ISSN: 0973-7510
E-ISSN: 2581-690X
Microbial cellulase enzyme, endoglucanase, performs a significant role in various industries, including textiles, paper pulp, biofuel production, agriculture, and pharmaceuticals. The aim of this research was focused on extracting, purifying, and investigating the microbial enzyme produced by Bacillus subtilis strain R2A (accession number PP088049). The cellulase-producing Bacillus strain was isolated from agricultural fields near Gwalior in Madhya Pradesh. The bacterial isolate of Bacillus species was preliminarily identified via cultural characteristics and biochemical characterization, using selective media culture. B. subtilis isolate exhibited significant cellulase enzyme activity under optimized conditions (pH 4.0-9.0, 37 °C, 120 rpm). The cellulase enzyme was refined via salting out, utilizing Diethylaminoethyl Cellulose-52 matrix column chromatography to purify microbial cellulase enzymes. The microbial cellulase was refined up to a 5.44 enrichment factor and had an catalytic activity of 137.95 U/mg protein. Microbial cellulase was a single-unit enzyme possessing an estimated protein molecular weight of 51.4 kDa, as evaluated through SDS-PAGE. This indicates the potential of B. subtilis isolate as a potent resource for biotechnological and industrial processes, particularly in producing sustainable fuels and cellulose-derived bioproducts. Microbial cellulases play an essential role in drug discovery by initiating the biotransformation of complex carbohydrates into secondary metabolites, as a result expediting the development of novel therapeutics, and improving medication efficacy. Future studies could focus on upscale production and exploring its application in diverse industrial settings.
Cellulase, Bacillus subtilis Isolate, Column Chromatography, SDS-PAGE
Cellulose constitutes 50% of plant biomass by dry weight and 50% of the secondary biomass sources, in the form of agro-industrial waste.1-5 Cellulose is considered one of the most resistant fibers and non-crystalline polysaccharides that are resistant to hydrolysis and insoluble in water.6-8 It consists of monomeric units of D-glucose, which are bonded through β-1,4 carbohydrate linkages.9-11 Cellulase enzymes can break down these β-1,4 carbohydrate linkages, resulting in the synthesis of smaller oligosaccharides and ultimately glucose.12-15 Lignocellulose biomass is the primary structural constituent of flora and is a sustainable energy source within the biosphere.16-22
Although carboxymethyl cellulase (endoglucanases) from Bacillus species are unable to degrade crystalline cellulose, certain polysaccharide-degrading enzymes, such as alkaline cellulases, have shown promising results, and have been extensively used in detergent.23-25 Bacteria are considered efficient producers of cellulase enzymes because they can secrete substantial amounts extracellularly, which simplifies the extraction and purification processes.26-30 Among them, B. subtilis has been extensively investigated for its remarkable cellulase production capabilities.31,32 Various strains, for example, Bacillus subtilis JBS250, B. subtilis strain LFS3, CBS31 strain of Bacillus subtilis, Bacillus halodurans IND18, and BC1 strain of Bacillus subtilis, have demonstrated significant potential in biomass fermentation, fiber modification, and pharmaceutical applications.33-36
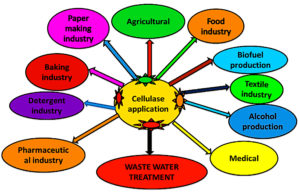
This investigation aimed to obtain, isolate, produce, and purify microbial cellulase from cellulose-degrading bacteria, with a specific focus on a Bacillus subtilis isolate, followed by characterization of the enzyme for potential large-scale applications.37 The examination is concentrated on maximizing production, enzyme separation through DEAE-Cellulose 52-column chromatography methods, and determining its physiochemical properties using SDS-PAGE.38 Microbial cellulase enzymes have various applications in the pharmaceuticals, brewing, textiles, food, paper, and pulp industry (Figure 1).39
Materials
Folin-Ciocalteu (phenol) reagent, bovine albumin, and DEAE Cellulose-52 were obtained from HiMedia. Acrylamide, Bis-acrylamide derivative TEMED, diammonium persulfate (APS) was procured from Genetix Biotech Asia Pvt. Limited. The other chemicals, all of LG grade, were obtained from Sigma-Aldrich Corporation.
Isolation of microorganisms
Cellulase-degrading bacterial samples were obtained from five cellulose degradation sites, including agricultural fields. The two differential media used in the streak-plate method were: Carboxymethyl cellulose (CMC) agar media: Peptone (10 gm/litre), K₂HPO₄ (2 gm/litre), Carboxymethyl cellulose (10 gm/litre), MgSO4.7H2O (0.3 gm/litre), gelatin (2 gm/litre), and (NH4)2 SO4 (2.5 gm/litre), and agar (15.0 gm/litre). Neutral pH was maintained, and Petri dishes were kept at 37 °C for 72 hours for incubation.
Sucrose agar media (SAM): yeast extracts (2.0 gm/litre), MgSO4.7H2O (0.25 gm/litre), K2HPO4 (0.5 gm/litre), sucrose (2.0 gm/litre), and agar (15.0 gm/litre). Similarly, neutral pH was maintained and the Petri dishes were kept at 37 °C for 72 hours for incubation.39,40
Characterization of Cellulolytic bacteria
Cultural characteristics
Microorganisms were characterized based on their colony characteristics, form, aroma, edge, texture, and pigmentation. Gram staining is a microbiological technique that categorizes microorganisms into Gram-positive (+) and Gram-negative (-) microorganisms.40,41
Biochemical analysis
After the phenotypic and cultural properties of the bacterial strains had been recorded, further confirmation based on their biochemical analysis includes IMViC, catalase, fermentation, and Simmon’s citrate test using standard biochemical methods.41
Microbial cellulase enzyme is produced using the submerged fermentation method
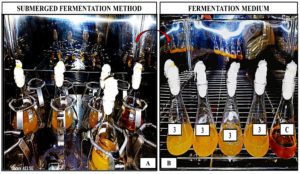
Bacterial cultures were analyzed for cellulolytic enzyme synthesis through submerged culture techniques. CMC broth was prepared by dissolving 1% peptone, 0.3% beef extract, 3% carboxymethylcellulose, and 0.5% NaCl in 100 mL of deionized water, and sterilized at 121 °C for 15 minutes in an autoclave to ensure sterility. Sucrose broth media (SBM) was composed of 0.2% yeast extract, 0.2% Sucrose, 0.05% KH2PO4, and 0.025% MgSO4.7H2O in 100 ml of deionized water and autoclaved at 121 °C for a duration of 15 min. Post-sterilization, 3.0 ml of microbial isolate broth was combined with the culture broth and maintained at 37 °C for 2 days with an intermittent shaking of 120 cycles/min (Figure 2). Cellulase enzymatic activity was measured at 24 hrs intervals. The crude enzyme source was the supernatant after centrifugation.42
Figure 2. Submerged fermentation for cellulase enzyme production using Bacillus subtilis R2A. (A) Fermentation medium inoculated with Bacillus subtilis placed in a biological shaker incubator at 37°C and 120 rpm for 48 hours. (B) Post-incubation culture in conical flask showing enzyme production in submerged fermentation broth after 2 days
Quantification of microbial cellulase production using the DNSA method
The dinitrosalicylic acid (DNSA) method is a colorimetric technique used to quantify cellulase activity.43
Purification of cellulase enzyme from bacterial isolates
Ammonium sulfate fractionation (salting out)
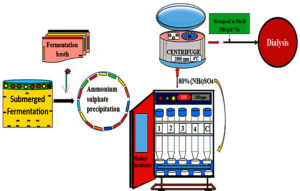
To precipitate proteins from the crude extract, solid ammonium sulfate (NH4)2SO4 was added to achieve 80% saturation. The solution was continuously agitated at 4 °C overnight using a mechanical stirrer to ensure uniformity and stability. After incubation, the sample was subjected to centrifugation, and the resulting precipitate was resuspended in 50 mM PBS (pH 7.0) for subsequent fractionation and analysis. The Semi-concentrated microbial cellulase was then subjected to dialysis against the same phosphate buffer to remove excess salts (Figure 3).44
Figure 3. Ammonium Sulfate Precipitation of crude enzyme obtained from submerged fermentation. Proteins in the crude enzyme extract were precipitated by gradual addition of solid ammonium sulfate (NH₄)₂SO₄ to 80% saturation. The mixture was stirred at 4 °C overnight, centrifuged, and the pellet resuspended in 50 mM phosphate buffer (pH 7.0). The semi-purified cellulase was then dialyzed against the same buffer to remove residual salts. Adapted from Ibrahim et al.41
DEAE cellulose -52 column chromatography
Diethylaminoethyl Cellulose -52 Column (1.5 × 15 cm) was vertically mounted and prepared for protein purification at a steady flow of 200 µl min-1. Four-bed vol. of 50 mM Tris-HCl buffer was used to equilibrate the column. Unbound proteins were eliminated by thoroughly washing the chromatographic system. The matrix-bound proteins were released through a stepwise NaCl elution gradient (0 to 0.5 M) in Tris-HCl buffer. Six equal-volume fractions (2 ml each) were obtained at a consistent flow rate. Protein concentrations in each fraction were calculated using the Folin-Ciocalteu assay, with purified albumin derived from bovine serum as the standard.45
Electrophoretic analysis of purified microbial cellulase
The relative molar mass of the isolated microbial cellulase was evaluated by electrophoretic analysis. In addition to the enzyme sample, standard protein markers were added. The electrophoresis procedure was performed following Bracewell’s protocol. The enzyme was combined with an equivalent aliquot of 2X Laemmli sample buffer, which included 4% SDS in 0.125 M Tris-HCl (pH 6.8), and underwent thermal denaturation at 95 °C for 10 minutes. The microbial cellulase enzymes were further analyzed using Polyacrylamide Gel Electrophoresis (PAGE), with a separating gel of 12% concentration and a stacking gel of 4% concentration, gel electrophoresis was carried at room temperature for 2 hrs at 100 V using the Mini-Protein II System (Bio-Rad, USA).
Identification and molecular analysis of cellulolytic microorganisms from soil samples
Microbial cellulases are extensively studied as industrially relevant biocatalysts owing to their capacity to hydrolyze cellulose, major component of plant biomass. Due to their broad applications in industries such as biofuel production, and paper, there is a continuous demand for novel bacterial strains that exhibit enhanced cellulase activity and produce enzymes with greater stability under diverse conditions.45,46 Microbial cellulases play a crucial role in drug discovery by facilitating the biotransformation of complex carbohydrates into secondary metabolites. This process accelerates the development of novel therapeutics, supports textile processing, enables efficient biomass degradation, and improves medication efficacy.47 A cellulase-producing Bacillus strain was isolated from agricultural fields near Gwalior in Madhya Pradesh. The bacterial isolate exhibited rod-shaped morphology and Gram-positive staining. The colony morphology was irregular, opaque, creamy white or pale yellow, with uneven edges. Based on these characteristics, the isolate was preliminarily identified as a Bacillus species. Further subculturing on selective media such as Carboxymethyl Cellulose (CMC) Agar and Sucrose Agar Media (SAM), along with positive biochemical tests including Voges-Proskauer (VP), Simmons’ citrate, fermentation, casein hydrolysis, and catalase, and negative results for methyl red (MR) and indole production, confirmed the identity of the bacterium as Bacillus subtilis.48,49
Extraction and identification of microbial cellulase enzyme
The B. subtilis isolate exhibited significant cellulase enzyme activity under optimized conditions (pH 4.0-9.0, 37 °C, 120 rpm).50 The crude enzyme extract had a concentration of 0.383 mg/ml and showed a total cellulolytic activity of 9.7 U/ml.51,52 The microbial cellulase produced in this study exhibited impressive pH stability ranging from 4.0 to 9.0, which is particularly advantageous for industrial applications where varying pH conditions are often observed.53 Compared with cellulase derived from Bacillus pumilus and Bacillus licheniformis, our enzyme demonstrated superior pH tolerance.54,55 The broad pH stability of our enzyme makes it highly suitable for diverse industrial bioprocessing applications, including biofuels, pharmaceuticals, and waste treatment. Microbial cellulases, particularly those derived from Bacillus subtilis R2A, have attracted considerable attention due to their broad applicability across various industries including biofuel production, and emerging roles in the biomedical and pharmaceutical sectors. The cellulase produced in this study exhibited exceptional purification efficiency, high catalytic activity, and significant stability, surpassing several microbial cellulases previously reported in the literature.
Figure 4. Impact of Enzyme Efficiency After Salting Out (Ammonium Sulfate Fractionation): Max. efficiency was observed at 80% saturation
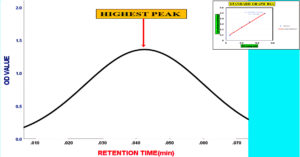
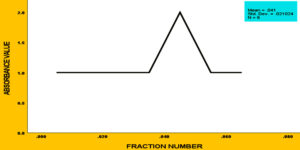
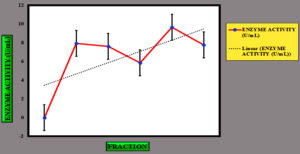
Figure 5. Elution profile of cellulase enzyme using column chromatography. The retention time is represented on the X-axis, while the OD at 660 nm is shown on the Y-axis. Analysis conducted with IBM SPSS Statistics Version 27 indicates that Fraction 4M exhibits the highest OD value, forming the peak of the Gaussian curve
Figure 6. Elution profile of cellulase enzyme using column chromatography. The X-axis corresponds to the fraction number, while the optical density (OD) at 660 nm is plotted on the Y-axis. The analysis was performed using IBM SPSS Statistics, Version 27, highlighting the peak in the cellulase enzyme’s purification curve
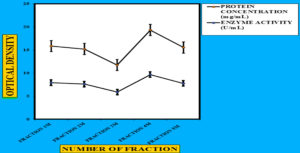
Figure 7. Separation profile of the microbial enzyme produced by Bacillus subtilis isolate on Diethylaminoethyl Cellulose-52, showing protein content (mg/mL) and cellulolytic activity (U/mL). The dual-axis graph was generated using Microsoft Excel, representing the correlation between protein elution and enzymatic activity across different fractions
Purification
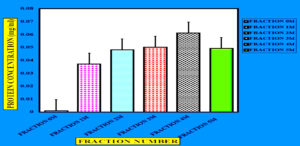
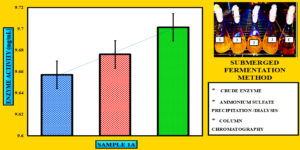
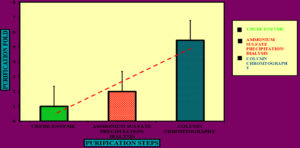
To precipitate proteins from the crude extract, solid ammonium sulfate (NH4)2SO4 was added to achieve 80% saturation (Figure 4). The catalytic activity of the purified microbial cellulase enzyme was recorded at 137.95 U/mg, a value significantly higher than cellulases produced by other Bacillus subtilis strains. For instance, Msakni et al.53 reported moderate cellulase activity following partial purification. In contrast, our study achieved a 99.54% yield with a 5.44-fold enrichment factor using a systematically optimized multi-step purification strategy. This strategy included ammonium sulfate precipitation, dialysis, and DEAE-cellulose column chromatography. During the DEAE-cellulose chromatography step, multiple fractions were collected from Sample 3 (Figure 5). Among these, Fraction 4M exhibited the highest cellulase activity, indicating successful enrichment of the active enzyme (Figure 6). A distinct peak observed at Fraction 4M represented the maximum elution of the target enzyme, confirming its successful purification and separation from impurities (Figure 7). The high recovery rate and enzyme stability achieved through this method highlight the superior efficiency of our purification process, enhancing its effectiveness compared to other methods (Figure 8). In contrast, Dikbas et al.54 reported only 20% cellulase recovery from Bacillus pumilus ND8, primarily due to significant enzyme loss during purification stages. Such losses are often attributed to denaturation or enzyme adsorption to matrix surfaces common issues in enzyme purification. However, in our study, the multi-step purification protocol minimized these problems, ensuring both high enzyme purity and stability with minimal degradation during processing (Figure 9). The purification efficiency achieved in this study surpasses the results obtained by Wijayanti et al.55 through a single-step ammonium sulfate precipitation method used for cellulase extraction from Bacillus licheniformis P12. Although their process retained 38% of enzyme activity, the enrichment factor was relatively low at 0.34, underscoring the limitations of relying solely on precipitation for enzyme purification at an industrial scale (Figure 10).55 In contrast, our multi-step chromatographic approach resulted in a 5.44-fold enrichment factor and 99.54% yield with minimal loss of enzyme activity, demonstrating a significantly more efficient and scalable purification process (Figure 11).
Figure 8. Elution profile of protein obtained through column purification. The X-axis shows the fraction number, while the Y-axis represents the protein amount (mg/mL). A distinct peak at fraction-4M indicates the maximum elution of the target enzyme, confirming its successful purification and separation from impurities, as analyzed using Microsoft Excel
Figure 9. Purification of Microbial Cellulase Enzyme: Comparative Analysis of Crude Extract, Ammonium Sulfate Precipitation, and Column Chromatography Fractions. The X-axis represents purification stages, while the Y-axis represents cellulolytic activity. Elution profile of cellulase produce by Bacillus subtilis R2A on DEAE Cellulose -52 by Microsoft Excel.4
Figure 10. Chromatographic separation of cellulase produce by Bacillus subtilis R2A on Diethylaminoethyl Cellulose -52 by Microsoft Excel
Figure 11. Comparison of purification fold at various stages of cellulase purification. The X-axis represents the various stages of purification, whereas the Y-axis indicates the purification fold. A progressive increase in purification fold was observed, reaching 5.44-fold after column chromatography.55
Molecular weight and structural characteristics
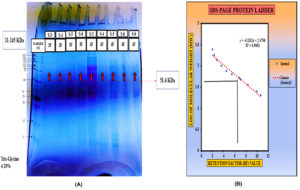
SDS-PAGE analysis revealed that the purified microbial cellulase had a molecular weight of approximately 51.4 kDa, consistent with cellulases from the GH5 family, which are known for their endoglucanase activity. SDS-PAGE was performed on the refined microbial cellulase enzyme, and its molecular weight was measured to be approximately 51.4 kDa by comparing its migration pattern to that of a protein ladder (Figure 12A). The standard curve plotting log (MW) versus relative mobility (Rf) used for molecular weight determination is shown in Figure 12B.51-56
Figure 12. (A) Purified cellulase from B. subtilis isolate was analysed using SDS-PAGE at pH 7.0 on a 12% PAGE. Subsequently, Coomassie Brilliant Blue R-250 staining. M represents the protein marker (51.4 kDa). (B) Standard curve plotting log (MW) versus relative mobility (Rf) for molecular weight determination of unknown proteins.55
This study successfully identified and characterized B. subtilis isolate as a potent cellulase-producing bacterium from various sources. The bacterium demonstrated optimal cellulase production under specific conditions (neutral pH, 37 °C, and 120 rpm), with the enzyme achieving a 5.44-fold increase and a catalytic activity of 137.95 U/mg. Validation of enzymatic activity was made via zymogram analysis. In conclusion, the relative molecular weight (MW) of the isolated microbial-derived enzyme was measured to be approximately 51.4 kDa through SDS-PAGE analysis. These results demonstrate the potential of Bacillus subtilis isolate as a valuable resource for biotechnological and industrial processes, particularly in the sustainable generation of bioenergy and other cellulose-derived bioproducts, which aligns with recent studies published by independent researchers. Future studies could focus on enhancing production efficiency and investigating its broader industrial applications to fully realize its potential.
ACKNOWLEDGMENTS
The authors sincerely thank the Director and staff of ICAR-CIRG, Makhdoom, for research facilities and guidance, and Dr. Ashok Kumar and Dr. A.K. Mishra for their support. The authors are grateful to Dr. Ashok Chauhan (President, RBEF), Dr. Aseem Chauhan (Additional President, RBEF & Chairman, AUMP), Lt. Gen. V.K. Sharma, AVSM (Retd., Pro-Chancellor, AUMP), and Prof. (Dr.) Rajesh Singh Tomar (Vice-Chancellor, AUMP) for their encouragement, Prof. (Dr.) Vinay Dwivedi, Director, Amity Institute of Biotechnology, AUMP for continuous guidance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the Madhya Pradesh Council of Science and Technology (MPCST) – an autonomous organization of Govt. of M.P., vide Endorsement No. 886/CST/FTYS/2024.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Sakon J, Irwin D, Wilson DB, Karplus PA. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4(10):810-818
Crossref - Cavaco-Paulo A. Mechanism of cellulase action in textile processes. Carbohydr Polym. 1998;37(3):273-277
Crossref - Bhat M. Cellulases and related enzymes in biotechnology. Biotechnol Adv. 2000;18(5):355-383
Crossref - Chandara SK, Snishamol C, Prabhu NG. Cellulase production by native bacteria using water hyacinth as substrate under solid state fermentation: Malaysian. J Microbiol. 2005;1:25-29
Crossref - Cho KM, Hong SJ, Math RK, et al. Cloning of two cellulase genes from endophytic Paenibacillus polymyxa GS01 and comparison with cel 44C man 26A. J Basic Microbiol. 2008;48(6):464-472
Crossref - Doi RH. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann New York Acad Sci. 2008;1125(1):267-279
Crossref - Immanuel G, Dhanusha R, Prema P, Palavesam A. Effect of different growth parameters on endoglucanase enzyme activity by bacteria isolated from coir retting effluents of estuarine environment. Int J Environ Sci Technol. 2006;3(1):25-34
Crossref - Ibrahim AS, El-diwany AI. Isolation and identification of new cellulases producing thermophilic bacteria from an Egyptian hot spring and some properties of the crude enzyme. Aust J Basic Appl Sci. 2007;1(4):473-478.
- Alokika, Kumar V, Singh B. Biochemical characteristics of a novel ethanol-tolerant xylanase from Bacillus subtilis subsp. subtilis JJBS250 and its applicability in saccharification of rice straw. Biomass Conv Bioref. 2023;13(1):1937-1949
Crossref - Reverbel-Leroy C, Pages S, Belaich A, Belaich JP, Tardif C. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J Bacteriol. 1997;179(1):46-52
Crossref - Nakamura K, Kitamura K. Isolation and identification of crystalline cellulose hydrolyzing bacterium and its enzymatic properties. Journal of Fermentation Technology. 1982;60(4):343-348.
- Gyalai-Korpos M, Nagy G, Mareczky Z, Schuster A, Reczey K, Schmoll M. Relevance of the light signaling machinery for cellulase expression in Trichoderma reesei (Hypocrea jecorina). BMC Res Notes. 2010;3:1-0
Crossref - Creuzet N, Berenger JF, Frixon C. Characterization of exoglucanase and synergistic hydrolysis of cellulose in Clostridium stercorarium. FEMS Microbiol Lett. 1983;20(3):347-350
Crossref - Ogawa K, Toyama D, Fujii N. Microcrystalline cellulose-hydrolyzing cellulase (endo-cellulase) from Trichoderma reesei CDU-11. J Gen Appl Microbiol. 1991;37(3):249-259
Crossref - Beguin P, Aubert JP. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13(1):25-58
Crossref - Bhat MK, Bhat S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv. 1997;15(3-4):583-620
Crossref - Zhu L, Guan L, Wang K, et al. Recent trends in extraction, purification, structural characterization, and biological activities evaluation of Perilla frutescens (L.) Britton polysaccharide. Front Nutr. 2024;11:1359813
Crossref - El-Shora HM, Abo-Elmaaty SA, El-Sayyad GS, Al-Bishri WM, El-Batal AI, Hassan MG. Immobilization of purified pectinase from Aspergillus nidulans on chitosan and alginate beads for biotechnological applications. Microb Cell Fact. 2025;24(1):5
Crossref - Sharma V, Nargotra P, Sharma S, et al. Purification and biochemical characterization of an ionic liquid tolerant cellulase from Aspergillus assiutensis VS34 for potential biomass conversion applications. Environmental Sustainability. 2024;7(3):325-338
Crossref - Thakur N, Nath AK, Sharma A. Optimization of production conditions, isolation, purification, and characterization of tannase from filamentous fungi. Folia Microbiologica. 2024 Oct;69(5):1123-1135
Crossref - Tran DM, Nguyen TH, Huynh TU, Pentekhina I. Recombinant expression and characterization of the family 5 cellulase from Bacillus velezensis in Escherichia coli BL21-CodonPlus (DE3)-RIPL. Biochem Biophys Rep. 2025;41:101898
Crossref - Pérez-Contreras S, Avalos-de la Cruz DA, Lizardi-Jiménez MA, Herrera-Corredor JA, Baltazar-Bernal O, Hernández-Martínez R. Production of ligninolytic and cellulolytic fungal enzymes for agro-industrial waste valorization: Trends and applicability. Catalysts. 2024;15(1):30.
Crossref - Adab FK, Yaghoobi MM, Gharechahi J. Enhanced crystalline cellulose degradation by a novel metagenome-derived cellulase enzyme. Sci Rep. 2024;14(1):8560
Crossref - Delgado-Garcia M, Campos-Muzquiz LG, Castillo-Godina RG, et al. Xylanases: Sources, Production, and Purification Strategies. In: Chowdhary P, Yadav D, Anand G, Gaur RK, eds. Microbial Enzymes: Production, Purification and Industrial Applications. Wiley 2024;1:1-29
Crossref - Moodley D, Botes A. A carboxymethyl cellulase from the yeast Cryptococcus gattii WM276: Expression, purification and characterisation. Protein Expr Purif. 2025;225:106594
Crossref - Ugras S, Bicen HE, Emire Z. Determination of Cellulase enzyme produced by Bacillus cereus DU-1 Isolated from Soil, and its effects on Cotton Fiber. Braz Arch Biol Technol. 2024;67:e24230391.
Crossref - Zhang W, Yu X, Xin L, Xu S. Production of Aspergillus niger cellulase using defatted rice bran and its use in lignocellulosic saccharification reaction. J Environ Chem Eng. 2025;13(1):115003
Crossref - Doan CT, Tran TN, Pham TP, et al. Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii. Polymers. 2024;16(14):2037
Crossref - Vijayaraghavan P, Vincent SGP. Purification and characterization of carboxymethyl cellulase from Bacillus sp. isolated from a paddy field. Pol J Microbiol. 2012;61(1):51-55
Crossref - Bhatt B, Bhatt K, Lal S, Bhatt V. Production of a novel cellulase by Bacillus amyloliquefaciens OKB3 isolated from soil: Purification and characterization. International Journal of Biological Macromolecules. 2024;282:137454.
Crossref - Ji R, Wang Z, Kuang H. Extraction, purification, structural characterization, and biological activity of polysaccharides from Schisandra chinensis: A review. Int J Biol Macromol. 2024;271(Part 1):132590
Crossref - Rajesh R, Gummadi SN. Purification and biochemical characterization of novel α-amylase and cellulase from Bacillus sp. PM06. Prep Biochem Biotechnol. 2024;54(6):796-808
Crossref - Neog PR, Saini S, Konwar BK. Purification, and characterization of detergent-compatible serine protease from Bacillus safensis strain PRN1: A sustainable alternative to hazardous chemicals in detergent industry. Protein Expr Purif. 2024;219:106479
Crossref - Shanmugapriya K, Saravana PS, Krishnapriya, Manoharan M, Mythili A, Joseph S. Isolation, screening and partial purification of cellulase from cellulase producing bacteria. Int J Adv Biotechnol Res. 2012;3(1):509-514.
- Rawat R, Tewari L. Purification and characterization of an acidothermophilic cellulase enzyme produced by Bacillus subtilis strain LFS3. Extremophiles. 2012;16:637-644
Crossref - Regmi S, Choi YS, Kim YK, et al. Endoglucanase produced by Bacillus subtilis strain CBS31: Biochemical characterization, thermodynamic study, enzymatic hydrolysis, and bio-industrial applications. Biotechnol Bioprocess Eng. 2020;25:104-116
Crossref - Demissie MS, Legesse NH, Tesema AA. Isolation and characterization of cellulase producing bacteria from forest, cow dung, Dashen brewery and agro-industrial waste. Plos One. 2024;19(4):e0301607
Crossref - Aneja KR (eds.). Experiments in microbiology, plant pathology and biotechnology. Reprint edition. New Age International; 2015.
- Asha BM, Sakthivel N. Production, purification and characterization of a new cellulase from Bacillus subtilis that exhibit halophilic, alkalophilic and solvent-tolerant properties. Ann Microbiol. 2014;64:1839-1848
Crossref - Barr BK, Hsieh YL, Ganem B, Wilson DB. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35(2):586-592
Crossref - Ibrahim AN, Ahmed FH, Ibrahim MMK, Arafa MAI. Precepitation and purification of amylase enzyme produced by Streptomyces aureofaciens. Arch Pharm Res. 1990;13:28-32
Crossref - Rahman UU, Qasim S, Zada NS, et al. Evaluation of agricultural wastes as a sustainable carbon source for the production of β-glucosidase from Bacillus stercoris, its purification and characterization. Pak J Agric Sci. 2023;60(2):367-375
Crossref - Badoni V, Rana GS, Dubey A, Verma AK. b Glucosidase Production and Its Applications. Microbial Enzymes: Production, Purification and Industrial Applications. 2025;2:437-476
Crossref - Battisti JA, Rocha GB, Rasbold LM, et al. Purification, biochemical characterization, and biotechnological applications of a multifunctional enzyme from the Thermoascus aurantiacus PI3S3 strain. Sci Rep. 2024;14(1):5037
Crossref - Wibowo NA, Fatimah S. Morphological and biochemical characterization of cellulase bacterial from bearcat. AIP Conf Proc. 2024;2957
Crossref - Ezima EN, Adegbesan BO, Osonuga IO, et al. Purification and characterization of cellulase from May beetle (Phyllophaga errans) gut. Asian J Trop Biotechnol. 2024;21(1)
Crossref - Parmar TS, Ahirwar R, Sahay S. Isolation, purification and characterization of carboxymethyl cellulase (CMCase) from psychrotolerant yeast Rhodotorula mucilaginosa BPT1. Mater Today: Proc. 2023;72(5):2768-2772
Crossref - Saha S, Roy RN, Sen SK, Ray AK. Characterization of cellulase producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac Res. 2006;37(4):380-388
Crossref - Cherry JR, Fidantsef AL. Directed evolution of industrial enzymes: an update. Curr Opin Biotechnol. 2003;14(4):438-443
Crossref - Deka D, Bhargavi P, Sharma A, Goyal D, Jawed M, Goyal A. Enhancement of cellulase activity from a new strain of Bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enzyme Res. 2011; 2011(1):151656
Crossref - Bangoria P, Patel A, Shah AR. Thermotolerant and protease-resistant GH5 family β-mannanase with CBM1 from Penicillium aculeatum APS1: purification and characterization. 3 Biotech. 2023;13(3):107
Crossref - Afe AE, Lawal OT, Oyelere BR, Bamidele OS, Sanni DM. Optimization, Isolation, Purification, and Characterization of a Thermally Stable, Acidophilic Cellulase from Aspergillus awamori AFE1 for Industrial and Biotechnological Applications. Res Sq. 2023
Crossref - Msakni S, Demirkan E, Tanik NA. Production, optimization, partial purification, and characterization of a novel cellulase from Bacillus subtilis 171ES and its potential for use in textiles. J Sci Ind Res. 2024;83(6):677-687
Crossref - Dikbas N, Al Dahluz WSS, Alm S, Ucar S. Partial Purification and Biochemical Characterization of Cellulase from Bacillus pumilus ND8 Isolated from Garden Waste. Turkish Journal of Nature and Science. 2024;13(3):62-66
Crossref - Wijayanti SD, Oliviani K, Kusnadi J, Putri RA. Characterization of crude cellulase enzyme produced by Bacillus licheniformis P12 isolate. IOP Con Ser.: Earth Environ Sci. 2020;475(1):012085
Crossref - Ameen F. Purification and characterization of xylanase produced by Aspergillus fumigatus isolated from the northern border region of Saudi Arabia. Fermentation. 2023;9(7):595
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.