, raw materials and products of animal origin.
Test materials used in the study. Isolates of Enterobacteriaceae: 13 strains of E. coli and 18 strains of S. enteritidis. In particular: isolates of E. coli – 7 strains «1/44», «9/97», «10/98», «11/99», «12/100», «13/101», «14/102» isolated from mastitis cow’s milk; 2 strains «5/73», «6/77» isolated from dog’s and elephant’s faeces; 3 strains «7/80», «15/124» і «S14/58» isolated from pathological material from poultry (chickens, quails) and pig; 1 strain «16/175» isolated from animal feed samples (granulated soybean meal); isolates of S. enteritidis – 8 strains «1/3», «26/145», «29/171.а», «37/245», «48/284», «45/265», «48/284», «49/286», «58/347», «59/348» isolated from food (chicken filet, milk-containing cheese product, ready to eat fish; shawarma from chicken fillet; read to eat rise; Eclair cakes, ready to eat chicken); 3 strains «57/346», «71/422» and «51/300» isolated from poultry samples (pathological material, meconium and faeces) and 6 strains «9/20», «14/71», «16/78», «22/114», «46/275», «47/276» isolated from environmental objects (chick box papers, dust from the poultry building). Specialists of the State Research Institute selected isolates of Enterobacteriaceae for passive monitoring during 2019 for Laboratory Diagnostics and Veterinary Sanitary Examination (Kyiv).
Mechanisms of setting the disc-diffusion method to determine the general profiles of antibiotic resistance, acquired mechanisms of resistance with confirmation of ESBL products, quality control of studies and screening of experimental isolates for the presence of plasmid DNA. Determination of general profiles of antibiotic resistance was performed by disco-diffusion method (DDM) using disks with antibiotics: tetracycline (30 μg / disk), nalidixic acid (30), ampicillin (10), cefoxitin (30), gentamicin (10, 10 ); ciprofloxacin (5), chloramphenicol (30), imipenem (10), sulfamethoxazole (25), ceftazidime (10), cefotaxime + clavulanic acid (30/10), cefotaxime (5 and 30), cefepime (30), ceftazidime + clavulanic acid (30/10), aztreonam (30), trimethoprim (5), ceftriaxone (30), amikacin (30), amoxiclav (10). Diameters of growth retardation zones (mm) of experimental cultures of Еnterobacteriaceae were measured and interpreted taking into account the recommendations of the EUCAST5,19.
DDM (disco-diffusion method) with cephalosporin diagnostic markers in the combination recommended by EUCAST, ceftriaxone (30) and ceftazidime (10) was used to determine ESBL products in experimental Enterobacteriaceae5.
Quality control of the studies was carried out with a test culture of E. coli ATCC 25922, recommended by EUCAST as a control strain for studies of isolates of the Enterobacterales genus.
In order to screen bacteria for the presence of plasmid DNA, we used the method of DNA imaging according to Eckhardt (1978) and preparative isolation of plasmid DNA. For preparative obtaining plasmid DNA preparations, we used the method of alkaline lysis by Birnboim and Dolly (1979). The obtained plasmid DNA was visualized by agarose gel electrophoresis and stored at -20°C. Visualization of plasmid DNA preparations and DNA fragments using the method of electrophoresis in 1% agarose gel and Tris-borate buffer solution. After electrophoresis, the gel was stained in buffer with 0.5 μg/ml ethidium bromide, washed with water and the DNA was observed under ultraviolet light.
In order to identify epidemiologically significant mechanisms of antibiotic resistance to beta-lactams in isolates of E. coli and S. enteritidis – pathogens of zoonoses isolated from the environment, pathological material, raw materials, products of animal origin and poultry industry under passive monitoring, the identified isolates were studied comprehensively for sensitivity to antibiotics. Table 1 shows the antibiotic resistance profiles of E. coli field isolates.
Table (1):
Antibiotic profile of field isolates of E. coli strains.
The name of E. coli strain |
Tetracycline |
Ceftazidime |
Nalidixic acid |
Ampicillin |
Cefoxitin |
Gentamicin |
Ciprofloxacin |
Chloramphenicol |
Imipenem |
Sulfamethoxazole |
|---|---|---|---|---|---|---|---|---|---|---|
«1/44» |
R |
S |
R |
R |
S |
S |
S |
S |
S |
Х |
«5/73» |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
«6/77» |
S |
S |
R |
S |
S |
S |
S |
S |
S |
R |
«7/80» |
R |
S |
R |
R |
S |
S |
R |
S |
S |
R |
«9/97» |
R |
R |
S |
S |
S |
S |
S |
S |
S |
|
«10/98» |
R |
MR |
R |
S |
R |
S |
S |
S |
S |
S |
«11/99» |
S |
S |
R |
S |
Х |
S |
S |
S |
S |
S |
«12/100» |
R |
R |
S |
S |
S |
S |
S |
S |
S |
S |
«13/101» |
R |
S |
R |
S |
S |
S |
S |
S |
S |
S |
«14/102» |
R |
S |
R |
S |
R |
S |
S |
S |
S |
S |
«15/124» |
R |
S |
R |
R |
Х |
S |
S |
S |
S |
R |
«16/175» |
S |
MR |
S |
S |
Х |
S |
S |
S |
S |
S |
S «14/58» |
Х |
R |
Х |
R |
S |
R |
R |
R |
S |
Х |
Note. R- resistant; S-sensitive; MR – moderately resistant; Х − not tested.
Analysis of the study results showed that among the field isolates of E. coli 50% were strains polyresistant to antibiotics (resistance up to 3 or more varieties of antibiotics). Among the studied strains of Escherichia in 3 of them – “9/97”, “12/100” and S «14/58» , resistance (R) to ceftazidime was detected and in 2 more strains “10/98”, “16/175” the value of the diameters of the zones of growth inhibition was within the range of moderate resistance (MR).
Owing to the fact that according to EUCAST, resistance of Enterobacteriaceae, including E. coli, to most cephalosporins is an indicator of the presence of acquired mechanisms of resistance, we were interested in deeper studies to confirm the production of extended-spectrum beta-lactamases in ceftazidime-resistant strains of E. coli “9/97” and “12/100” and check the moderate sensitivity to ceftazidime strains of E. coli for possible ESBL products. Therefore, following the EUCAST methodology for the detection of resistance mechanisms in Еnterobacteria of the I-st group (E. coli, Salmonella spp., Klebsiella spp., Shygella spp., P. mirabilis), in particular the production of carbapenemases of extended-spectrum (ESBL), we conducted an ESBL screening using a cephalosporin test with a combination of ceftriaxone and ceftazidime, as shown in Table 2.
Table (2):
Screening for ceftazidime-resistant E. coli isolates for confirmation of ESBL-producers.
| No | Type of experimental isolate and ID number (species/general) | ESBL Screening | Screening results | |
|---|---|---|---|---|
| cephalosporin test marker:(combination cefotaxim and ceftazidime) | ||||
| Cefotaxim (30 μg/disk) |
Ceftazidime (10 μg/disk) |
ESBL production was confirmed by the values of the diameters of the zones of growth inhibition ceftriaxone (<23) and ceftazidime (<22), yes / no |
||
| According to the EUCAST documents range of resistance for the control strain of E. coli АТСС 25922 | < 21 | < 22 | no | |
| E. coli АТСС 25922 (actual test results) | 21 | 20 | non-ESBL-product (negative quality control) no |
|
| 1. | E. coli strain «9/97» | 19 | 10 | ESBL-product |
| 2. | E. coli strain «12/100» | 14 | 13 | ESBL-product |
| 3. | E. coli strain «S14/58»» | 0 (self-growth) | 20 | ESBL-product |
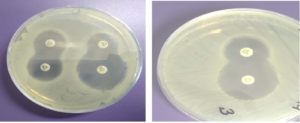
Analysis of the ESBL screening results showed that E. coli strains “9/97”, “12/100” and S «14/58» are resistant to ceftazidime, which was determined by the general profile of antibiotic resistance and after screening testes with cefatoxime, to confirm the ESBLs it was found that the designated strains have acquired resistance mechanisms and perhaps produce beta-lactamases of extended-spectrum. After conducting targeted studies to confirm ESBL production by test bacteria using the double- disc synergy test (DDST) with cephalosporins – ceftazidime, cefotaxime, cefepime and clavulanic acid, in all cases the “keyhole” effect was revealed: inhibition zones around any of cephalosporin disks are augment in the direction of the disk containing clavulanic acid, which confirmed the production of such an enzyme (Fig. 1).
Fig. 1. Double-disc synergy test (DDST) with cephalosporins – ceftazidime, cefotaxime, cefepime and clavulanic acid, the “keyhole” effect , E. coli strain «9/97».
Table 3 shows data on the general profiles of antibiotic resistance of ceftazidime- resistant field isolates of S. enteritidis.
Table (3):
Antibiotic profile of ceftazidime- resistant field isolates of S. enteritidis.
| Strain’s name | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| «1/3» | «9/20» | «14/71» | «16/78» | «22/114» | «26/145» | «29/171а» | «37/245» | «45/265» | «46/275» | «47/276» | «48/284 | «49/286» | «51/300» | «57/346» | «58/347» | «59/348» | «71/422» | |||||
| Ciprofloxacin | S | S | S | R | S | S | S | S | S | S | S | S | S | S | R | MR | MR | R | ||||
| Ampicillin | S | S | S | R | S | S | R | S | S | S | R | S | S | S | Р | S | S | R | ||||
| Trimethoprim | S | S | S | S | S | S | S | S | S | R | R | S | S | S | S | S | S | S | ||||
| Ceftazidime | S | S | S | S | S | S | MR | S | S | S | MR | R | S | R | R | MR | MR | R | ||||
| Amikacin | S | S | S | R | S | S | S | S | S | S | S | S | S | S | S | R | S | R | ||||
| Cefepime | R | S | S | S | R | S | R | S | S | S | R | S | S | R | R | R | R | R | ||||
| Gentamicin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | ||||
| Chloramphenicol | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | R | R | ||||
| Imipenem | S | S | S | R | S | R | R | S | S | S | S | S | S | S | S | S | S | S | ||||
| Aztreonam | S | S | S | S | R | R | S | S | R | R | R | S | S | S | R | R | R | R | ||||
| Nalidixic acid | R | R | R | R | R | R | R | R | R | R | R | R | R | R | Х | Х | Х | Х | ||||
| Amoxiclav | R | R | R | R | R | R | R | R | R | R | R | R | R | R | Х | Х | Х | Х | ||||
| Cefoxitin | S | S | S | S | S | R | R | S | S | S | R | S | S | S | R | R | R | R | ||||
Note. R – resistant; S-sensitive; MR – moderately resistant, X – not tested.
Analysis of the research findings of the general antibiotic resistance profiles of field isolates of S. enteritidis showed that 72.2% of strains were polyresistant (3 or more) to the applied antibiotics. Attention has to be paid to the absence of any effect of amoxiclav and nalidixic acid on S. enteritidis isolates, i.e. they have 100.0% resistance to these antibiotics.
Resistance of Enterobacteriaceae of the first group (E. coli, Salmonella spp., Klebsiella spp., Shygella spp., P. mirabilis) to most cephalosporins is an indicator of the presence of acquired resistance mechanisms, and according to the analyzed results of the antibioticogram of S. enteritidis strains “51/300”, «57/346» and «71/422» proved to be resistant to ceftazidime. To confirm ESBLs in ceftazidim-resistant S. enteritidis, we performed an ESBL screening, the results of which are shown in Table 4.
Table (4):
Screening for ESBL-products in S. enteritidis isolates with cephalosporin markers.
| No | Type of experimental isolate and ID number (species / general) | ESBL screening | Screening results | |
|---|---|---|---|---|
| cephalosporin test marker: (combination of cefotaxim and ceftazidime) | ||||
| Cefotaxim (30 μg/disk) |
Ceftazidime (10 μg/disk) |
ESBL-producers confirmed by the diameter size of the zones of growth inhibition ceftriaxone (<23) and ceftazidime (<22), yes/no |
||
| According to EUCAST documents, the range of resistance for the control strain of E. coli АТСС 25922 |
< 21 | < 22 | no | |
| E. coli АТСС 25922 (actual test results) | 30 | 25 | non- ESBL-product (negative quality control) no | |
| 1. | S. enteritidis strain «51/300» | 30 | colonies in the zone of growth inhibition | no |
| 2. | Salmonella enteritidis strain «57/346» | 20 | 16 | ESBL-product |
| 3. | Salmonella enteritidis strain «71/422» | 20 | 17 | ESBL-product |
Analysis of the experimental results confirmed the acquired resistance in S. enteritidis strains “57/346” and “71/422”, because after the application of a specific cephalosporin test in combination with ceftazidime and ceftriaxone, these strains had showed resistance to these antibiotics. Ceftazidime-resistant strain S. enteritidis “51/300” had showed sensitivity to ceftriaxone, therefore ESBL production was not confirmed.
While analyzing results of DDM quality control with cephalosporin markers in combination of ceftriaxone (30 μg / disk) and ceftazidime (10 μg / disk), we confirmed the absence of ESBLs in the test culture of E. coli ATCC 25922, which meets the EUCAST recommendations and allows to recognize reliable the test results on detection of ESBLs in field isolates of S. enteritidis and E. coli.
It is known that the vast majority of beta-lactamases, in particular ESBL, are products of expression of plasmid-associated genes for antibiotic resistance of R-plasmids. Therefore, it was important to study the presence of plasmids in examined field isolates of bacteria.
Studies have shown that the method of alkaline lysis is an effective method for the preparative isolation of plasmids from isolates of S. enteritidis and E. coli. Eckhard’s method is suitable for screening on the presence of plasmids directly only during the agarose gel electrophoresis.
As shown in Table 5, plasmid DNA was detected in the field isolate strains of S. enteritidis and E. coli.
Table (5):
Results of plasmid DNA screening in field isolate strains of S. еnteritidis and E. coli.
| Enterobacteriaceae type |
Strain′s name | The presence of plasmid DNA described by : | |
|---|---|---|---|
| Eckhard’s method | alkaline lysis method (Birnboim and Destiny’s) | ||
| S. еnteritidis | «1/3» | – | – |
| «9/26» | + | + | |
| «14/71» | – | – | |
| «16/78» | + | + | |
| «22/114» | + | + | |
| «26/145» | + | + | |
| «29/171a» | + | + | |
| «37/245» | – | – | |
| «45/265» | + | + | |
| «46/275» | + | + | |
| «47/276» | + | + | |
| E. coli | «1/44» | + | + |
| «5/73» | – | – | |
| «6/77» | – | – | |
| «7/80» | + | + | |
| «9/97» | + | + | |
| «10/98» | + | + | |
| «11/99» | + | + | |
| «12/100» | + | + | |
| «13/101» | + | + | |
| «14/102» | + | + | |
| «15/124» | + | + | |
| «16/175» | – | – | |
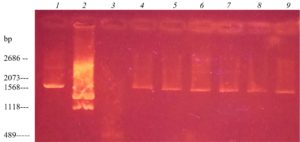
The results showed that among the isolates of S. enteritidis eight strains were plasmid containing (Fig. 1). Since the studied plasmids were low molecular weight, it was assumed that they belong to the class of R-plasmids.
Analysis of plasmid-containing field isolates of S. enteritidis showed the presence of seven strains of this species of Salmonella, carbapenemase and the absence of all plasmid-containing strains of ESBLs. Thus, we can assume that the identified plasmids carry genes that encode the synthesis of carbapenemases in Salmonella and therefore there is a need to develop methods for molecular genetic typing of plasmid-associated genes encoding the synthesis of such enzymes.
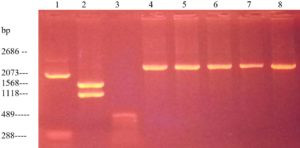
Regarding the screening of antibiotic resistance plasmids in 12 isolates of E. coli, Fig. 2 shows that nine strains had low molecular weight plasmids.
Fig. 2. Electrophoregram of DNA markers and isolated DNA plasmid of field isolates of S. enteritidis: 1– plasmid marker pUC19, digested with the enzyme Mva I (2686, 2073,288 bp); 2 – plasmid marker pUC19, digested with the enzyme Bgl I (1568, 1118 bp); 3 – plasmid marker pUC19, digested with with the enzyme Msp I (489 BP); 4 – S. enteritidis 16/78; 5 – S. enteritidis 22/114; 6 – S. enteritidis 26/145; 7 – S. enteritidis 29 / 171a; 8 – S. enteritidis 45/265
Such isolates of Еnterobacterial pathogens, due to their polyresistance, pose a potential danger to humans and animals in the spread and acquisition among identical populations of microorganisms sensitive to antibiotics, plasmid-associated genes of antibiotic resistance. It was found that among the epidemiologically significant beta-lactamases studied in E. coli field isolates there were four plasmid-containing strains, of which – two ESBL-producing and two – produce other acquired beta-lactamases. The other five plasmid-containing strains of E. coli are potential carriers of non-β-lactamase-encoding antibiotic resistance genes.
Fig. 3. Electrophoregram of DNA markers and isolated DNA plasmid by Birnboim-Dolis’ alkaline lysis method from E. coli field isolates: 1 – plasmid marker pUC19, digested with the enzyme Mva I (2686, 2073 BP); 2 – plasmid marker pUC19, digested with the enzyme Bgl I (1568, 1118 BP); 3 – plasmid marker pUC19, digested with the enzyme Msp I (489 BP); 4 – E. coli 1/44; 5 – E. coli 7/80; 6 – E. coli 9/97; 7 – E. coli 10/98; 8 – E. coli 12/100; 9 – E. coli 13/101
Resistance of Enterobacteriaceae, including E. coli and Salmonella spp., to a number of beta-lactam antibiotics is rapidly spreading and is becoming a serious problem in veterinary and human medicine22,23. It is becoming obvious that the effectiveness of detecting resistance to these antibiotics using traditional methods remains quite low. This has been confirmed by numerous studies, as well as in our case20, because the detection of resistance in microorganisms by determining only the general profile of antibiotics does not allow to determine whether such microorganisms have already acquired plasmid-mediated genes of antibiotic resistance.
ESBL-producing E. coli isolates are increasingly identified in farm animals, in food products of animal origin, which confirms the hypothesis that animals are natural resistant sources of infection with ESBL-producing Enterobacteriaceae22,23. In this case, this hypothesis is confirmed by the results of our studies, because ESBL-producing strains of E. coli “9/97” and “12/100” were isolated from mastitis cow’s milk and strain S « 14/58» from pathological material from pig. Thus, 13 strains of E. coli, mono- and polyresistant to ADR, were isolated from livestock products, 23,1% of which had acquired resistance mechanisms, which was confirmed by ESBL products during the screening with cephalosporin markers and the presence of plasmid-mediated resistance genes in these strains, according to the results of antibiotic resistance plasmids screening.
Screening results for ESBL production in ceftazidime-resistant field isolates of S. enteritidis showed the presence of this enzyme in strains “57/346”, “71/422”, which was 11.1% of the tested isolates.
Infections caused by zoonotic pathogens resistant to antibiotics are a serious problem for veterinary and humane medicine; because the effectiveness of treatment is reduced or absent, there is a risk of life-threatening conditions in animals and humans, increasing the duration of treatment with more expensive antibiotics or finding alternative treatment methods. This leads to significant budget expenditures in the economics and social sphere24.
Almost all pathogens of the most common infections circulating in Ukraine produce beta-lactamases and are a factor of resistance to many, and in some cases – to all, antibiotics, especially to widely used in the treatment of these infections, which limits or eliminates the possibility of their further application25. In addition, ESBL-producing strains of E. coli can become one of the links in the food chain in animal and human nutrition, which already carries biological risks for society as a whole.
Beta-lactamases are most often known to produce gram-negative microorganisms, although gram-positive bacteria Staphylococcus spp. also produce them26.
The mechanism of beta-lactamase impact on ADR lies the binding of beta-lactamase with beta-lactam antibiotic, which triggers the hydrolysis of the amine bond of the lactam ring, which leads to inactivation of antibiotics.
The production of extended-spectrum beta-lactamases (ESBL) is one of the most common clinically significant mechanisms of resistance. ESBL includes a significant number of enzymes that are capable of cleaving oxyimino-beta-lactams, are third and fourth generation cephalosporins, aztreonam and penicillins, and are sensitive to inhibitors – clavulanic acid, sulfabactam and tazobactam23,27.
Some scientists, to overcome the resistance of microorganisms to antibiotics, suggest developing and implementing new antibiotics in combination with beta-lactamase inhibitors28,29.
The catastrophic situation with the spread of antibiotic resistance requires new approaches to monitoring bacterial resistance30,31. A comprehensive study of the resistome will help to be one step ahead of bacteria in the “race of antibiotics”32. Modern approaches to fattening and treatment of animals have created conditions of high selective pressure of various antibiotics, which leads to the formation of dangerous, in terms of antibiotic resistance, pool of plasmids of antibiotic resistance (R-plasmids) in probiotic and transient microflora of animals.
It is known that plasmids of the IncI2 group carrying Bla CTX-M genes can undergo interspecific migration among potential bacterial pathogens from E. coli33,34. Molecular characterization of plasmids in field isolates of the most common pathogens of the genus Enterobacteriaceae – E. coli and S. enteritidis is important for understanding the local and global distribution of their resistance to the most common beta-lactam antibiotics33.
Genes encoding ESBL are located on plasmids. Thus, the prevalence of extended-spectrum β-lactamases (ESBL) producing E. coli and Salmonella spp. in animal populations35 and diversity in ESBL genotypes, including CTX-M blah, SHV blah, TEM blah, OXA in sewage Еnterobacteriaceae36.
Our study has shown the presence of plasmids in the examined field isolates of S. enteritidis (among 13 isolates, eight had plasmins, which accounted for 72.7% of the studied material) and E. coli (of the 13 studied – 9 plasmid-containing, which accounted for 75.0% of the studied material). In our studies, such plasmids were classified as R-plasmids, according to their size. The scientists also attributed the plasmid DNA of Enterobacteriaceae to R-plasmids, according to these characteristics. In addition, the increase in bacterial mass of S. enteritidis and E. coli for plasmid screening was carried out in antibiotic-free environment, as the latter are known inducers of increased copies of R-plasmids in bacterial cells. Therefore, even in the absence of antibiotics in the culture environment, plasmid preparations with a high concentration of plasmid DNA were obtained, which testified in favor of multicopy of the studied plasmids and allows attributing them on this basis to the class of R-plasmids34.
Among S. enteritidis field isolates carrying plasmids, seven strains showed the presence of carbapenemases and the absence of ESBLs in all plasmid-containing strains. Among the examined E. coli isolates there were four plasmid-containing strains, two of which were ESBL-producing and two ones were producing other acquired beta-lactamases. The other five plasmid-containing strains of E. coli are potential carriers of antibiotic resistance genes that are not associated with β-lactamase encoding. They are likely to encode tetracyclines, as these strains have this type of antibiotic resistance, which may be related to their spread in beef and chicken production37.
Therefore, the obtained data indicate the need to continue the research aimed at identifying the mechanisms of antibiotic resistance to beta-lactams of other types in isolates of Enterobacteriaceae (E. coli, Salmonella spp., Ps. aeruginosa, and others) and the development of methods for molecular genetics of genetic type synthesis of carbapenemases in zoonotic pathogens isolated from pathogenic and biomaterials, raw materials and products of animal origin and environmental objects.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Lehtinen S, Blanquart F, Lipsitch M. Fraser C, with the Maela Pneumococcal Collaboration. On the evolutionary ecology of multidrug resistance in bacteria. PLoS Pathogens. 2019;15(5):e1007763.
Crossref - Zhang J, Chua QW, Mao F, et al. Effects of activated carbon on anaerobic digestion-Methanogenic metabolism, mechanisms of antibiotics and antibiotic resistance genes removal. Bioresource Technology Reports. 2019;5:113-120.
Crossref - WHO. Global action plan on antimicrobial resistance, 2015. https://apps.who.int/iris/bitstream/handle/10665/193736/9789241509763_eng.pdf?sequence=1
- WHO. The FAO Action Plan on Antimicrobial Resistance 2016-2020, 2016. http://www.fao.org/3/a-i5996e.pdf
- The European Committee on Antimicrobial Susceptibility Testing MIC distributions and epidemiological cut-off value (ECOFF) setting EUCAST SOP 10.1 (2019). http://www.eucast.org/documents/sops/
- Aenishaenslin C, Hasler B, Ravel A, Parmley J, Stark K, Buckeridge D. Evidence needed for antimicrobial resistance surveillance systems. Bull World Health Organ. 2019;97:283-289.
Crossref - Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics. 2019;8(2):37-41.
Crossref - Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88(1):26-40.
Crossref - Kukhtyn M, Kravcheniuk K, Beyko L, Horiuk Y, Skliar O, Kernychnyi S. Modeling the process of microbial biofilm formation on stainless steel with a different surface roughness. East-Eur J Enterp Technol. 2019;2/11(98):14−21.
Crossref - Lomovskaya O, Nelson K, Rubio-Aparicio D, Tsivkovski R, Sun D, Dudley MN. Impact of intrinsic resistance mechanisms on potency of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob Agents Chemother. 2020;64(6):e00552-20.
Crossref - Liao X, Ma Y, Daliri EBM, et al. Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens. Trends Food Sci Technol. 2020;95:97-106.
Crossref - Pemberton OA, Noor RE, Kumar V, Sanishvili R, Kemp MT, Kearns FL, Chen Y. Mechanism of proton transfer in class A β-lactamase catalysis and inhibition by avibactam. PNAS USA. 2020;117(11): 5818-5825.
Crossref - Sampaio JLM, Gales AC. Antimicrobial resistance in Enterobacteriaceae in Brazil: focus on β-lactams and polymyxins. Braz J Microbiol. 2016;47:31-37.
Crossref - Kristoffersson AN, Bissantz C, Okujava R, et al. A novel mechanism-based pharmacokinetic-pharmacodynamic (PKPD) model describing ceftazidime/avibactam efficacy against β-lactamase-producing Gram-negative bacteria. J Antimicrob Chemother. 2020;75(2):400-408.
Crossref - Bradford PA, Kazmierczak KM, Biedenbach DJ, Wise MG, Hackel M, Sahm DF. Correlation of β-lactamase production and colistin resistance among Enterobacteriaceae isolates from a global surveillance program. J Antimicrob Chemother. 2016;60(3):1385-1392.
Crossref - Smet A, Martel A, Persoons D, et al. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. 2010;34(3):295-316.
Crossref - Pinilla-Redondo R, Cyriaque V, Jacquiod S, Sorensen SJ, Riber L. Monitoring plasmid-mediated horizontal gene transfer in microbiomes: recent advances and future perspectives. Plasmid. 2018;99:56-67.
Crossref - Bimboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513-1523.
Crossref - Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis. 2010;10(1):43-50.
Crossref - Lynch III JP, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases). Expert Opin Pharmacother. 2013;14(2):199-210.
Crossref - Rezai MS, Bagheri-nesami M, Hajalibeig A, Ahangarkani F. Multidrug and cross-resistance pattern of ESBL-producing Еnterobacteriaceae agents of nosocomial infections in intensive care units. Journal of Mazandaran University of Medical Sciences. 2017;26(144):39-49.
- Koga VL, Maluta RP, da Silveira WD, et al. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of South Brazil. BMC microbiology. 2019;19:174.
Crossref - Meini S, Tascini C, Cei M, Sozio E, Rossolini GM. AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection. 2019;47(3):363-375.
Crossref - Wilke MS, Lovering AL, Strynadka NC. β-Lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol. 2005;8(5):525-533.
Crossref - Horiuk YV, Kukhtyn MD, Stravskyy YS, Havrylianchyk RY, Horiuk VV, Fotina HA. Comparison of the minimum bactericidal concentration of antibiotics on planktonic and biofilm forms of Staphylococcus aureus: Mastitis causative agents. Res J Pharm Biol Chem Sci. 2018;9(6):616−622.
- Wencewicz TA. Crossroads of antibiotic resistance and biosynthesis. J Mol Biol. 2019;431(18):3370-3399.
Crossref - Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-686.
Crossref - Livermore DM, Mushtaq S. Activity of biapenem (RPX2003) combined with the boronate β-lactamase inhibitor RPX7009 against carbapenem-resistant Enterobacteriaceae. J Antimicrob Chemother. 2013;68(8):1825-1831.
Crossref - Drawz SM, Papp-Wallace KM, Bonomo RA. New β-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother. 2014;58(4):1835-1846.
Crossref - Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol. 2013;39(1):79-101.
Crossref - Hammoudi D, Moubareck CA, Sarkis DK. How to detect carbapenemase producers? A literature review of phenotypic and molecular methods. J Microbiol Methods. 2014;107:106-118.
Crossref - Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15(7):422-434.
Crossref - Castellanos LR, Donado-Godoy P, Leon M, et al. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS One. 2017;12(1):e0170777.
Crossref - Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53(6):2227-2238.
Crossref - Lalruatdiki A, Dutta TK, Roychoudhury P, Subudhi PK. Extended-spectrum β-lactamases producing multidrug resistance Escherichia coli, Salmonella and Klebsiella pneumoniae in pig population of Assam and Meghalaya, India. Veterinary Worl. 2018;11(6):868-873.
Crossref - Li Q, Chang W, Zhang H, Hu D, Wang X. The role of plasmids in the multiple antibiotic resistance transfer in ESBLs-producing Еscherichia coli isolated from wastewater treatment plants. Front Microbiol. 2019;10:633.
Crossref - Shevchenko LV, Dobrozhan YV, Mykhalska VM, Osipova TY, Solomon VV. Contamination of hen manure with nine antibiotics in poultry farms in Ukraine. Regul Mech Biosyst. 2019;10(4):532-537.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.