Nosocomial or hospital acquired infections (NI or HAI) pose significant danger to immunocompromised individuals. Many pathogens involved in NI evade early detection during the initial testing since they are misdiagnosed as contaminants and are also resistant to various first-line antibiotics. Few of the common pathogens that cause NIs or HAIs include various strains of E. coli, Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella sp., etc. Kocuria sp. previously considered to be an opportunistic pathogen, has recently been identified as the causative agent for a variety of NIs affecting neonates and immunocompromised; cholecystitis, peritonitis, endocarditis, etc. being some of the symptoms caused. Hence, this genus has received more research attention to devise novel strategies for diagnosis and effective treatments of infections. The present review focuses on various features of the genus Kocuria, different types of infections caused, case studies highlighting various promising treatment strategies and the antimicrobial resistance shown by the members of this genus.
Nosocomial, Kocuria, Antibiotic Resistance, Catheter-related Infections, Horizontal Gene Transfer, Biofilm
Nosocomial or hospital acquired infections (NI or HAI) are those which occur in a patient within 48-72 hours after admission in a hospital, but were not present at the time of entry. Such infections often arise because a patient’s immune system is weakened while fighting pre-existing illnesses. The most commonly encountered NI affects the respiratory, urinary, gastrointestinal and circulatory tracts and also surgical sites.1 The main pathogens which cause NIs include various strains of Klebsiella species, Staphylococcus aureus, E. coli, Pseudomonas aeruginosa, etc. These are notorious as most of them are resistant to many of the commonly used antibiotics.2 In a global study on the prevalence of NIs in various countries, it was found that they are on the rise, with a global annual rate of increase of 0.06%.3 Kocuria belongs to a bacterial genus which has been observed to cause more NIs recently. They are considered potential pathogens affecting neonates, immunocompromised and those receiving urgent medical attention.4 The commonly observed symptoms include peritoneal dialysis associated infections, meningitis, skin infections, catheter-associated bacteremia, brain abscess, canaliculitis, cholecystitis, keratitis, peritonitis, endophthalmitis, endocarditis and necrotizing mediastinitis.5,6 Once considered merely opportunistic pathogens, Kocuria species are now known to cause a range of serious infections. This review discusses the different Kocuria species, their associated infection types, and the current strategies for disease management.
Nosocomial infections are the leading cause of preventable harm to hospital patients and pose a huge drain on healthcare resources. They are typically caused by several antibiotic-resistant bacteria, and the management of nosocomial infections many a times promotes the development of resistant bacteria. Understanding the intricate interplay of factors that contribute to nosocomial infection is the first step in improving patient outcomes. Most of the Kocuria infections reported were from patients who were affected by severe underlying illnesses, such as solid tumors, catheter-related bacteraemia, hematologic malignancies, or metabolic disorders.7,8 Though many species have been reported in this genus, 5 of them have been found to be opportunistic pathogens.5 Future research must prioritize understanding Kocuria infections, given the lack of significant knowledge gaps in epidemiology.9
Features of members of genus Kocuria
The word Kocuria is adopted from the name of a Slovak microbiologist, Miroslav Kocúr. Kocuria spp. are coccoid Actinobacteria that are Gram-positive, coagulase-negative, catalase-positive, belonging to the family Micrococcaceae, order Micrococcales and class Actinomycetes.5 They are found to be organized as irregular clusters, in pairs, short chains and tetrads.5 To date, 26 species of Kocuria have been described. Among these, K. rosea is a significant representative and K. coralli is the most recently discovered.10 Initially, this bacteria was classified as Micrococcus, but taxonomic changes were introduced in accordance with 16S rDNA sequences and composition of amino acids in peptidoglycan, following phylogenetic analysis.5,11 They are coagulase-negative and catalase-positive in nature, often wrongly identified as coagulase-negative Staphylococci, which might cause misjudgment of the infections caused in patients.12,13 On nutrient agar, Kocuria rosea has pink colonies which are smooth and circular. It is aerobic, non-spore-forming, catalase-positive and oxidase-negative. Also, in blood agar it forms non-hemolytic colonies and is non-capsulated, non-motile, non-spore forming and gives a positive reaction for Voges-Proskauer test and non-acid fast test. Various Kocuria species reacts differently to biochemical tests including nitrate reduction, gelatinase, citrate, urease, amylase, phosphate, oxidase, arabinose, and N-acetyl-L-glutamic acid tests.5 Kocuria spp. form round and large colonies with diffusible golden pigmentation on tryptic soy agar (TSA). Menaquinones MK-7(H2) and MK-8(H2): lysine-based A3a-type peptidoglycan, and saturated branched fatty acids are the predominant components that are found in the cell envelopes of Kocuria.11
The close relationship between Micrococcus and Arthrobacter species was noted before the results of 16S rDNA sequence analyses became available. In 1974 it was pointed out that “Micrococcus species should be regarded as degenerate forms, locked into the coccoid stage of the Arthrobacter life cycle”. The scientist Stackebrandt et al.5 proposed the genus Kocuria, which has 26 species discovered till now. Many members of Kocuria species show differences in their biochemical tests and hence identification may not be accurate in many instances. More advanced techniques like 16S rRNA and MALDI-TOF MS could give more precise taxonomic identification of the members of this genus,14 though many times, taxonomic reframing of species of Kocuria has been observed.15
Only three of these known species viz. K. kristinae, K. rosea, and K. rhizophila, have been recognized to be clinically important infectious agents. All three species are connected to catheter-related bacteremia.16-18 Some of the important species coming under the genus Kocuria, with their characteristic features are listed in Table 1.
Table (1):
Different species of Kocuria and their characteristic features
Species of Kocuria |
Characteristic features |
|---|---|
K. rosea |
Spherical cells, circular, pink or red and smooth (occasionally rough): obligate aerobe, mesophile or psychrophile based on strain.5 |
K. aegyptia |
Pink, circular, opaque colonies, non-motile coccoid cells.19 |
K. carniphila |
Coccoid cells, non-acid-fast, non-motile, opaque, colonies of circular shape.20 |
K. rhizophila |
Non-endospore-forming, appear as packets measuring radius of 0.5 to 0.75 micrometre, non-acid-fast.21 |
K. palustris |
Light-yellowish pigmented colonies. Spherical cells, 0.5 to 0.75 micrometre radii, non-endospore forming, non-acid-fast.21 |
K. kristinae |
Spherical cells (radius 0.35-0.55 µm), colonies seen includes smooth or rough. |
K. flava |
Gram-positive, yellowish colonies, distinguished from K. turfanensis by ~6 mol% difference in DNA G+C values.22 |
K. polaris |
Isolated from Antarctic cyanobacterial mat, orange colonies, cells are of 0.5-0.75 µm in radius and are coccoid cells.23 |
K. himachalensis |
Circular, reddish orange colonies with 0.5-0.75 µm in radius.24 |
K. turfanensis |
Gram-positive, yellowish to orange colonies.22 |
K. koreensis |
Light orangish colonies which are opaque. Cells are coccoid with a radius of 0.5-0.75 µm.25 |
K. gwangalliensis |
Aerobic, non-motile, non-endospore forming, pink–orange pigmented colonies, spherical 0.3-0.6 µm in radius.26 |
K. atrinae |
Light yellowish colonies, coccoid cells with a radius of 0.5-0.75 micrometer.25 |
K. halotolerans |
Light yellowish colonies, cells are coccoid with 0.3-0.5 µm in radius.27 |
K. dechangensis |
Beige-yellow colonies, Gram-positive & coccoid cells (0.4-0.65 µm in radius).28 |
K. indica |
Gram-positive, lemon-yellow colonies, aerobic, non-motile coccoid.29 |
K. salsicia |
Lemon-yellow colonies, opaque, circular coccoid cells, measuring 0.5-0.75 µm in radius.30 |
K. marina |
Halotolerant cells- tolerates upto 16% NaCl, non-motile, aerobic, coccoid cells.31 |
K. varians |
Recorded from milk, spherical cells (radius of 0.45-0.75 µm); yellowish colonies, glistening smooth colonies.5 |
K. arsenatis |
Arsenic resistant endophyte, aerobic, cells measure 0.1-0.3 µm in radius, non-endospore producing. Opaque, circular, smooth yellow colonies.19 |
K. tytonis |
Non-lipophilic, non-motile, elevation is pulvinate, 0.5 mm radius, dry, non-viscous, non-sporulating cocci.32 |
K. pelopila |
Mangrove-rhizosphere isolate, Gram-positive, spherical, non-motile, aerobic, measuring approximately 0.5 µm in radius, circular colonies.33 |
K. subflava |
Marine sediment isolate, Gram-positive, catalase positive, oxidase negative, aerobic, non-motile cocci, 0.25-0.45 mm in radius.34 |
K. oceani |
Coccoid, circular, convex, pinkish orange.35 |
K. salina |
Gram-positive, non-motile, oxidase-negative, pastel orange colored colonies, catalase positive, starch hydrolysis positive.36 |
K. coralli |
Non-motile, aerobic. Spherical cells with 0.35-0.45 µm in radius, circular colonies, polar lipids include cardiolipin, phosphatidylglycerol.10 |
Pathogenic species of Kocuria and disease symptoms
Scientists describe members belonging to the genus Kocuria as saprophytes since they are commonly found in soil, water, and the atmosphere.5 Some of them are also found in the human skin epidermis and oral flora.15,21,37 In 1974, it was initially recognized as an origin of infection of the urinary tract and the name Micrococcus kristinae was given.20 Kocuria spp. have been found in the oral cavity, but their pathogenicity is diminished, since, when extracted from clinical specimens, they are usually handled as lab contaminants and given little consideration. The late twentieth century saw a rise in cases of diseases caused by Kocuria species. Identification of phenotypes may not be able to distinguish between every member of the species due to the variability of the results of carbon assimilation and biochemical tests in different Kocuria species.38 Some of the important pathogenic species of the genus Kocuria and their clinical manifestations are listed in Table 2.
Table (2):
Important pathogenic species of the genus Kocuria and their clinical manifestations
Name of the pathogen |
Disease caused |
Manifestation |
Symptoms |
|---|---|---|---|
Kocuria kristinae |
Cholecystitis |
Inflammation of the gallbladder |
Pain in the right upper abdomen and right shoulder, nausea, vomiting, fever.18 |
Kocuria rhizophila |
Dacryocystitis |
Infection of the lacrimal sac |
Pain, redness, and swelling in the inner corner of the eye.39 |
Kocuria marina |
Peritonitis |
Infection of peritoneum |
Abdominal pain, turbid effluent, nausea, mild fever.40 |
Kocuria rosea |
Catheter associated bacteremia |
Bacteremia originating from an intravenous catheter |
Tachycardia, elevated fever and rate of respiration.41 |
Kocuria koreensis |
Infectious keratitis |
Conjunctival allergic disease with atopic facial dermatitis |
Eye redness, blurred vision, eye pain.42 |
The most common symptoms of Kocuria infection include sepsis and enhanced levels of platelets, leucocytes and CRP.7 Despite the lack of knowledge on the epidemiology and pathogenicity qualities, it has been suggested that biofilms have a role in adhesion, colony formation and infection.43 Although K. rosea is a very uncommon human pathogen and a commensal of the epidermis and oropharynx, its significance in infection may be underappreciated due to probable misidentification and the perception of micrococci as contaminants.38 These bacteria cause multiple kinds of infections, most of them occurring in immunocompromised hosts with serious underlying diseases.5,8
The usage of N-acetyl-L-glutamic acid, inulin, urease, oxidase, amylase, phosphatase and nitrate reduction tests, among other standard biochemical tests, have all been found to cause distinct reactions in different species of Kocuria.8 This may be the cause of the incorrect identification provided by automated and conventional technologies for bacterial identification. But previous studies have shown that Kocuria can still be distinguished from Micrococcus and Staphylococcus with morphology, culture identification and antibiotic discs in the absence of molecular and diagnostic approaches. Kocuria spp. are resistant to nitrofurantoin and lysostaphin but sensitive for bacitracin and lysozyme.44,45
As an opportunistic pathogen, K. rosea was found in many infections of urinary tract, endocarditis and in people receiving peritoneal dialysis, bacteremia, as well as disease related to medical equipment in immunocompetent patients and those with underlying conditions.18,46,47 K. kristinae has also been linked to acute cholecystitis.18 K. varians, another species, was found in a patient with a brain oedema.48 K. kristinae has been linked to various intravascular diseases in immunocompromised hosts, an illness aggravated by septic pulmonary emboli caused by suppurative thrombosis. In this case, a catheter-related BSI that resulted in such problems was notable.7
Nosocomial infections – case studies
The importance of Kocuria in transmitting illnesses picked up in hospitals has been addressed.49 The same study also stated that Kocuria spp. should be regarded as possible pathogens in immunocompromised patients, those receiving critical care treatment, and newborns, even though they coexist with people, animals, and the environment. More than 50% of patients in a research involving twelve adolescents who had underlying, severe illnesses such as cancer and were born from preterm delivery developed invasive infections with Kocuria spp.50 There is an increasing trend of reported illnesses brought on by Kocuria spp. in previously healthy and immunocompetent people.
A 58 year-old woman with descending necrotizing mediastinitis, who was also taking medication for hypertension and gout, was found to have K. rosea.49 A 10 year-old female patient developed endocarditis from K. rosea despite being healthy, although she had undergone surgery to treat congenital heart problems.51 The strain was collected from a case of peritonitis and tested negative for biofilm formation by Kocuria species.44 The isolation of K. marina in a 7 year-old patient on epoprostenol therapy, tolerant to highly alkaline conditions, serves as a warning about the potential of Kocuria spp. for opportunistic and nosocomial infections.46,52
In a study conducted from June 2019 to June 2021, 7 out of 261 bacteremia positive cases contained Kocuria isolates. The study was conducted on pediatric patients between the age of 4 months to 10 years. These patients either had urinary tract related diseases or symptomatic bacteraemia, showing fever within 24 hours of hospitalization. Five cases of K. kristinae, one case of K. rhizophila, and one case of K. rosea were detected. These species exhibited 100% resistance to ceftazidime, gentamicin, and amikacin. The low frequency of Kocuria species isolations may possibly be caused by the fact that these species are members of a class of bacteria that are usually overlooked while diagnosing. Additionally, it might be underrated, given how much they resemble coagulase-negative Staphylococci.53 These results are in line with previous research conducted on K. kristinae urinary tract infections.54-56
A 55 year-old African American woman with sickle cell pain episodes in her arms, legs, and chest was later diagnosed with a nosocomial infection by K. rosea.57 Standard anti-staphylococcal drugs were ineffective because of the resistance of Kocuria to lysostaphin and nitrofurantoin. The infection, which began as a catheter-associated case, progressed to bacteremia.
K. varians induced central venous catheter (CVC) infection could also be successfully treated with CVC salvage.58
In a study of 12 pediatric patients with Kocuria infection in blood, one child had acute leukemia and six were premature infants, all of whom had central venous catheters. Fever, hypothermia, apnea, or bradycardia were the symptoms; there was no other evident illness and sepsis treatment was given.59,60 Such cases call for the need to correctly identify the bacteria to prevent such cases not just in patients with impaired immune systems, but also in healthy people.61
Successful treatment strategies for Kocuria infections
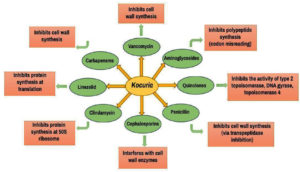
Given that the infections caused by Kocuria species are rare, clinicians may misinterpret and classify them as culture contaminants and hence due care has to be taken in patients with medical implants.9,62 Even in people with known risk factors, infections by Kocuria species are uncommon, yet they can occur in patients without these characteristics as well.62 The resistance to nitrofurantoin/furazolidone displayed by Kocuria species is one of the main requirements for first phenotypic identification. Susceptibility to beta-lactams, quinolones, lincosamides and cotrimoxazole and resistance to kanamycin (among the aminoglycosides) has been observed. Few of the prominent antibiotics used against Kocuria and their modes of action are depicted in Figure. Reports of resistance to fusidic acid, rifampicin, linezolid, or streptogramins have not been reported yet. It is known that Gram-positive bacteria inherit resistance to colistin and polymyxin; however, Kocuria species vulnerable to polymyxin are unusual.38 To assess the susceptibility profile of the Gram-positive Kocuria species, a few case studies have been conducted.
Figure. Mode of action of antibiotics commonly used to treat infections caused by pathogenic species of Kocuria
In a study of 20 isolates of K. kristinae, four patients (3 with catheter-related bacteremia and 1 with infective endocarditis) had implanted catheters. Removal of these catheters helped in reducing the infection. Four K. kristinae isolates had MICs up to 4 mg/L for Oxacillin and hence were deemed resistant. One isolate of K. marina was also identified in this study.63
In a 31 year-old patient who received complete parenteral feeding due to propionic acidemia (via the central venous catheter, CVC) and on hemodialysis, K. kristinae was identified in the blood culture. In a situation of uncomplicated CRBSI (Catheter-related bloodstream infection): combination treatment and 10-14 days of antibiotic delivery through the colonized catheter was advised, along with antibiotic lock therapy as advised by the IDSA (Infectious Diseases Society of America) as per the revised guidelines issued in 2011. This was the first successful treatment to preserve the catheter.64
A 52 year-old patient with comorbidities was infected with K. varians causing brain abscess. After receiving third-generation cephalosporins intravenously for four weeks, the patient was transitioned to taking them orally for two weeks. While erythromycin, amikacin, amoxicillin, ceftriaxone, and cefuroxime were reported to be effective against the majority of Kocuria and Micrococcus strains, third-generation cephalosporins were utilized due to breach of blood-brain-barrier and penetration of the abscess capsule.65 In a similar case of brain abscess caused by K. rosea, antibiotic therapy with cefepime was administered for four weeks, gradually improving mental capabilities and resulting in a normal neurological evaluation at release. A month later, lesions were no longer present in the brain MRI.62
In the case of Kocuria ocularis which caused dacrocystitis infection in a patient aged 74 years, the patient was given 1 g amoxicillin and 125 mg clavulanic acid via oral route twice a day combined with topical application of tobramycin dexamethasone ophthalmic ointment (Alcon) and it was observed that the patient’s symptoms promptly decreased and did not reappear 15 months post surgery. By disc diffusion analysis, the isolate was found to be sensitive to a wide range of antimicrobials (like benzylpenicillin, oxacillin, cefoxitin, moxalactam, novobiocin, linezolid, erythromycin, lincomycin, pristinamycin, rifampicin, ofloxacin, vancomycin, teicoplanin, kanamycin, gentamicin, tobramycin, rifampicin, tetracycline, fosfomycin, cotrimoxazole, and fusidic acid) and antibiotics like beta-lactamase (breakpoints of Staphylococcus spp. were used).66
The first instance of K. rhizophila causing bacteremia with infective endocarditis occurred in 2021, recorded in a 81 year-old patient with comorbidities. The strain was sensitive to Cefoxitine, kanamycin, clindamycin, and trimethoprim/sulfamethoxazole but was resistant to erythromycin and norfloxacin (according to the sixth edition of EUCAST-European Committee for Antimicrobial Susceptibility Testing, staphylococcus-species interpretation criteria for the agar disk diffusion assay). Third-generation cephalosporins were administered to the patient in compliance with the EUCAST section “PK/PD (non-species related) breakpoints”, sensitivity profile, and categorization rearrangement in order to prevent clinical failure. Currently, Kocuria has no established treatment plans or standards for antibiotic susceptibility and hence most often Staphylococcus breakpoints are used in most publications.9
A recent case was reported on total hip replacement surgery performed on a 74 year old male patient due to pain on the right side of hip. After 6 weeks of surgery he had symptoms like erythema, swelling, tenderness over the wound and problem with weight-bearing. On examination, it was found that debridement and implant retention had to be performed. Further, the patient had to be put on intravenous Vancomycin and Piperacillin-Tazobactam. The culture test revealed 1 out of 5 samples isolated from lesser trochanter as K. rhizophila and the patient was put on IV Vancomycin for a period of 12 weeks.67
In Nigeria during the COVID-19 pandemic, a 72 year old male presented with symptoms of COVID-19 like cough, loss of taste, sore throat, fever and furthermore respiratory distress and urinary incontinence was later diagnosed with the presence of S. aureus and Kocuria species (which was earlier misidentified for coagulase-negative Staphylococci) showing 97.87% match to the DNA sequence of K. rosea. Both the strains showed high resistance to multiple drugs upon subjecting them to antibiotic susceptibility testing. After 5 days, the patient died while the treatment was ongoing due to certain complications.68
A 74 year-old man with stomach pain and cloudy peritoneal effluent who has been undergoing peritoneal dialysis for 32 months (previously diagnosed with anuric kidney failure) was found to be infected with K. salsicia, which was reported to be sensitive to aminoglycosides, trimethoprim-sulfamethoxazole, erythromycin, clindamycin, linezolid, and glycopeptides but resistant to penicillin, oxacillin, and quinolones. Initially he was treated with intraperitoneal vancomycin (2 g dose given once) and ceftazidime (1 g dose given everyday at overnight exchange) alternatively switching with dialysis 4 times each day. Later due to the unresponsive nature of the infection, the prescribed course of antibiotics was adjusted to include 600 mg of rifampicin taken orally every day, 350 mg of daptomycin, and 50 mg of tobramycin exchanged overnight. No improvement was observed which could possibly be due to the formation of biofilm on the surface of the catheter by the bacteria, the type of strains involved or the type of antibiotics used for the treatment.69
Addressing a prospective one-year observational research conducted at a hospital from January 2021-December 2021, samples (10 CSF, pus, peritoneal fluid, extraventricular drainage tip, and BAL) were collected from 14 patients. On examination and confirmation by Vitek 2 and MALDI-TOF, 6 isolates were identified as K. kristinae and 6 as K. rosea and 2 as K. rhizophila. Kocuria was not isolated from any of the adult patients throughout the observation period; the antibiograms of these 3 Kocuria species also were varied. Given that there are no established guidelines for Kocuria in CLSI, utilizing the disc diffusion method, antibiotic susceptibility testing was carried out, and zone interpretation was predicated on Staphylococcus.70
Another case of single-center retrospective analysis in China, which is considered as the largest case series, a range of diseases in children brought about by the Kocuria species was examined. Out of the 36 patients, 29 people contracted the infection from K. kristinae, 4 by K. rosea, 2 by K. varians and 1 by K. rhizophila (bloodstream infection was identified in a total of 26 patients, 6 with pneumonia related to ventilator use, and 1 with urinary tract infection, purulent meningitis, cholangitis and empyema related to catheter use). While most of these patients were immunocompromised, some were immunocompetent; the study set included 4 children too, with early onset before the age of one. While all Kocuria species exhibited resistance to oxacillin and penicillin, they were all vulnerable to linezolid, vancomycin, and tigecycline. Majority of the cases were resolved using suitable antimicrobial medications.71
Vancomycin (47%):cephalosporins (39.5%):quinolones (36.6%):linezolid (17%):aminoglycosides (14%):penicillin (7.9%):aminopenicillin (6.9%):clindamycin (5.9%):carbapenems (5%): and daptomycin (2%): were the most often used antimicrobials for treating Kocuria infections. In 36.6% of cases, surgery was combined with antimicrobial therapy. The overall death rate was 5.9%, of which 4.9% was directly related to the infection.72 Thus, the treatment strategy to cure Kocuria infections depends on a number of factors like accurate and timely diagnosis, resistance profile of the strain , age of patient, comorbidities, days of infection etc.
Antimicrobial resistance, resistance genes and possible mechanisms
Initially members of Kocuria species were considered non-pathogenic but recently there has been an increase in the infections with symptoms like endocarditis, peritonitis, meningitis, osteomyelitis, and brain abscess.65 Due to the limited number of cases reported for infections caused by Kocuria, there are no particular therapeutic treatments and administrative measures for this pathogen. At the same time, they show resistance to antimicrobials like ampicillin and erythromycin.38 Antimicrobial resistance in bacteria poses a huge global problem. Our ability to treat infectious diseases and make improvements in health and medicine is impaired due to the antimicrobial resistance.5 To avoid the transmission of antimicrobial-resistant bacterial strains, it is important to use antimicrobials more appropriately and wisely.13
A French study demonstrated the ability of Kocuria spp. to grow even after simultaneous administration of penicillin and oxacillin. Kocuria spp. was found to be resistant to erythromycin to which it was earlier sensitive.73 Kocuria genus was found to possess 5 types of ARGs (antibiotic resistance gene) including fosmidomycin (rosA):bacitracin (bacA):quinolone (qepA):MLS (macB) and multidrug (mdtF, mexF and oprC).74 A study conducted on pasteurized milk in Brazil, showed multidrug resistance (MDR) of more than half of the isolates of Kocuria. The major resistance was found to be against tetracycline, penicillin and clindamycin.75
K. rosea resides on our skin as a normal flora but it is noticed that this gram positive bacteria is capable of causing blood infections associated with the central line in patients who are catheterized and immunocompromised.61,76 K. rosea demonstrated resistance to various classes of antibiotics like macrolides, cephalosporin, fluoroquinolones, ciprofloxacin and ceftriaxone.76 In a study to detect mecA gene in different species of bacteria by PCR, an isolate of K. rosea was found with the mecA gene, which was previously not reported. On consecutive analysis, it was concluded and confirmed by the NCBI database that the mecA gene in K. rosea was a result of horizontal gene transfer from S. aureus.77 In a study, from catheter related bacteraemia, though, K. rosea was found to be sensitive to vancomycin in vitro, no changes in the condition of the patient was noticed after drug administration until the removal of the catheter. According to a hypothesis, the bacteria gained protection against the antibiotics by the formation of biofilm on the device.41 The index of adhesiveness of Kocuria spp. is high, which explains their function in the first stage of biofilm development.78 The microorganism organizes itself in a three dimensional arrangement with high synergism which helps withstand external hindrances like harsh environmental factors, immune system factors and antimicrobial agents, all these being traits conferred to biofilm forming bacteria.
Isolates of K. kristinae, which is another species of Kocuria show frequent resistance to penicillins, gentamicin and erythromycin.79
A recent in silico study identified presence of many antibiotic resistance genes in 5 strains of Kocuria.80
K. kristinae were found to be associated with invasive infections in very young children and in patients suffering from malignancies or who are immunocompromised.81 For the isolate, K. kristinae_LC, seven genes were implicated in antibiotic resistance. Furthermore, this isolate also showed resistance to bacitracin. Gene_1280 was identified in the PHI database which produces multidrug-resistance proteins and provides antimicrobial resistance to the bacteria; four genes that encode prevent-host-death proteins were revealed after genome analysis. Overexpression of these genes could have contributed to antibiotic resistance and increased rate of biofilm formation.82
Isolates of K. varians were collected in Spain from cheese samples made out of raw milk. Two isolates were highly resistant to oxacillin and furazolidone and showed resistance to more than five antibiotics.83 Meletis et al. reported the resistance of K. varians to levofloxacin in a patient receiving continuous ambulatory peritoneal dialysis (CAPD).43 An isolate of K. rhizophila from a 3 year-old patient with a catheter showed resistance to erythromycin and ciprofloxacin.84 K. rhizophila isolated from the blood of another pediatric sepsis patient was found to be resistant only to norfloxacin.17 The genome of K. rhizophila strain DC2201 revealed the presence of 13 proteins that are likely to be involved in a multidrug efflux mechanism. These proteins facilitate the active transport of various unrelated molecules from the cytoplasm to the extracellular environment. It is presumed that during this process, the toxic organic compounds are transported outside. Interestingly, one of these 13 efflux proteins shows homology with Xanthomonas albilineans pump, which is a plant pathogen. This could be possibly due to some evolutionary association of the similar niches shared by these 2 bacterial ancestors. The extent of efflux activity on providing drug resistance and affecting drug sensitivity is still unclear.76
Recent research on antibiotic resistance of Kocuria has thrown light on some serious pointers as indicated by infective endocarditis caused by K. kristinae,85 relapsing dialysis associated peritonitis due to K. rhizophila86 and catheter associated bacteremia in a pediatric cancer patient.87 In a recent study on clinical samples collected from 2 countries viz. Egypt and Iraq, macrolide resistance of K. kristnae was very evident.88 An even more pertinent problem lies in the spread of these resistant Kocuria in the environment. In a recent study, raw milk samples have been shown to harbour multidrug-resistant (Lindamycin and Ampicillin) Kocuria with biofilm forming ability.89 Hence measures should be initiated to have in depth studies on the environmental spread of such resistant strains and their impact on public health.
When compared to major Gram-positive (S. aureus and Clostridium difficile) and Gram- negative (Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterobacter, Acinetobacter baumannii) nosocomial pathogens, the number of cases and severity of infections caused by Kocuria are lower, with catheter related sepsis in immunocompromised and post surgery patients being a major event in multiple cases; mostly resulting in peritonitis, endocarditis and colitis.41,84,87
The genome of Kocuria indica DP-K7 was sequenced via k-mer-based method in PATRIC platform; the genome was found to carry antimicrobial resistance genes like rpoB, folA, gyrA, fabL-like, dxr, S10p, efG, Iso-tRNA, fabG, gyrB, folP, rho, efTu, dfr, alr, htdX, gidB, kasA, mtrA, rpoC, erm(X):mtrB, ddl, pgsA, GgdpD, murA and lpqB.90 Table 3 elaborates the prominent AMR mechanisms of few important pathogenic bacterial species.
Table (3):
Important antimicrobial resistance genes in pathogenic bacteria and their mechanisms
Gene |
AMR Gene family |
Mechanism |
Ref. |
|---|---|---|---|
mecA |
Methicillin-resistant
PBP2 |
Antibiotic target replacement- Antibiotic resistance arises from the replacement or substitution of the antibiotic action target (β-lactams). It codes for PBP2a which allows the continuous production of bacterial cell wall due to low affinity of PBP2a to the β-lactam antibiotics like methicillin. Thought to have entered Kocuria by HGT from S. aureus |
91-93 |
rosA |
major facilitator superfamily antibiotic efflux pump |
Antibiotic efflux-resistance from antibiotics through transport of antibiotics out of the cell |
94 |
macB |
ATP-binding cassette (ABC) antibiotic efflux pump |
macB-responsible for antibiotic resistance in both Gram-negative and positive bacteria through antibiotic ejection. Genome analysis and resistance phenotypes suggest the presence of direct homologs of these genes in Kocuria. The gene is responsible for formation of the MacA-MacB-TolC assembly that transports macrolide antibiotics. These antibiotics are responsible for inhibiting protein synthesis in the bacteria. |
95-97 |
ErmX |
Erm 23S ribosomal RNA methyl- transferase |
Antibiotic target modification-antibiotic resistance was induced by mutational changes or enzymatic modifications. Its methyltransferase activity on 23S ribosomal RNA provides resistance to lincosamides, macrolides and Streptogramin b (MLSB phenotype). |
98,99 |
bacA |
undecaprenyl pyrophosphate |
During cell wall biosynthesis, responsible for recycling the undecaprenyl pyrophosphate which provides resistance against bacitracin; also controls bac operon in P. aeruginosa which controls biofilm formation. |
100 |
MexF |
resistance-nodulation- cell division (RND) antibiotic efflux pump |
Antibiotic ejection-resistance to antibiotics by transporting them out of the cell. The gene and the pump is studied in detail when it comes to Gram-negative bacteria. Being an inner membrane protein it is able to throw out the antibiotics using the proton motive force. |
101 |
QepA |
major facilitator superfamily (MFS) antibiotic efflux pump |
Antibiotic ejection- resistance to antibiotics by transporting them out of the cell. The efflux pumps formed are plasmid born and are responsible for reducing the sensitivity against fluoroquinolone. The pump belongs to the major facilitator super-family (MFS). |
102 |
Kocuria spp., owing to its potential pathogenicity to cause a range of infections including urinary tract infections, endocarditis, meningitis, etc., has gained attention in recent years as a nosocomial pathogen. Species such as K. kristinae, K. rosea, etc. have been identified to be a threat to people who are immunocompromised and those with underlying medical conditions. Treatment methods vary, including surgical interventions and the use of antibiotic cocktails. However, the identification of the species has remained quite tricky due to its morphological similarities with other closely related species. This has in turn led to the initiation of inappropriate treatment approaches for the diseases associated with the pathogen. Kocuria species show resistance to several classes of antibiotics such as penicillin, erythromycin, cephalosporin and fluoroquinolones, making the treatment excessively difficult. Due research focus should be on biofilm mechanisms and ways of tackling them, genome characterization to identify virulence and resistance genes, identifying HGT events and also to pinpoint nosocomial transmission routes. Hence, it is important to prevent the transmission of such strains of bacteria by understanding the mode of disease transmission, enabling improved and early identification, enhanced patient care and by the use of appropriate antibiotics.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Department of Life Sciences, CHRIST University, Bangalore, for the help and support extended for this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SS conceptualized the study. SS, AG, LRP, KD and MG wrote the manuscript. SS reviewed the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Hazard D, von Cube M, Kaier K, Wolkewitz M. Predicting Potential Prevention Effects on Hospital Burden of Nosocomial Infections: A Multistate Modeling Approach. Value Health. 2021;24(6):830-838.
Crossref - Irek EO, Amupitan A, Obadare TO, Aboderin AO. A systematic review of healthcare-associated infections in Africa: An antimicrobial resistance perspective. Afr J Lab Med. 2018;7(2):796.
Crossref - Raoofi, S, Kan FP, Rafiei S, et al. Global prevalence of nosocomial infection: A systematic review and meta-analysis. PloS One. 2023;18(1):e0274248.
Crossref - Napolitani M, Troiano G, Bedogni C, Messina G, Nante N. Kocuria kristinae: An emerging pathogen in medical practice. J Med Microbiol. 2019;68(11):1596-160
- Stackebrandt E, Koch C, Gvozdiak OR, Schumann P. Taxonomic Dissection of the Genus Micrococcus: Kocuria gen. nov, Nesterenkonia gen. nov, Kytococcus gen. nov, Dermacoccus gen. nov, and Micrococcus Cohn 1872 gen. emend. Int J Syst Bacteriol. 1995;45(4):682-692.
Crossref - Dotis J, Printza N, Stabouli S, Papachristou F. Kocuria species peritonitis: although rare, we have to care. Perit Dial Int. 2015;35(1):26-30.
Crossref - Dunn R, Bares S, David MZ. Central venous catheter-related bacteremia caused by Kocuria kristinae: case report and review of the literature. Ann Clin Microbiol Antimicrob. 2011;10:31.
Crossref - Kiraz A, Durmaz S, Baykan A, Perçin D. Endocarditis and Bacteremia due to Kocuria rosea Following Heart Valve Replacement. Eur J Basic Med Sci. 2013;3(4):93-5.
Crossref - Pierron A, Zayet S, Toko L, Royer PY, Garnier P, Gendrin V. Catheter-related bacteremia with endocarditis caused by Kocuria Rhizophila. Infect Dis Now. 2021;51(1):97-98.
Crossref - Li J, Zhang S. Kocuria coralli sp. nov, a novel actinobacterium isolated from coral reef seawater. Int J Sys Evol Microbiol. 2020;70(2):785-789.
Crossref - Stackebrandt E, Frederiksen W, Garrity GM, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Sys Evol Microbiol. 2002;52(3):1043-1047.
Crossref - Kandi V, Palange P, Vaish R, Bhatti AB, Kale V, Kandi MR, Bhoomagiri MR. Emerging Bacterial Infection: Identification and Clinical Significance of Kocuria Species. Cureus. 2016; 10;8(8):e731.
Crossref - Organji SR, Abulreesh HH, Elbanna K, Osman GEH, Almalki MHK. Diversity and characterization of Staphylococcus spp. In food and dairy products: a foodstuff safety assessment. J Microbiol Biotechnol Food Sci. 2018;7(6):586-593.
Crossref - Parte AC, Carbasse JS, Meier-Kolthoff JP, Reimer LC, Goker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607-5612.
Crossref - Quadri SR. Survival Strategy, Metabolic Potential and Taxonomic Reframe of Kocuria polaris. J Pure Appl Microbiol. 2024;18(3):1620-1626.
Crossref - Basaglia G, Carretto E, Barbarini D, et al. Catheter-Related Bacteremia Due to Kocuria kristinae in a Patient with Ovarian Cancer. J Clin Microbiol. 2002;40(1):311-313.
Crossref - Becker K, Rutsch F, Uekotter A, et al. Kocuria rhizophila Adds to the Emerging Spectrum of Micrococcal Species Involved in Human Infections. J Clin Microbiol. 2008;46(10):3537-3539.
Crossref - Ma ES, Wong CL, Lai KT, Chan EC, Yam WC, Chan AC. Kocuria kristinae infection associated with acute cholecystitis. BMC Infect Dis. 2005;5:60.
Crossref - Li W, Zhang Y, Schumann P, et al. Kocuria aegyptia sp. nov, a novel actinobacterium isolated from a saline, alkaline desert soil in Egypt. Int J Sys Evol Microbiol. 2006;56(4):733-737.
Crossref - Tvrzova L, Schumann P, Sedlacek I, et al. Reclassification of strain CCM 132, previously classified as Kocuria varians, as Kocuria carniphila sp. nov. Int J Sys Evol Microbiol. 2005;55(1):139-142.
Crossref - Kovacs GM, Burghardt J, Pradella S, Schumann P, Stackebrandt E, Marialigeti K. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov, isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int J Sys Evol Microbiol. 1999;49(1):167-173.
Crossref - Zhou G, Luo X, Tang Y, et al. Kocuria flava sp. nov. and Kocuria turfanensis sp. nov, airborne actinobacteria isolated from Xinjiang, China. Int J Sys Evol Microbiol. 2008;58(6):1304-1307.
Crossref - Reddy GSN, Prakash JSS, Prabahar V, Matsumoto GI, Stackebrandt E, Shivaji S. Kocuria polaris sp. nov, an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int J Sys Evol Microbiol. 2003;53(1):183-187.
Crossref - Mayilraj S, Kroppenstedt RM, Korpole S, Saini HS. Kocuria himachalensis sp. nov, an actinobacterium isolated from the Indian Himalayas. Int J Sys Evol Microbiol. 2006;56(8):1971-1975.
Crossref - Park E, Roh SW, Kim M, Jung MH, Shin K, Bae J. Kocuria koreensis sp. nov, isolated from fermented seafood. Int J Sys Evol Microbiol. 2010;60(1):140-143.
Crossref - Seo YB, Kim D, Kim G, et al. Kocuria gwangalliensis sp. nov, an actinobacterium isolated from seawater. Int J Sys Evol Microbiol. 2009;59(11):2769-2772.
Crossref - Tang S, Wang Y, Lou K, et al. Kocuria halotolerans sp. nov, an actinobacterium isolated from a saline soil in China. Int J Sys Evol Microbiol. 2009;59(6):1316-1320.
Crossref - Wang K, Zhang L, Liu Y, et al. Kocuria dechangensis sp. nov, an actinobacterium isolated from saline and alkaline soils. Int J Sys Evol Microbiol. 2015;65(Pt-9):3024-3030.
Crossref - Dastager SG, Tang S, Krishnamurthi S, Lee J, Li W. Kocuria indica sp. nov, isolated from a sediment sample. Int J Sys Evol Microbiol. 2014;64(Pt-3):869-874.
Crossref - Yun J, Roh SW, Jung MH, et al. Kocuria salsicia sp. nov, isolated from salt-fermented seafood. Int J Sys Evol Microbiol. 2011;61(2):286-289.
Crossref - Kim SB, Nedashkovskaya OI, Mikhailov VV, et al. Kocuria marina sp. nov, a novel actinobacterium isolated from marine sediment. Int J Sys Evol Microbiol. 2004;54(5):1617-1620.
Crossref - Braun MS, Wang E, Zimmermann S, Wagner H, Wink M. Kocuria tytonis sp. nov, isolated from the uropygial gland of an American barn owl (Tyto furcata). Int J Sys Evol Microbiol. 2019;69(2):447-451.
Crossref - Hamada M, Shibata C, Tamura T, et al. Kocuria pelophila sp. nov, an actinobacterium isolated from the rhizosphere of a mangrove. Int J Sys Evol Microbiol. 2016;66(9):3276-3280.
Crossref - Jiang Z, Zhang WH, Yuan CG, et al. Kocuria subflava sp. nov, isolated from marine sediment from the Indian Ocean. Antonie van Leeuwenhoek. 2015;108(6):1349-1355. Crossref
- Zhang L, Xi L, Ruan J, Huang Y. Kocuria oceani sp. nov, isolated from a deep-sea hydrothermal plume. Int J Sys Evol Microbiol. 2017;67(1):164-169.
Crossref - Camacho M, Redondo Gomez S, Rodriguez-Llorente I, et al. Kocuria salina sp. nov, an actinobacterium isolated from the rhizosphere of the halophyte Arthrocnemum macrostachyum and emended description of Kocuria turfanensis. Int J Sys Evol Microbiol. 2017;67(12):5006-5012.
Crossref - Kloos WE, Tornabene TG, Schleifer KH. Isolation and Characterization of Micrococci From Human Skin, Including Two New Species: Micrococcus lylae and Micrococcus kristinae. Int J Syst Bacteriol. 1974;24(1):79-101.
Crossref - Savini V, Catavitello C, Masciarelli G, et al. Drug sensitivity and clinical impact of members of the genus Kocuria. J Med Microbiol. 2010;59(12):1395-1402.
Crossref - Ajeeba SA, Anand AR, Mukherjee B. Kocuria rhizophila dacryocystitis: Report of a rare causative organism in a common clinical condition. TNOA Journal of Ophthalmic Science and Research. 2022;60(1):57-59.
Crossref - Brandle G, L’Huillier AG, Wagner N, Gervaix A, Wildhaber BE, Lacroix L. First report of Kocuria marina spontaneous peritonitis in a child. BMC Infect Dis. 2014;14(1).
Crossref - Altuntas F, Yildiz O, Eser B, Gundoan K, Sumerkan B, Cetin M. Catheter-related bacteremia due to Kocuria rosea in a patient undergoing peripheral blood stem cell transplantation. BMC Infect Dis. 2004;4(1):62.
Crossref - Inada N, Shoji J, Yamagami S. Atopic keratoconjunctivitis complicated by Kocuria koreensis keratitis: the first case. Allergy Asthma Clin Immunol. 2017;13(1):6.
Crossref - Meletis G, Gogou V, Palamouti M, et al. Catheter-related relapsing peritonitis due to Kocuria varians in a patient undergoing continuous ambulatory peritoneal dialysis. Nefrologia. 2012;32(4):541-542.
Crossref - Purty S, Saranathan R, Prashanth K, et al. The expanding spectrum of human infections caused by Kocuria species: a case report and literature review. Emerg Microb Infect. 2013;2(10):e71.
Crossref - Ben-Ami, R, Navon-Venezia S, Schwartz D, Carmeli Y. Infection of a Ventriculoatrial Shunt with Phenotypically Variable Staphylococcus epidermidis Masquerading as Polymicrobial Bacteremia Due to Various Coagulase-Negative Staphylococci and Kocuria varians. J Clin Microbiol. 2003a;41(6):2444-2447.
Crossref - Dotis J, Printza N, Stabouli S, Papachristou F. Kocuria Species Peritonitis: Although Rare, we Have to Care. Perit Dial Int. 2015;35(1):26-30.
Crossref - Srinivasa KH, Agrawal N, Agarwal A, Manjunath CN. Dancing vegetations: Kocuria rosea endocarditis. BMJ Case Reports. 2013;2013:bcr2013010339.
Crossref - Saldaña-Ruiz MA, Chávez-García JM, Ortiz-Alonso F, Ortiz-Arce CS, Espinosa-Mora JE, Cortés-Cárdenas JR. Kocuria kristinae neuroinfection in an immunocompetent patient: a case report and review of the literature. IJID Reg. 2023;9:117-119.
Crossref - Lee MK, Choi SH, Ryu DW. Descending necrotizing Mediastinitis caused by Kocuria rosea: a case report. BMC Infect Dis. 2013;13:475.
Crossref - Chen HM, Chi H, Chiu NC, Huang FY. Kocuria kristinae: a true pathogen in pediatric patients. J Microbiol Immunol Infect. 2015;48(1):80-84.
Crossref - Moreira JS, Riccetto AGL, da Silva MTN, dos Santos Vilela MM. Endocarditis by Kocuria rosea in an immunocompetent child. Braz J Infect Dis. 2015;19(1):82-84.
Crossref - Horiuchi A, Kubota N, Hidaka E, et al. Notable alkaline tolerance of Kocuria marina isolate from blood of a pediatric patient with continuous intravenous epoprostenol therapy. J Infect Chemother. 2015;21(9):680-686.
Crossref - Taher NM. Kocuria Species: Important Emerging Pathogens in Pediatric Patients. J Pure Appl Microbiol, 2022;16(4):2874-2879.
Crossref - Tewari R, Dudeja M, Das AK, Nandy S. Kocuria kristinae in catheter associated urinary tract infection: A case report. J Clin Diagn Res. 2013;7(8):1692-1693.
Crossref - Kandi V. A case of urinary tract infection caused by Kocuria species and identified by conventional methods. Perspectives in Medical Research. 2016;4(2):64-66.
- Wojno KJ, Baunoch D, Luke N et al. Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology. 2020;136:119-126.
Crossref - Nudelman BG, Ouellette T, Nguyen KQ, Schmaus WH, Chokshi RR. Kocuria rosea Bacteremia in a Sickle Cell Patient: A Case Report. Cureus. 2022;14(9):e28870.
Crossref - Mathew A, Nath JR, Modaweb A, Lone R, Abuhammour W. A Rare Case of Pediatric Central Venous Catheter-Related Bloodstream Infection With Kocuria Varians. Cureus. 2021;13(9):e18200.
Crossref - Goldstein BGB, Giroir B, Randolph A. International Consensus Conference on Pediatric Sepsis. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2-8.
Crossref - Haque KN. Definitions of bloodstream infection in the newborn. Pediatr Crit Care Med. 2005;6(3 Suppl):S45-9.
Crossref - Hecht SM, Ardura MI, Yildiz VO, Ouellette CP. Central Venous Catheter Management in High-risk Children With Bloodstream Infections. Pediatr Infect Dis J. 2020;39(1):17-22.
Crossref - Montoya JEM, Moran MAM, Ardila JAB, Henao PGC, Rodriguez EEM, Meza GAM. Brain abscess by Kocuria rosea: Case report and literature review. Interdiscip Neurosurg. 2017;7:59-61.
Crossref - Lai C, Wang JY, Lin SH, et al. Catheter-related bacteremia and infective endocarditis caused by Kocuria species. Clin Microbiol Infect. 2011;17(2):190-192.
Crossref - Kimura M, Kawai E, Yaoita H, Ichinoi N, Sakamoto O, Kure S. Central Venous Catheter-Related Blood stream Infection with Kocuria kristinae in a patient with Propionic Acidemia. Case Rep Infect Dis. 2017;2017(1):1254175.
Crossref - Tsai C-Y, Su S, Cheng Y-H, Chou Y, Tsai T-H, Lieu A-S. Kocuria varians infection associated with brain abscess: a case report. BMC Inf Dis. 2010;10(102):102.
Crossref - Domont F, Fleche-Mateos AL, Bremond-Gignac D, Hamdad F. Kocuria dacryocystitis infection, caused by Kocuria ocularis sp. JMM Case Reports. 2014;1(2):1-4.
Crossref - McAleese T, Ahmed A, Berney M, O’Riordan R, Cleary M. Kocuria rhizophila prosthetic hip joint infection. J Surg Case Rep. 2023;(8):rjad484.
Crossref - Fowora MA, Omolopo IA, Aiyedogbon A, et al. Multidrug-Resistant and Methicillin-Resistant Co-Infection in a Nigerian Patient with COVID-19: A Case Report. Am J Case Rep. 2023;24:e938761.
Crossref - Giron FF, Ramos JO, Martin JMS, Numancia GMT,
Chaparro CG, Diez ID. Kocuria salsicia peritonitis in a peritoneal dialysis patient. Ren Fail. 2023;45(1):2210683.
Crossref - Biswal D, Kaur R, Satija S, Seth A, Rathi A, Kant L.. The Expanding Spectrum of Human Infections Caused By Kocuria Species In Pediatric Patients: One Year Observational, Prospective Hospital-Based Study. Science Direct. 2023;130(Suppl 2):S64.
Crossref - Zhang Y, Hu Q, Li Z, Kang Z, Zhang L. Kocuria species: an underappreciated pathogen in pediatric patients-a single-center retrospective analysis of 10 years’ experience in China. Diagn Microbiol Infect Dis. 2023;107(4):116078.
Crossref - Ziogou A, Giannakodimos I, Giannakodimos A, Baliou S, Ioannou P. Kocuria Species Infections in Humans-A Narrative Review. Microorganisms. 2023;11(9):2362.
Crossref - Perrin-Guyomard A, Soumet C, Leclercq R, Doucet-Populaire F, Sanders P. Antibiotic susceptibility of bacteria isolated from pasteurized milk and characterization of macrolide-lincosamide-streptogramin resistance genes. J Food Prot. 2005;68(2):347-352.
Crossref - Liang H, Wang F, Mu R, et al. Metagenomics analysis revealing the occurrence of antibiotic resistome in Salt Lakes. Sci Total Environ. 2021;790:148262.
Crossref - Machado MAA, Ribeiro WA, Toledo VS, Ramos GLPA, Vigoder HC, Nascimento JS. Antibiotic resistance and biofilm production in catalase-positive gram-positive cocci isolated from Brazilian pasteurized milk. Journal of Food Quality and Hazards Control. 2020;7(2):67-74.
Crossref - Takarada H, Sekine M, Kosugi H, et al. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J Bacteriol. 2008;190(12):4139-4146.
Crossref - Abdulla Z, Barzani K. Characterization and detection of mecA gene in different species of Staphylococcus, Streptococcus and Kocuria which isolated from thalassemia patients in Erbil City. Proceedings of the 4th International Scientific Conference of Cihan University-Erbil on Biological Sciences. 2017.
- Stykova E, Nemcova R, Valocky I, Novotny F, Guba P. Adherence of bacteria to mucus collected from different parts of the reproductive tract of heifers and cows. Can J Microbiol. 2013;59(11):720-725.
Crossref - Liebl W, Kloos WE, Ludwig W. Plasmid-borne macrolide resistance in Micrococcus luteus GenBank accession number for the sequence reported in this paper is AF462611. Microbiology. 2002;148(8):2479-2487.
Crossref - Pleshko EM, Zhurina MV. Distribution of Antibiotic Resistance Genes in Kocuria Species. Antibiotics, 2025;14(10), 1041.
Crossref 1 - Robles-Marhuenda A, Romero-Gomez MP, Garcia-Rodriguez J, Arnalich-Fernandez F. Native valve endocarditis caused by Kocuria kristinae. Enfermedades Infecciosas y Microbiologia Clinica. 2016;34(7):464-465.
Crossref - Meng X, Chen F, Xiong M, Hao H, Wang K-J. A new pathogenic isolate of Kocuria kristinae identified for the first time in the marine fish Larimichthys crocea. Front Microbiol. 2023;14:1129568.
Crossref - Rodriguez-Alonso, P, Fernandez-Otero C, Centeno JA, Garabal JI. Antibiotic resistance in lactic acid bacteria and micrococcaceae/staphylococcaceae isolates from artisanal raw milk cheeses, and potential implications on cheese making. J Food Sci. 2009;74(6):M284-93.
Crossref - Moissenet D, Becker K, Merens A, Ferroni A, Dubern B, Vu-Thien H. Persistent bloodstream infection with Kocuria Rhizophila related to a damaged central catheter. J Clin Microbiol. 2012;50(4):1495-1498.
Crossref - Poyser TA, Gbadebo D, Krebs J, Brock JM, Robinson E. Kocuria kristinae Induced Infective Endocarditis: Unveiling an Emerging Threat in Clinical Practice. Cureus. 2024;16(4):e58979.
Crossref - Nakata M, Kuji H, Toishi T, et al. Relapsing peritoneal dialysis associated peritonitis due to Kocuria rhizophila: A case report. Case Rep Nephrol Dial. 2024;14(1):10-14.
Crossref - Kachhwaha A, Gupta A, Kumar K, Omar BJ, Nath UK. Infection Control Challenge: Kocuria rhizophila Bacteremia from A Peripherally Inserted Central Venous Catheter in A Pediatric Oncology Patient. Mathews Journal of Cancer Science. 2024;9(1):49.
Crossref - Al-Khamesi MB, Ibrahim MK, El-Shatoury EH, El-Kholy IMA, Mikawye EE, Tolba STM. Antibiotic resistance and molecular characteristics of upcoming pathogenic Kocuria spp. isolated from Iraq and Egypt. Baghdad Science Journal. 2024;22(6):1560-1567.
Crossref - Mathew M, Athulya B, Chaithanya MV, et al. Antibiotic resistance and biofilm formation of thermoduric Kocuria and Micrococci isolated from the milk samples of healthy cows. Proc Indian Natl Sci Acad. 2025.
Crossref - Kumaran S, Ngo ACR, Schultes FPJ, Tischler D. Draft genome sequence of Kocuria indica DP-K7, a methyl red degrading actinobacterium. 3 Biotech. 2020;10(4):175.
Crossref https://doi.org/10.1007/s13205-020-2136-3 - Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8(6):747-763.
Crossref - Cuny C, Layer, F, Strommenger B, Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PloS One. 2011;6(9):e24360.
Crossref - Khosravi AD, Hoveizavi H, Farshadzadeh Z. The Prevalence of Genes Encoding Leukocidins in Staphylococcus aureus Strains Resistant and Sensitive to Methicillin Isolated from Burn Patients in Taleghani Hospital, Ahvaz, Iran. Burns. 2012;38 (2):247-51.
Crossref - Bengoechea JA, Skurnik M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol. 2000;37(1):67-80.
Crossref - Xu Y, Sim SH, Nam KH, et al. Crystal structure of the periplasmic region of MacB, a noncanonic ABC transporter. Biochemistry. 2009;48(23):5218-5225.
Crossref - Fitzpatrick AWP, Llabres S, Neuberger A, et al. Structure of the MacAB-TolC ABC-Type Tripartite Multidrug Efflux Pump. Nat Microbiol. 2017;2:17070.
Crossref - Kobayashi N, Nishino K, Hirata T, Yamaguchi A. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Letters. 2003;546(2-3):241-246.
Crossref - Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for Macrolide and Macrolide-Lincosamide-Streptogramin B Resistance Determinants. Antimicrob Agents Chemother. 1999;43(12): 2823-30.
Crossref - Rosato AE, Lee BS, Nash KA. Inducible macrolide resistance in Corynebacterium jeikeium. Antimicrobial agents and chemotherapy, 2001;45(7):1982-1989.
Crossref - Shaaly A, Kalamorz F, Gebhard S, Cook GM. Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. The Journal of antimicrobial chemotherapy, 2013;68(7):1583-1593.
Crossref - Köhler T, Epp SF, Curty LK, Pechère JC. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. Journal of bacteriology, 1999;181(20), 6300–6305
Crossref - Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrobial agents and chemotherapy, 2007;51(9): 3354-3360.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.