ISSN: 0973-7510
E-ISSN: 2581-690X
Legumain an asparginyl endopeptidase expressed by both tumor cells and cells present in tumor microenvironment is an ideal therapeutic target for development of cancer therapies due to its correlation with high metastasis and invasion in various cancers. Microbial derivatives have demonstrated many pharmacological properties such as antioxidant, anti-inflammatory, anti tumor and immunostimulatory activities.In the current study, 541 microbial derivatives were screened for their potential to inhibit legumain using Lib dock .Out of 541 compounds screened we have identified 55 microbial derivatives which showed binding to legumain by docking. Molecular interaction analysis of top five docked derivatives revealed the interaction of derivatives with the catalytic residues of legumain. These compounds need to be further evaluated in vitro and in vivo for Legumain inhibition and ultimately cancer regression.
In silico, Legumain, Lib dock, Microbial derivatives
Legumain (LGMN) also known as asparaginyl endopeptidase (AEP) is implicated in various cancer such as prostrate, breast, colon, lung, ovarian, central nervous system (CNS) related cancers, melanoma and lymphoma1.LGMN expression has also been reported in Tumor associated macrophages (TAM) also called as M2 macrophages2. LGMN is sparsely expressed by the normal tissues1. LGMN undergoes series of maturation steps from its pro-enzyme form to become proteolytically active3. LGMN expression has been correlated with low apoptosis and high invasion and metastasis of cancer cells both in vitro and in vivo1.LGMN is expressed not only in tumor cells but also found in the cells present in tumor microenvironment. Hence it holds the potential of serving as a prognostic factor and as a therapeutic target in cancer1,2,4.
Microbial derivatives have shown promising results in the development of therapies for cancer5. Bacterial Azurin produced from Pseudomonas aeruginosa has demonstrated cytotoxicity towards cancer cell lines such as Melanoma (UISO-Mel-2)6 and breast cancer (MCF-7)7 cell lines in vitro. It has also shown to increase apoptosis mediated by stabilising p53 and increasing the expression of pre-apoptotic protein Bax6,7,8. Trichostatin produced from Streptomyces hygroscopicus is a well-known Histone deacetylase(HDAC)inhibitor, a validated target for the development of antitumor therapies9. Thiocoraline bioactive compound isolated from Micromonospora marina, has shown selective cytotoxicity against lung and colon cancer cell lines as well as melanoma10. Macrolactin-A a major metabolite of Noctilucascintillans is reported to inhibit B16-F10 murine melanoma cancer cells11. Borophycina boron-containing metabolite, isolated from Nostoclinckia and N. spongiaeforme var. tenue, marine cyanobacterial strains has exhibited cytotoxicity against human epidermoid carcinoma (LoVo) and human colorectal adenocarcinoma (KB) cell lines 12,13.
As evidenced by the literature about the potential of microbial derivatives in the development of antitumor therapies, the current study employs the use of in silico tools for screening and identification of LGMN inhibitors. In silico methods have been efficient and quicker for the virtual screening of compounds with a known target protein. Molecular docking is one of the in silico approaches which plays a major role in computer aided drug designing by predicting the binding of lead compounds in the active sites of target proteins.
In the current study we have screened 541 microbial derivatives for their potential to inhibit LGMN by using Lib dock14 module available in Accelrys Discovery Studio 3.5 (San Diego, CA, USA ).
Selection of LGMN structure from Protein Data Bank:The Crystal structure of active LGMN in complex with YVAD-CMK at pH 5.0 15 was retrieved from Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB ID: 4AWA) (http://www.rcsb.org/pdb). All bound water molecules, other hetero atoms and ligands were removed manually from the PDB file prior to docking. The protein was prepared using “Prepare Protein”module available in discovery studio 3.5.
Generation of ligand dataset: The structures of 541 microbial derivatives (ligands) were collected from PubChem compound database (https://pubchem.ncbi.nlm.nih.gov/). Prior to docking, the ligands were prepared using the ‘‘prepare ligand’’ module available in Discovery studio 3.5.
Active site analysis of 4AWA structure: Prediction of active site is crucial step in molecular docking studies for identification of potent inhibitors.As per the literature LGMN harbours a catalytic triad consisting of three amino acid residues (Cys189-His148-Asn42)15.A receptor grid was created around the binding cavity (active sites) of protein by specifying the key amino acid residues (Cys 189, His 148 and Asn42 ). Binding site sphere was set and 35.78,24.36 and -7.80 are the dimensions of X,Y and Z respectively.
Molecular Docking using Discovery Studio 3.5: To identify new compounds that could potentially inhibit LGMN through binding to the catalytic triad pocket, a virtual screening is carried out using Lib dock module of Discovery Studio 3.514. Lib dock docks ligand into the active site by calculating hot spots and using polar and a polar probes and these hot spots are further used to align ligands to form interactions 16.The default lib dock protocol available in the module was used for the docking. Details of successful and failed ligands are available in the “docked ligands” and “failed ligands” sections respectively of the result file. Different Poses of protein-ligand complex were obtained after successful docking process with their specific lib dock score displayed on it. The interactions between the ligand and the protein molecules were investigated using “Analyze ligand poses” and “2D diagram” of docked receptor-ligand complexes. This analysis gives better idea of interactions between the key residues of protein and complimentary groups/atoms of ligands.
The crystal structure of LGMN (PDB ID:4AWA) was retrieved from protein data bank and was prepared using prepare protein module. Active site pocket was created using catalytic residues of LGMN (Cys189-His148-Asn42).
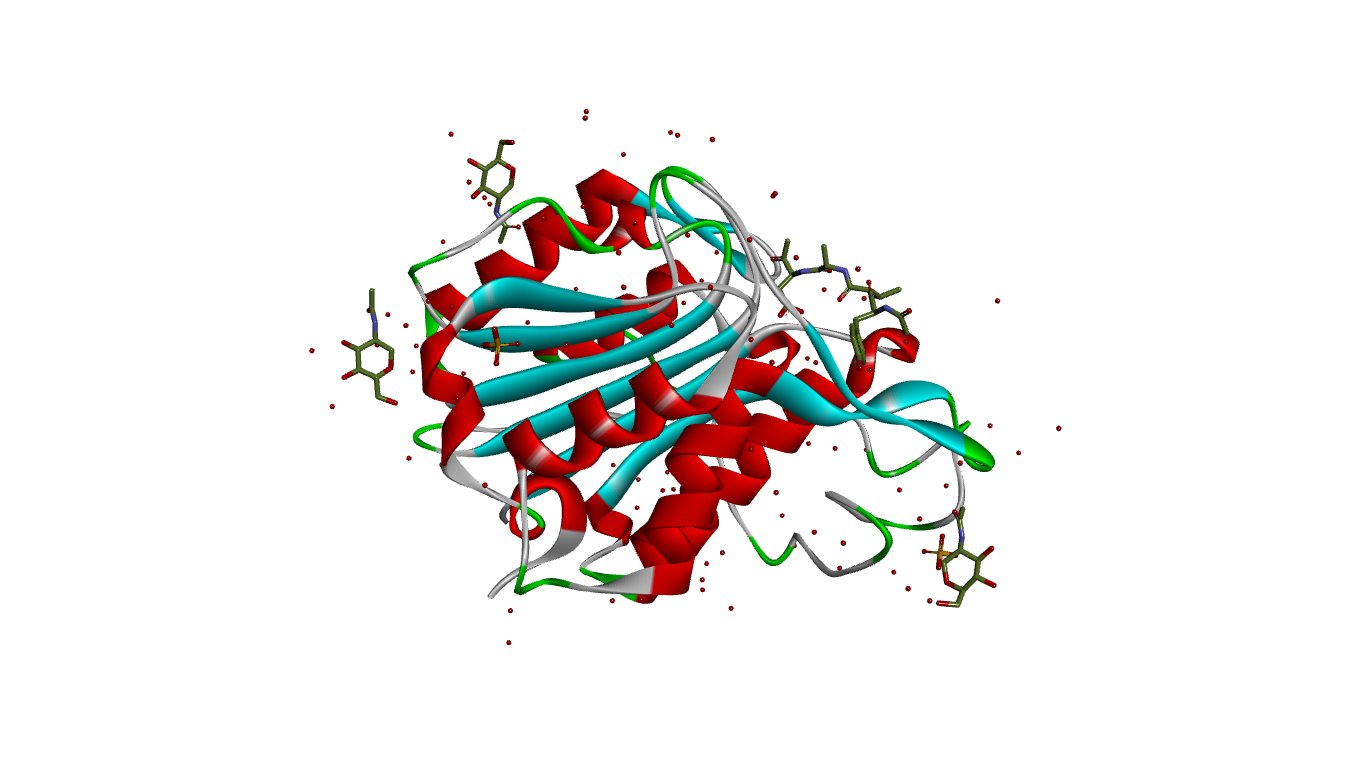
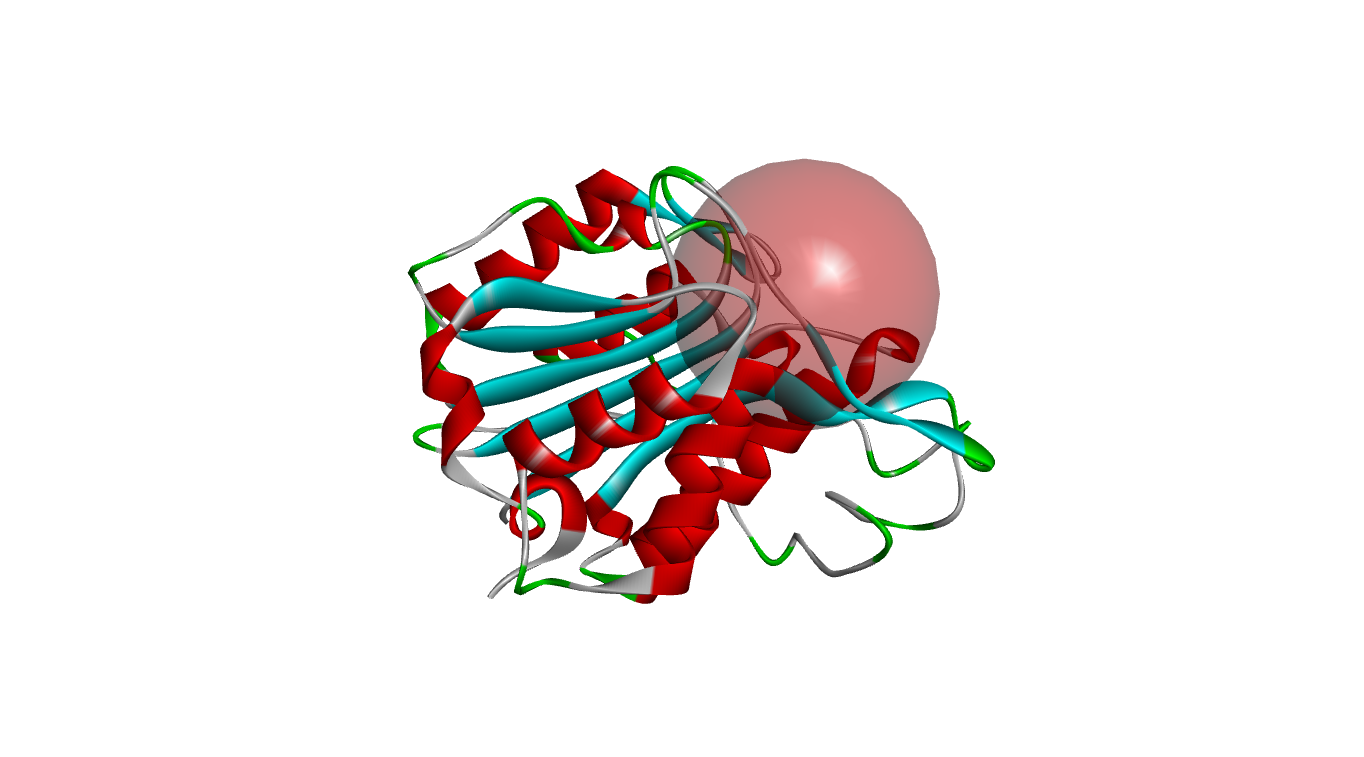
Fig. 1A depicts the 3D structure of LGMN retrieved from PDB. Fig.1B illustrates the prepared structures of the protein after removal of hetero atoms, ligands and water molecules with a sphere around the active site.
A total of 541 microbial derivatives were docked at the catalytic site of LGMN using Lib Dock. Among the derivatives docked,55 compounds demonstrated successful docking at the catalytic site of LGMN. All the docked poses were ranked by the Lib dock score. The list of compounds docked successfully with their respective lib dock score has been given in Table 1.
Table (1):
List of 55 successfully docked microbial derivatives at the active site of LGMN.
S.No |
Name of the compounds |
Pubchem ID |
Lib dock score |
|---|---|---|---|
1 |
Blasticidin S hydrochloride |
356629 |
117.08 |
2 |
Bicyclomycin benzoate |
91618023 |
99.62 |
3 |
α-Zearalenol |
5284645 |
89.35 |
4 |
Sinefungin |
65482 |
85.47 |
5 |
9-Methylstreptimidone |
6373950 |
85.15 |
6 |
Cerulenin |
5282054 |
83.99 |
7 |
Mycophenolic acid |
446541 |
83.39 |
8 |
4-Hydroxyalternariol |
118797633 |
82.72 |
9 |
LL Z1640-2 |
46882176 |
81.18 |
10 |
Tetradecanoyl-L-homoserine lactone |
58122267 |
79.71 |
11 |
Dodecanoyl-L-homoserine lactone |
11565426 |
79.70 |
12 |
Epitetracycline hydrochloride |
54686189 |
78.96 |
13 |
Tetracycline |
54675776 |
78.96 |
14 |
Tetracycline hydrochloride |
54704426 |
78.96 |
15 |
Thiamphenicol |
27200 |
78.08 |
16 |
Toyocamycin |
11824 |
76.78 |
17 |
Toxoflavin |
66541 |
76.77 |
18 |
Deacetylanisomycin |
11790817 |
76.45 |
19 |
Bestatin |
72172 |
76.20 |
20 |
21-Hydroxyoligomycin A |
3016254 |
75.98 |
21 |
Corynecin III |
101131598 |
75.95 |
22 |
Terrein |
6436830 |
75.02 |
23 |
RK-682 |
54678922 |
74.51 |
24 |
TAN 1364B |
54690140 |
74.51 |
25 |
Sancycline |
54688686 |
74.16 |
26 |
Sancycline hydrochloride |
54712662 |
74.16 |
27 |
Octanoyl-L-homoserine lactone |
6914579 |
73.99 |
28 |
Chloramphenicol succinate sodium |
656833 |
73.87 |
29 |
Methacycline |
54675785 |
73.42 |
30 |
Methacycline hydrochloride |
54685047 |
73.42 |
31 |
Avenaciolide |
11747526 |
72.96 |
32 |
Anisomycin |
253602 |
72.64 |
33 |
LL Z1640-4 |
57370130 |
72.57 |
34 |
Clavulanate potassium |
23665591 |
71.69 |
35 |
Germicidin B |
86169826 |
71.30 |
36 |
Florfenicol amine |
156406 |
70.43 |
37 |
Corynecin IV |
133562649 |
69.99 |
38 |
Brefeldin A |
5287620 |
69.84 |
39 |
Germicidin A |
102106080 |
69.39 |
40 |
Clindamycin hydrochloride |
16051951 |
68.16 |
41 |
Dihydroaeruginoic acid |
5381954 |
67.72 |
42 |
Tenuazonic acid |
54683011 |
66.72 |
43 |
Roquefortine E |
5326324 |
64.45 |
44 |
Cycloechinulin |
16088234 |
64.05 |
45 |
Butyryl-L-homoserine lactone |
10130163 |
62.80 |
46 |
Moniliformin |
40452 |
62.21 |
47 |
acetyl-L-homoserine lactone |
10012012 |
61.35 |
48 |
Chloramphenicol acetate |
83940 |
60.58 |
49 |
Hexanoyl-L-homoserine lactone |
10058590 |
60.45 |
50 |
Simvastatin |
54454 |
59.49 |
51 |
Aphidicolin |
457964 |
58.92 |
52 |
Chloramphenicol |
5959 |
57.79 |
53 |
Butyrolactone I |
7302 |
51.49 |
54 |
Roquefortine C |
5935070 |
51.38 |
55 |
Cellocidin |
10971 |
39.88 |
The top 5 derivatives with highest lib dock scores were further used to evaluate the interactions with LGMN.
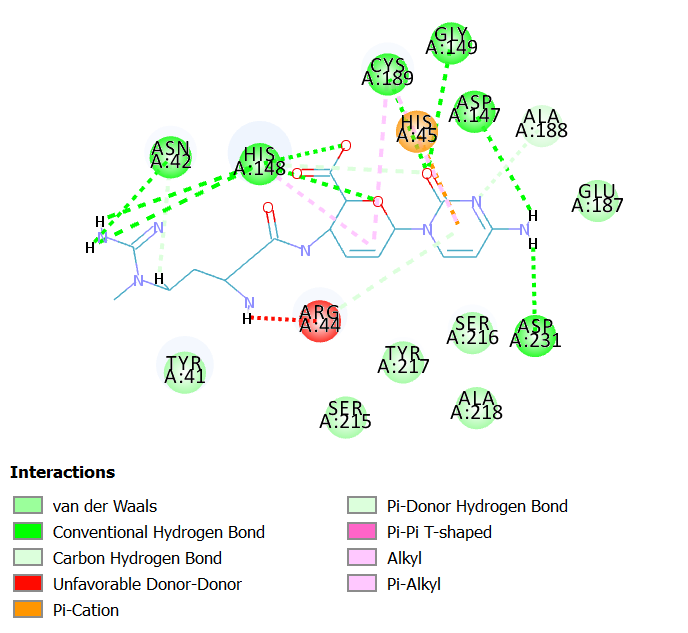
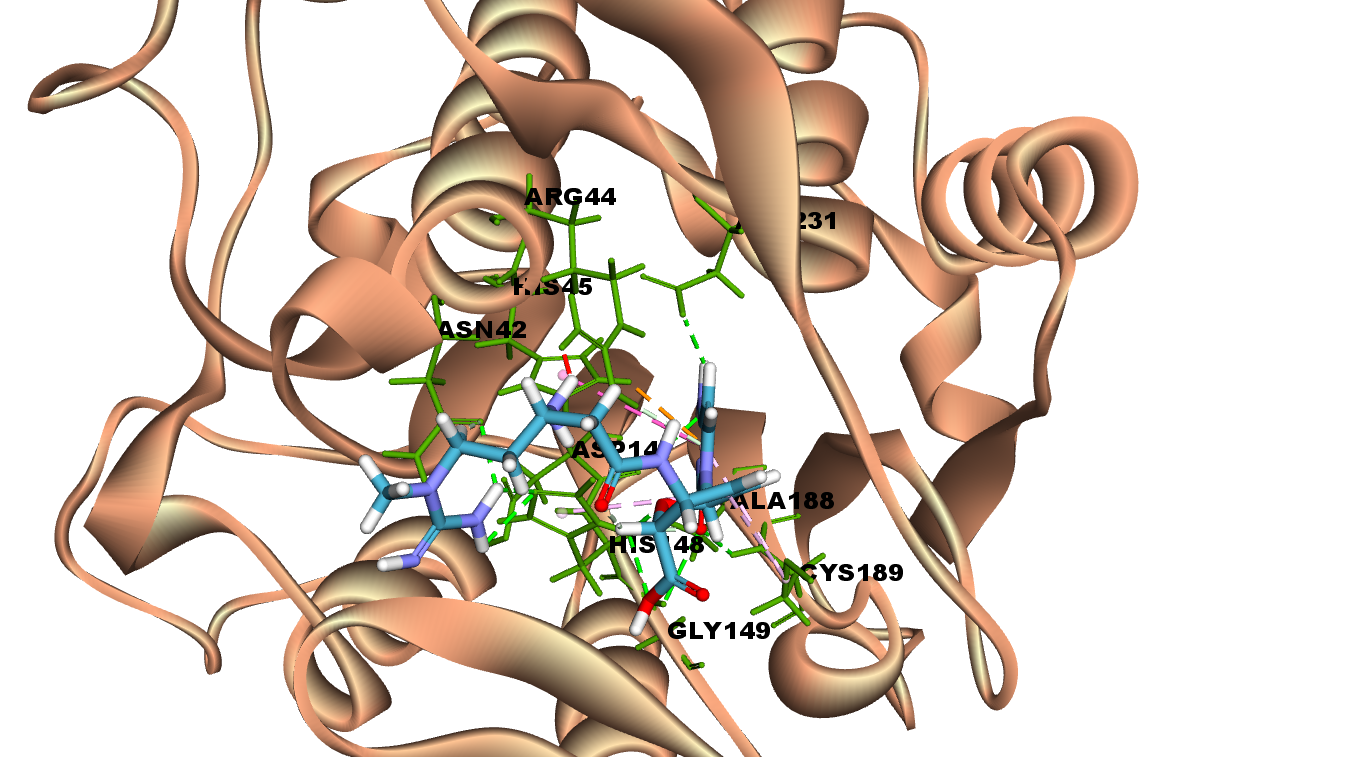
Interactions of Blasticidin S hydrochloride at LGMN catalytic site
Blastocidin S hydrochoirde is a salt of Blasticidin S a nucleoside antibiotic, produced by Streptomyces species. Blasticidin S HCl acts as a DNA and protein synthesis inhibitor17,18.
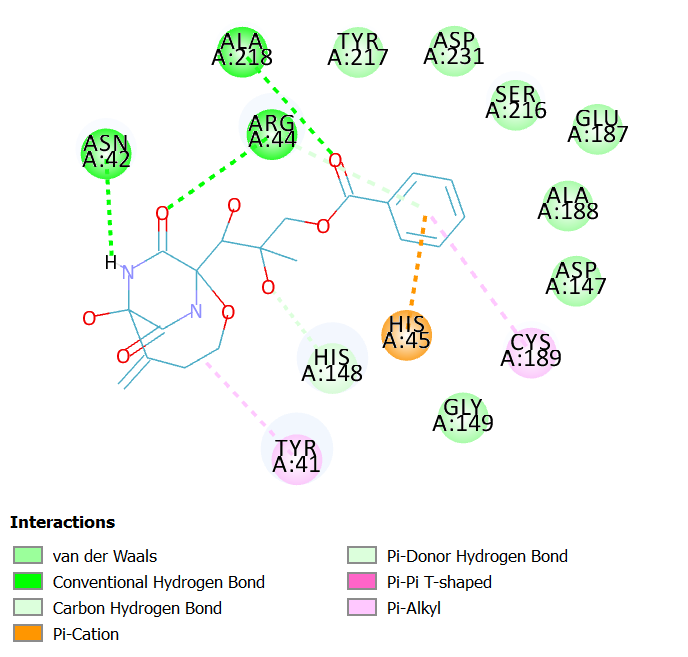
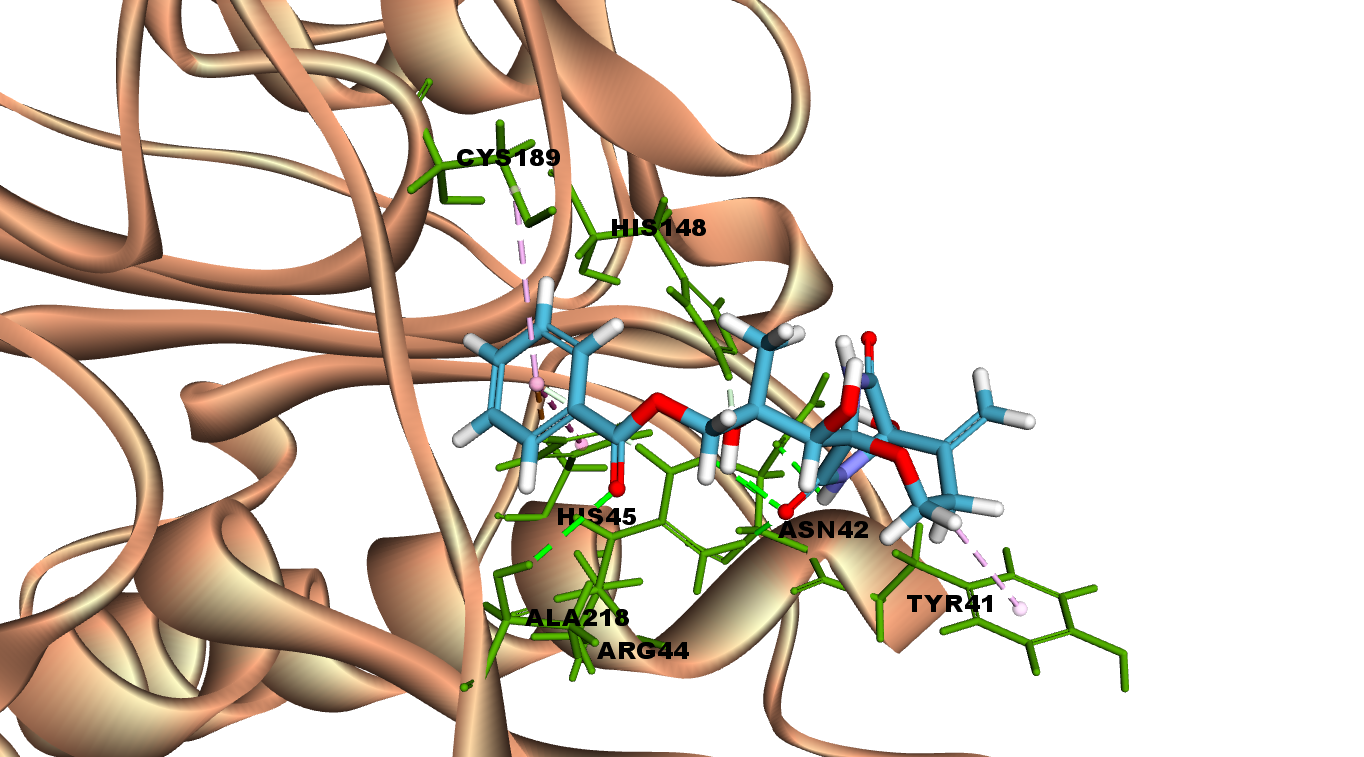
Blasticidin S hydrochloride interacted with all the three amino acids of catalytic residues Cys 189, His 148 and Asn 42 by forming hydrogen bonds. In addition, it has also interacted with Asp 231,Gly 149, Asp 147 with hydrogen bonding. The molecular interaction analysis indicates Blasticidin S hydrochloride as potent inhibitor of LGMN owing to its interaction with the catalytic triad amino acids residues and nine hydrogen bonds at the active site. Fig 2A illustrates 2D diagram of interactions of Blasticidin S hydrochloride at the LGMN catalytic site and Fig 2B shows the 3D diagram of interactions of Blasticidin S hydrochloride at the LGMN catalytic site.
Interactions of Bicyclomycin benzoate at LGMN catalytic site
Bicyclomycin benzoate is an antibiotic produced by Streptomyces sapporonensis and it inhibits gram-negative bacteria.
Bicyclomycin benzoate interacts with LGMN at the active site by forming hydrogen bonds with Asn 42 (catalytic aminoacid), Arg 44 and Ala 218. In addition, other interactions such as van der Waals, pi-Alkyl and pi-cation are also observed in the 2D diagram.
Fig 3A illustrates 2D diagram of interactions of Bicyclomycin benzoate at the LGMN catalytic site and Fig 3B shows the 3D diagram of interactions of Bicyclomycin benzoateat the LGMN catalytic site.
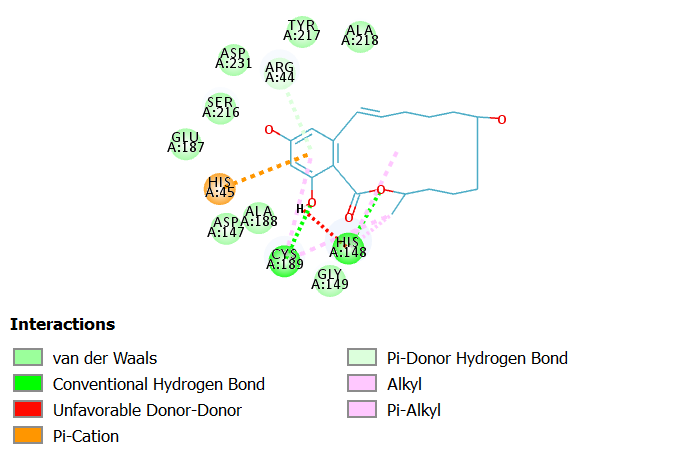
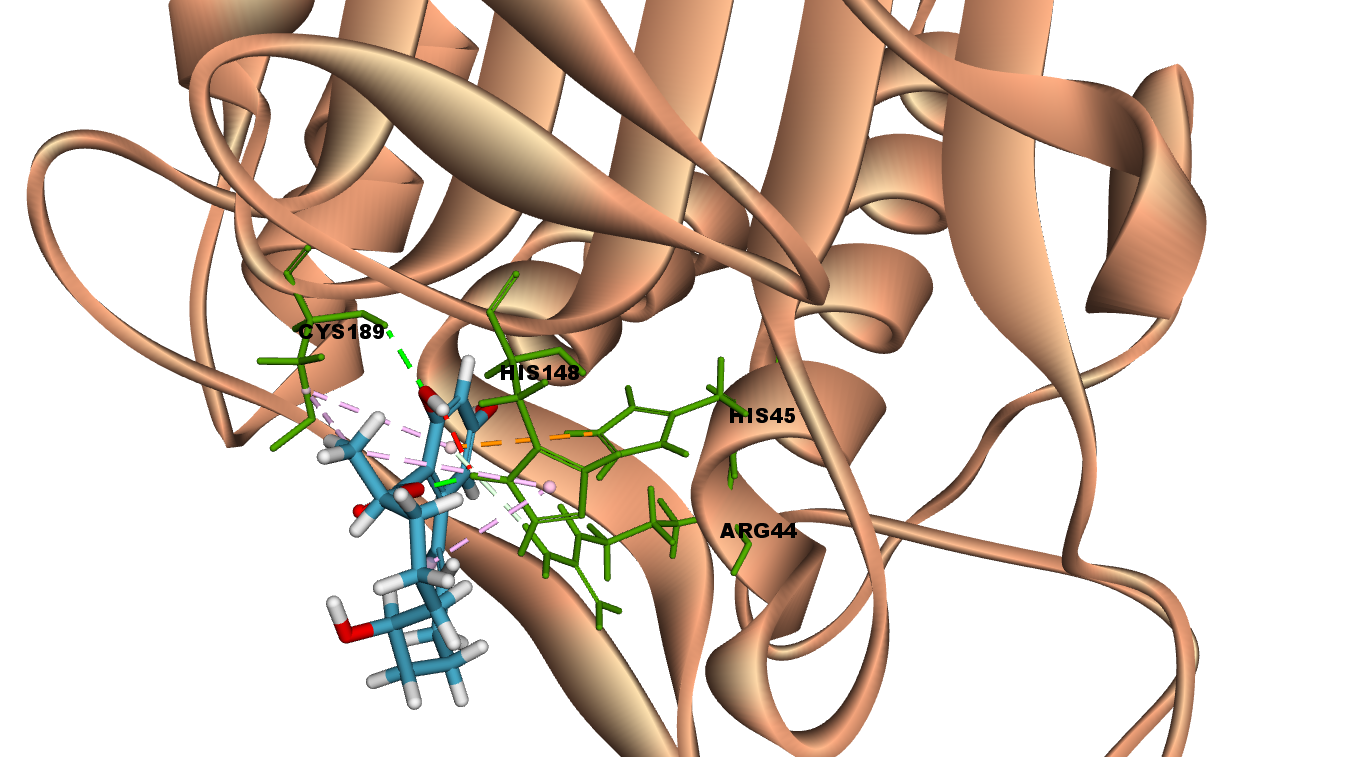
Interactions of a-Zearalenol at LGMN catalytic site
a-Zearalenol is an oestrogenic mycotoxin produced by several species of Fusarium that contaminate cereal crops19.
a-Zearalenol interacts with LGMN at the active site by forming two hydrogen bonds with catalytic amino acids Cyst 189 and His 148. In addition, other interactions such as van der Waals, pi-Alkyl and pi-cation are also observed in the 2D diagram.
Fig 4A illustrates 2D diagram of interactions of a-Zearalenol at the LGMN catalytic site and Fig 4B shows the 3D diagram of interactions of a-Zearalenol at the LGMN catalytic site.
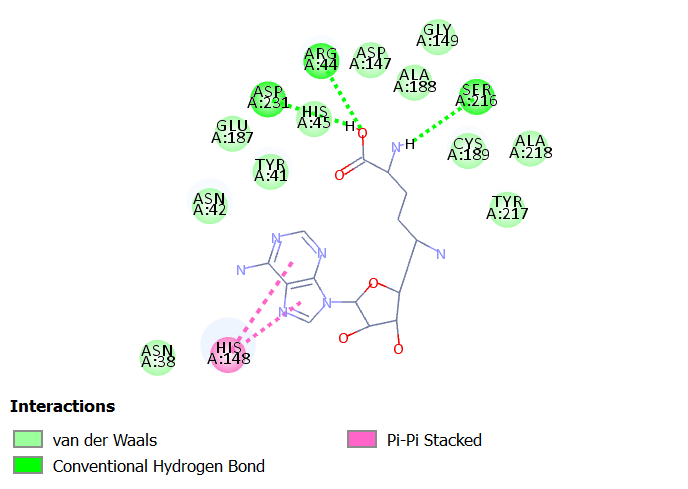
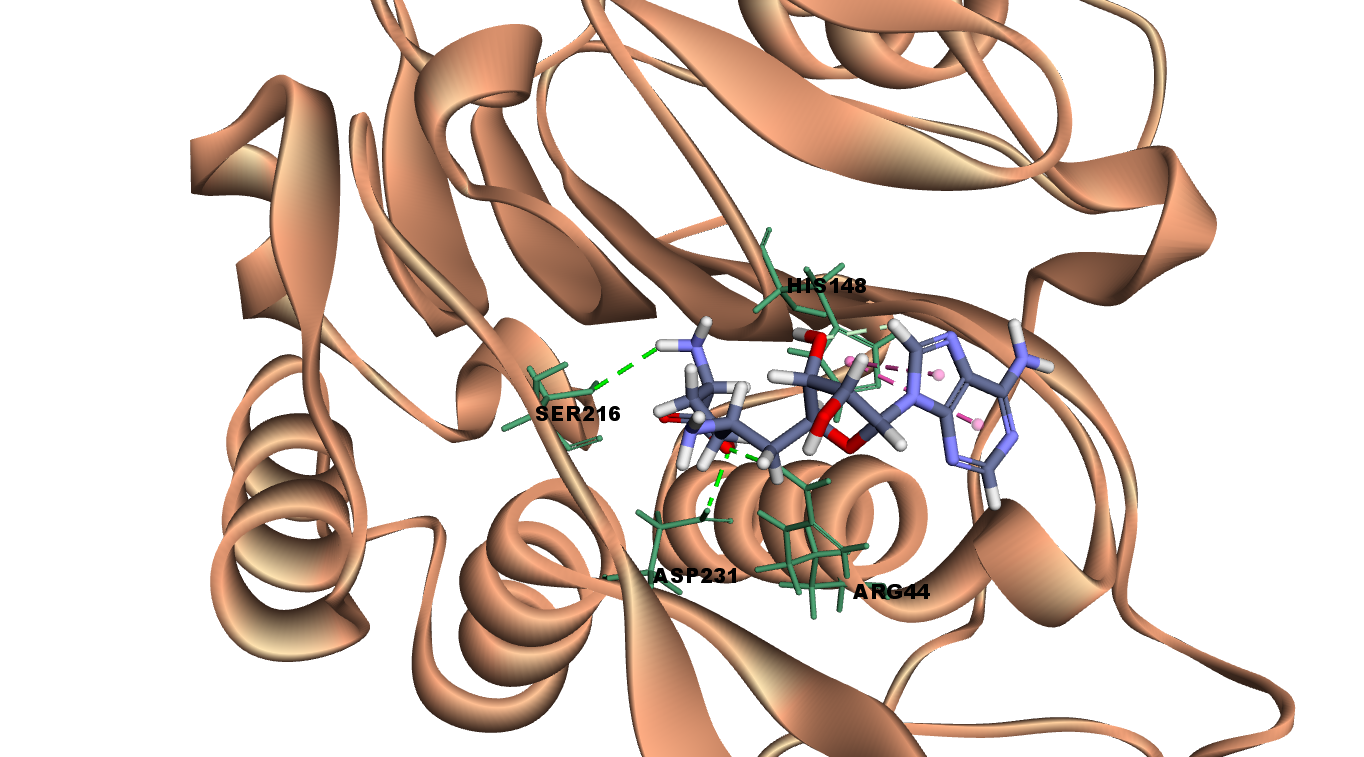
Interactions of Sinefungin at LGMN catalytic site
Sinefungin is an inhibitor of transmethylation reactions associated to DNA, RNA and Proteins. It is a natural nucleoside with antifungal, antiviral and antiprotozoal activities20,21
Sinefungin interacts with LGMN at the active site by forming three hydrogen bonds with Arg 44, Ser 216 and Asp 231. It interacts with the catalytic residues such as Asn 42 with van der Waal and His 148 with Pi-Pi stacked interactions.
Fig 5A illustrates 2D diagram of sinefungin at the LGMN catalytic site and Fig 5B shows the 3D diagram of interactions of sinefungin at the LGMN catalytic site.
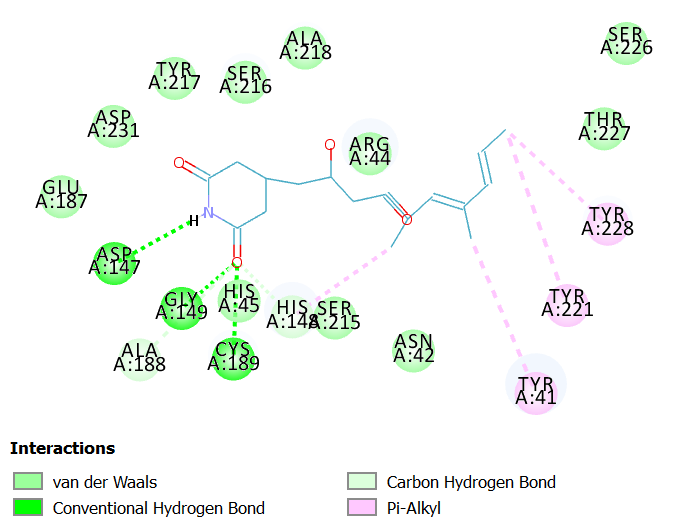
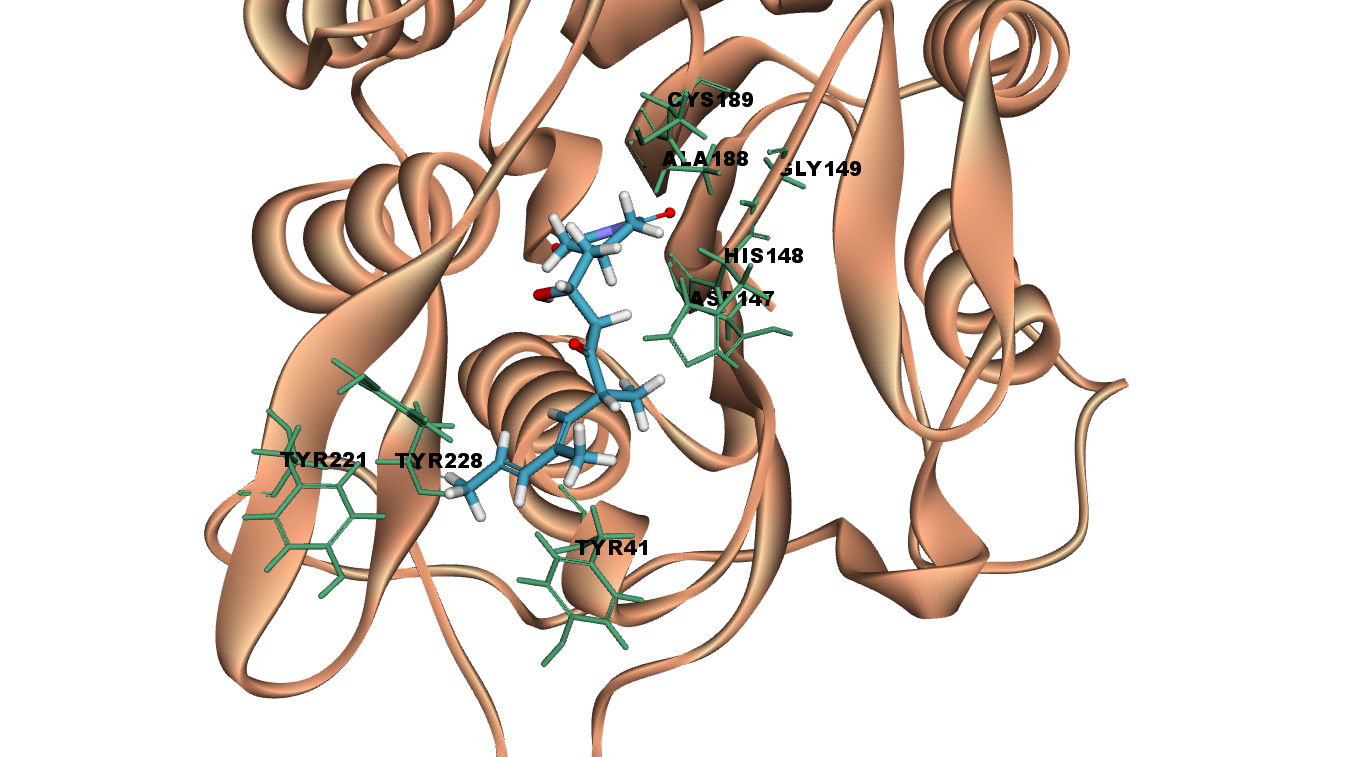
Interactions of 9-Methylstreptimidoneat LGMN catalytic site
9-Methylstreptimidone is isolated from Streptomyces species.
9-Methylstreptimidone exhibits antifungal and antiviral activity. Also known as an inhibitor of the nuclear factor, NF-kB22.
9-Methylstreptimidone interacts with LGMN at the catalytic site by forming three hydrogen bonds with Cys 189(catalytic amino acid), Asp 147 and Gly 149. It interacts with the other catalytic residues such as Asn 42 and His 148 with vander Waal interactions. Other interactions such as carbon-hydrogen bond and Pi alkyl stacked interactions are also observed.
Fig 6A illustrates 2D diagram of interactions between 9-Methylstreptimidone at the LGMN catalytic site and Fig 6B shows the 3D diagram of interactions between 9-Methylstreptimidone at the LGMN catalytic site.
The objective of the current study was to screen and identify microbial derivatives for their potential to inhibit LGMN activity using in silico approaches. Molecular docking of microbial derivatives has identified 55 potential LGMN inhibitors from 541 screened using Lib dock module. The results of this study not only demonstrate the probable binding mode of these derivatives with LGMN, but also encourage further evaluation of these microbial derivatives both in vitro and in vivo for LGMN inhibition and cancer regression.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the support of Department of Applied Microbiology, Sri Padmavati Mahila Visvavidyalayam (Women’s University), Tirupati, Andhra Pradesh, India without which the present study could not have been completed.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Liu C, Sun C, Huang H. Overexpression of Legumain in Tumors Is Significant for Invasion / Metastasis and a Candidate Enzymatic Target for Prodrug Therapy. 2003: 2957-2964.
- Luo Y, Zhou H, Krueger J, et al. JCI – Targeting tumor-associated macrophages as a novel strategy against breast cancer. 2006; 116(8). doi:10.1172/JCI27648
- Zhao L, Hua T, Crowley C, et al. Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 2014. doi:10.1038/cr.2014.4
- Mai C-W, Chung FF-L, Leong C-O. Targeting Legumain As a Novel Therapeutic Strategy in Cancers. Curr Drug Targets. 2017; 18(11). doi:10.2174/1389450117666161216125344
- Bernardes N, Seruca R, Chakrabarty AM, Fialho AM. Microbial-based therapy of Cancer Current progress and future prospects. Bioeng Bugs. 2010. doi:10.4161/bbug.1.3.10903
- Gupta DT. Bacterial redox protein azurin, tumour suppressor protein p53, and regression of cancer. Proc Natl Acad Sci U S A. 2002; 22: 14098-14103. doi:10.1073/pnas.222539699
- Yamada T, Hiraoka Y, Ikehata M, et al. Apoptosis or growth arrest: Modulation of tumor suppressor p53’s specificity by bacterial redox protein azurin. Proc Natl Acad Sci U S A. 2004. doi:10.1073/pnas.0400899101
- Apiyo D, Wittung-Stafshede P. Unique complex between bacterial azurin and tumor-suppressor protein p53. Biochem Biophys Res Commun. 2005. doi:10.1016/j.bbrc.2005.05.038
- Vigushin DM, Ali S, Pace PE, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res. 2001. doi:10.1016/s0092-8674(00)80211-1
- Sithranga Boopathy N, Kathiresan K. Anticancer drugs from marine flora: An overview. J Oncol. 2010. doi:10.1155/2010/214186
- Carté BK. Biomedical potential of marine natural products. Bioscience. 1996. doi:10.2307/1312834
- Banker R, Carmeli S. Tenuecyclamides A-D, cyclic hexapeptides from the cyanobacterium Nostoc spongiaeforme var. tenue. J Nat Prod. 1998. doi:10.1021/np980138j
- Davidson BS. New dimensions in natural products research: cultured marine microorganisms. Curr Opin Biotechnol. 1995. doi:10.1016/0958-1669(95)80049-2
- Diller DJ, Merz KM. High throughput docking for library design and library prioritization. Proteins Struct Funct Genet. 2001. doi:10.1002/1097-0134(20010501)43:2<113::AID-PROT1023> 3.0.CO;2-T
- Dall E, Brandstetter H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc Natl Acad Sci U S A. 2013. doi:10.1073/pnas.1300686110
- Zhou X, Yu S, Su J, Sun L. Computational Study on New Natural Compound Inhibitors of Pyruvate Dehydrogenase Kinases. 2016. doi:10.3390/ijms17030340
- Yamaguchi H, Tanaka N. Inhibition of protein synthesis by blasticidin S: II. studies on the site of action in e. coli polypeptide synthesizing systems. J Biochem. 1966;60(6):632-642. doi:10.1093/oxfordjournals.jbchem.a128489
- S.B. Sullia and D.H. Griffin. Inhibition of DNA synthesis by Cycloheximide and Blasticidin-S is Independent of their effect on protein synthesis. Biochim Biophys Acta, 475.
- Bennett JW, Klich M, Mycotoxins M. Mycotoxins. Clin Microbiol Rev. 2003. doi:10.1128/CMR.16.3.497
- Barbés C, Sánchez J, Yebra MJ, Robert-Geró M, Hardisson C. Effects of sinefungin and S-adenosylhomocysteine on DNA and protein methyltransferases from Streptomyces and other bacteria. FEMS Microbiol Lett. 1990. doi:10.1016/0378-1097(90)90073-Y
- Zheng S, Hausmann S, Liu Q, et al. Mutational analysis of Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase, structure of the enzyme bound to sinefungin, and evidence that cap methyltransferase is the target of sinefungin’s antifungal activity. J Biol Chem. 2006. doi:10.1074/jbc.M607292200
- Ishikawa Y, Tachibana M, Matsui C, Obata R, Umezawa K, Nishiyama S. Synthesis and biological evaluation on novel analogs of 9-methylstreptimidone, an inhibitor of NF-kappaB. Bioorg Med Chem Lett. 2009. doi:10.1016/j.bmcl.2009.01.107.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.