ISSN: 0973-7510
E-ISSN: 2581-690X
Loop-mediated isothermal amplification (LAMP) is a novel, high specific and sensitive method which amplifies nucleic acid under isothermal conditions. Salmonella is considered one of the threatening pathogens in food industries and these species are associated with distinct food poisoning called salmonellosis. Four primers (two outer and two inner primers) were designed to target six distinct regions on the target gene invA which is conserved in Salmonella species. The reaction was optimised for 60 mins at 65 ̊C. The sensitivity of the LAMP and PCR assay for Salmonella was 10 CFU/ml and 100 CFU/ml respectively. Artificial spiking of chicken meat shows detection of Salmonella even at dilution to extinction (<1 CFU/ml) immediately after spiking as well after 48hr enrichment. All the LAMP experiments were compared to PCR method. This study reports the development of a highly sensitive, specific and a rapid diagnostic assay for the detection of Salmonella from food. The developed method could be very useful for routine pathogens point of care (POC) diagnostics.
Food, invA gene, LAMP, Pathogens, PCR, Salmonella
Salmonellosis, one of the most common global foodborne bacterial diseases is caused by Salmonella spp.1 The two main etiological agents of Salmonellosis are Salmonella enterica serovar Enteritis and S. enterica ser. Typhimurium and poultry is a major source of the pathogen.2,-4 The prevalence and serotypes encountered vary considerably between localities, regions and countries. Many animal species, especially reptiles harbour Salmonella as their normal flora. Though these do not cause infections in reptiles, they can spread to humans and cause infection. In most food animal species, there is a significant risk of potential zoonosis associated with Salmonella. These infections may be clinically unnoticeable and of different durations than typical typhoid fever. These animals may be contributing in spread of infection between flocks and cause human foodborne infection. Therefore, identification and surveillance of Salmonella serotypes in humans and food animals especially poultry is essential to develop disease control measures.4 In case of human infection, contamination of food is only the possible cause when these pathogens enter the food chain. Therefore, product and environmental monitoring for these pathogens are very important in order to reduce or prevent Salmonella illnesses/outbreaks from contaminated animal or human food.5
Several methods with higher sensitivity and specificity for Salmonella detection from food have been developed, including immune magnetic separation, immunological methods, molecular methods, and bacteriophage detection systems.6 Molecular based techniques are more accurate, sensitive and faster, therefore efficiently used to diagnose pathogens from various sources. However, this technique requires expensive and sophisticated equipment and trained personnel, making it difficult to apply in field conditions. These difficulties can be overcome by the development of simpler and rapid techniques. One of such rapid and accurate method is loop-mediated isothermal amplification (LAMP). This method utilises a set of four specific primers and Bst DNA polymerase. Bst DNA polymerase not only synthesises DNA but has an auto-cycling strand displacement DNA synthesis activity. The primers set of four oligonucleotides recognizes six distinct regions on the target DNA.7 LAMP is a reliable, rapid and simple assay that provides high sensitivity and specificity results. Therefore, development and application of such reliable methods for the detection, identification, and characterization of Salmonella will provide a useful tool for monitoring and assessment of the food safety. The aim of this study was to develop and evaluate a Salmonella specific LAMP for the detection of this pathogen in chicken meat.
Bacteria and DNA extraction
S. enterica ser. Typhimurium ATCC 14028 (Himedia, India) was revived in Luria Bertani (LB) broth (Himedia, India) and incubated at 37°C for 18 hrs followed by extraction of genomic DNA using DNeasy kit (Qiagen, India) according to the manufacturer’s instruction.
Primer designing for LAMP assay
A specific set of primers (4) consisting of inner primers (SalFIP:5’-CCGGCTCTTCGGCACAAGTAATttttGGACTGATTGGCGATCTCG-3’ & SalBIP: 5’-GCTCAACTTGCGGAGCGTCTttt tAACAATACTTCCGGCAGGC-3’) in and outer primers (SalF3: 5’-GGAAAAAGAAGGGTCGTCGT-3’ & Sal B35’-ATGCTGTTATCGTCCAGGC-3’) were designed for the target gene invA (accession no M90846) using primer expolorerV4 (http://primerexplorer.jp/elamp4.0.0/index.html). SalFIP was designed as the F2 region at the 3′ end complementary to the F2c region, and the same sequence as the F1c region at the 5′ end with a TTTT spacer in between F2 and F1c. Similarly, SalBIP was designed as the B2 region at the 3′ end that is complementary to the B2c region, and the same sequence as the B1c region at the 5′ end with TTTT spacer. The primer sequences are available on request.
Optimization of LAMP conditions
Genomic DNA isolated from the reference strain was used for optimization of time and temperature for the LAMP assays. Optimization of reaction temperature involved monitoring the reaction at constant temperatures between 60°C and 65°C. The optimization of the reaction length of LAMP assays was done from 15 min and 60 min in optimized temperature with a difference of 15 min between each time point. The assay was carried out in 12.5µL reaction volume containing :1µLeach of inner primers (20pmol each), 0.5µl of F3 and B3, 6.25µL of 2X reaction mixture {40mM Tris HCl, 20mM KCl, 16mM MgSO4, 20mM (NH4)2SO4, 0.2% Tween 20, 1.6M betaine. 2.8 mM dNTPs each}, 0.5ul of Bst DNA polymerase, 1µL of target DNA at 60°C, 63°C and 65°C for 60 mins and the reaction termination was done at 80°C for 10 mins. The reaction time was optimized at 60 mins based on the clear amplification pattern of gel electrophoresis.
Specificity of LAMP
The specificity of the LAMP assays was ascertained with DNA template obtained from bacteria belonging to other genera of closely related species within the same family such as Proteus mirabilis ATCC 29906 (HiMedia, India), Escherichia coli ATCC 8739 (HiMedia, India), Shigella flexneri ATCC 12022 (HiMedia, India), Klebsiella pneumoniae ATCC 10031 (HiMedia, India) and Enterobacter aerogenes ATCC 13048 (HiMedia, India) (current taxonomy Klebsiella aerogenes). and performing the reaction at predetermined conditions.
Sensitivity of LAMP assay using artificial spiked samples
Once the LAMP conditions are optimized, the feasibility of the technique was checked by the ability of the method to detect the pathogens in artificially spiked samples. A single colony of S. enterica ser. Typhimurium ATCC 14028 was picked from a Nutrient agar plate and inoculated into 5ml Luria Bertani (LB) broth. The tube was incubated at 37°C with shaking up to 3 hours till the culture obtained OD600 of ~0.6 which corresponded to 3 x108 CFU/ml as previously determined by the spread plate method. The culture was serially diluted in physiological saline (0.85% sodium chloride) to achieve 3×103, 3x 102, 30, 3 and less than 0 CFU/ml culture dilutions. Chicken meat was procured from the local market and minced using a sterile blade. The meat was spread as a thin film on sterile petri dishes and subject to 15 minutes ultraviolet light treatment for both surfaces. One hundred µl of culture dilutions were added to 5-gram aliquots of the UV treated minced chicken meat (tests). The tests could absorb the culture at room temperature for 15 minutes. An unspiked control of the chicken mince was maintained. The test and the control were then transferred to culture bottles containing 45ml of Fluid Selenite cysteine (FSC) broth (Himedia, India). The bottles were vigorously shaken and 1ml pre-enrichment samples were drawn. The FSC bottles were incubated at 37°C for 18 hours and 1ml enrichment samples were drawn. Enrichment broths were then cultured on Xylose Lysine Deoxycholate (XLD) agar (Himedia, India) to detect Salmonella by the traditional culture method.8 The pre-enrichment and enrichment samples drawn were subject to DNA extraction followed by detection by PCR and LAMP. PCR detection was using the standard invA primers (Sal F -AACGTGTTTCCGTGCGTAAT; Sal-R – TCCATCAAATTAGCGGAGGC) in a reaction mixture of 15 µL consisting of 1.5 µL reaction buffer, 0.5 µL of 10mM dNTPs, 0.5 µL of 10mM of each primer and 0.2µL of Taq polymerase (5U/ µL) .Reaction conditions Random samples of chicken meat (n=30) were collected from different retail stores of the local market, DNA was extracted by crude method and kit (Quiagen, QIAmp DNA mini kit, Germany)without enrichment. The extracted DNA samples were used for further PCR and LAMP assay.
Assay conditions for Salmonella typhimurium detection
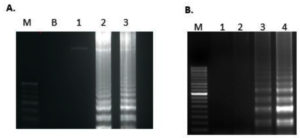
LAMP reaction optimisation for temperature and time of reaction was conducted using Salmonella typhimurium DNA template. Based on the clarity of the expected band size amplification time and temperature was optimized for 60 min at 65°C (Fig. 1) as no amplification was observed at 60°C for 60 mins. There was no amplification in 15 and 30 mins but the target gene amplified at 45 and 60 mins. However, the pattern of band formation at 60 mins was more prominent and clearer. Therefore, the reaction conditions were optimized as 65°C for 60 mins.
Fig. 1. Optimization of temperature and time for invA gene (A), Lane M: Molecular marker 100bp, B: Blank, 1: 60 °C, 2: 63 °C and 3: 65°C. (B) Lane M: Molecular marker 100bp, 1: 15 mins, 2: 30 mins, 3: 45mins and 4: 60 mins. All the products were run on 2.0% agarose gel and stained with ethidium bromide.
LAMP assay specificity
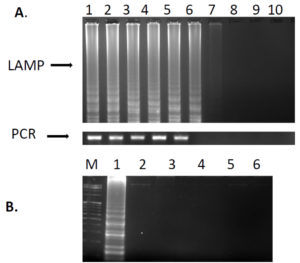
The specificity of LAMP reactions was examined with DNA extracted from closely related genera including Proteus mirabilis, Escherichia coli, Shigella flexneri, Klebsiella pneumoniae and Enterobacter aerogenes. After incubation for 65°C for 60mins, amplification was observed only for Salmonella whereas no amplification was seen for other isolates used reiterating its specificity for the detection of Salmonella (Fig. 2).
Fig. 2. A; Sensitivity of LAMP and PCR reaction for invA gene with 10-fold dilutions of DNA. Lanes: Lane 1: 10 ̄ 1, 2: 10 ̄ 2, 3: 10 ̄ 3, 4: 10 ̄ 4, 5: 10 ̄ 5, 6: 10 ̄ 6, 7 :10¯7, 8: 10 ̄ 8, 9: 10 ̄ 9, 10: 10 ̄ 10. Specificity of LAMP assay. Lane M: Molecular marker 100bp, 1: S. enterica ser. Typhimurium ATCC 14028, 2: Proteus mirabilis (ATCC 29906), 3: Escherichia coli ATCC 8739, 4: Shigella flexneri ATCC 12022, 5: Klebsiella pneumoniae ATCC 10031, 6: Enterobacter aerogenes ATCC 13048. All the products were electrophoresed on 2.0% agarose gel and stained with ethidium bromide.
Sensitivity of LAMP
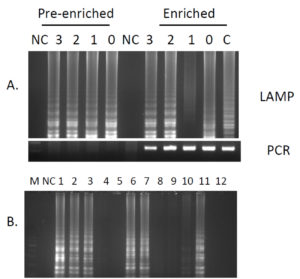
Based on the, 10-fold serial dilutions of the initial colony forming unit(cfu)/ml (3.8×108) was done and used for subsequent spiking of chicken meat (300 CFU, 30 CFU,3CFU and less than 3 CFU), LAMP could detect up to 3 CFU of the pathogen from the chicken meat directly while conventional PCR could detect the target gene only after 6hrs of enrichment in FSC broth (Fig. 3A and B).
Fig. 3. A). Detection of invA gene from pre-enriched and enriched spiked chicken meat samples. C- Control DNA from culture, NC-No template control, 0 : <3 CFU / gram , 1: ~ 3 CFU/gram, 2 : ~30 CFU/gram , 3: ~300 CFU/gram. Amplification observed in LAMP in pre-enriched samples but not by conventional PCR. B) Detection of Salmonella from random chicken meat in and around Mangalore. Lanes 1 to 12 represent different samples. All the products were electrophoresed on 2.0% agarose gel and stained with ethidium bromide.
The Food Safety Modernization Act (FSMA) regulations mandate that products and environment be routinely monitored for pathogens. This practice is critical in the prevention of Salmonella outbreaks/illnesses which account for 43% of foodborne illnesses worldwide.9 An ideal detection method should be a rapid method of high specificity and sensitivity yet simple and economical to handle.
LAMP is a novel nucleic acid amplification test developed a decade ago and has gained popularity compared to conventional PCR for the rapid and reliable detection of various fungal, bacterial, parasitic, and viral agents.7,10,11 The sensitivity of LAMP assay targeting Salmonella was reported and was much greater than PCR.12 Since then, many LAMP assays for Salmonella detection have been developed, with increasing broad applications in food and feed testing. The target gene for most the developed assay is invA which is present only in Salmonella spp.13-17 Four highly specific LAMP primers: two outer and two inner targeting the invA of Salmonella spp were designed. The conditions of developed assay optimized for detecting Salmonella DNA was at 65°C for 60 min. However, amplification of target gene was possible within 45 min at 65°C. This suggests that rapid detection of Salmonella is possible in a very short time (<60 min).
The specific binding of the four primers to the six distinct sequences in the gene of interest is the most critical factor in LAMP technique. The efficiency of LAMP is not hindered by the presence of non-target DNA which is highly recommended for a good diagnostic tool. Specificity of developed LAMP assay for detection of Salmonella from meat is high, the designed primers amplified only Salmonella spp and no other closely related genera including Shigella, Proteus or other members of Enterobacteriaceae. In addition, the reaction can be monitored as simple turbidity (data not shown) as LAMP synthesises a large amount of DNA and the specificity of the developed assay is found to be consistent with other studies of LAMP assays in bacterial detection.18-20
The Salmonella specific LAMP assay was found to be highly sensitive, as it could detect 1CFU in the chicken meat itself before enrichment, whereas by PCR, the detection from spiked samples was possible only after 6 hours of enrichment. This suggests that LAMP is more sensitive than the conventional PCR and overcomes the need for initial enrichment. In samples that contain very low concentrations of bacteria, LAMP is an alternative choice than PCR for diagnosis. In food samples, Salmonella should be completely absent in 25 grams for microbial permissibility.8
Salmonella has received attention as the prime global cause of foodborne illnesses. While many other pathogens have a permissible limit of 10 CFU/gram or 1 CFU/gram, the standard for food to remain microbiological safe is absence of Salmonella per 25 grams of food. To ensure that the food is completely Salmonella free, conventional methods require that two step enrichments in a minimum of two different medium in duplicates followed by selective enrichment on three different media is performed to ensure that the product is Salmonella free. This is very laborious and the overall turnaround time is more than 72 hours. Our study has shown that the developed LAMP method is able to detect Salmonella in the chicken meat directly with counts as low as 1 CFU. It was observed that the chicken meat which was pre-screened by conventional testing and found to be negative showed enrichment of invA gene which is specific for Salmonella after 6 hours of pre-enrichment by the LAMP method. Early studies on the rapid detection of pathogens using LAMP reported that this technique could detect up to 102 Salmonella in liquid suspensions.13 Recent validation of LAMP has report detection limits as low as 1 CFU Salmonella in animal feed.23 FDA has recently approved to screen animal feed by pre-enrichment followed by LAMP. Other reports of Salmonella specific LAMP have reported sensitivity to a minimum of 25 CFU24 and 1 CFU/250ml25 with enrichment while direct detection limits have been greater (>100 CFU).26,27 Modifications of LAMP with nanoparticles have poorer detection.28 The developed LAMP assay is highly specific, rapid and sensitive detection protocol for Salmonella spp. This study has been designed for the application of LAMP technique for the detection of Salmonella spp in chicken meat. The assay could detect lower abundance Salmonella spp in various parts of meat samples. The developed LAMP technique could be routinely applied in diagnostics and in surveillance in industries, food industries, farm cultures, so that bacterium-carrying meat can be segregated before it reaches the consumers.
ACKNOWLEDGMENTS
The authors would like to thank Nitte (Deemed to the University) for providing infrastructure and facilities for the smooth conduct of research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
GC designed the experiments. RV, JRM and GC performed the experiments. RV, JRM and GC analysed the data. RV wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was supported by grants SR/WOS-A/LS-1065/2014 (G) from the Department of Science and Technology, Govt. of India under DST women Scientist scheme (WOS-A).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All data generated or analysed during this study are included in the manuscript.
- Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015;3:12(12):e1001940.
Crossref - Andreoletti O, Budka H, Buncic S, et al. Microbiological risk assessment in feeding stuffs for food-producing animals Scientific Opinion of the Panel on Biological Hazards. European Food Safety Authority. 2008;720:1-84.
Crossref - Yang Q, Domesle KJ, Ge B. Loop-mediated isothermal amplification for Salmonella detection in food and feed: current applications and future directions. Foodborne Pathog Dis. 2018;15(6):309-331.
Crossref - Abd El-Aziz NK, Tartor YH, Gharieb RMA, et al . Extensive Drug-Resistant Salmonella enterica Isolated From Poultry and Humans: Prevalence and Molecular Determinants Behind the Co-resistance to Ciprofloxacin and Tigecycline. Front Microbiol. 2021;12:738784.
Crossref - Thomas M, Fenske GJ, Antony L, et al. Whole genome sequencing-based detection of antimicrobial resistance and virulence in non-typhoidal Salmonella enterica isolated from wildlife. Gut Pathogens. 2017;9:66.
Crossref - Ferguson BS. A look at the microbiology testing market. Food Safety Magazine. 2017;2017:14-15.
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63.
Crossref - Andrews WH,Wang H, Jacobson A, Ge B, Zhang G,Hammack T. BAM Chapter 5: Salmonella. https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella
- Ribera LA, Knutson RD. The FDA’s food safety modernization act and its economic implications. Choices. 2011;26(4):16-21.
Crossref - Stein CA, Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol Ther. 2017;25(5):1069-1075.
Crossref - Switzar L, Giera M, Niessen WMA. Protein digestion: an overview of the available techniques and recent developments. J Proteome Res. 2013;12(3):1067-1077.
Crossref - Li Y, Fan P, Zhou S, Zhang L. Loop-mediated isothermal amplification (LAMP): a novel rapid detection platform for pathogens. Microb Pathog. 2017;107:54-61.
Crossref - Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett. 2005;253(1):155-161.
Crossref - Ueda S, Kuwabara Y. The rapid detection of Salmonella from food samples by loop-mediated isothermal amplification (LAMP). Biocontrol Sci. 2009;14(2):73-76.
Crossref - Lu Y, Yang W, Shi L, et al. Specific detection of viable Salmonella cells by an ethidium monoazide-loop mediated isothermal amplification (EMA-LAMP) method. J Health Sci. 2009;55(5):820-824
Crossref - Ahn YC, Cho MH, Yoon IK, et al. Detection of Salmonella using the loop mediated isothermal amplification and real-time PCR. J Korean Chem Soc. 2010;54(2):215-221.
Crossref - Jiang K, Lv Q, Zhang D, et al. A novel, sensitive, accurate multiplex loop-mediated isothermal amplification method for detection of Salmonella spp., Shigella spp. and Staphylococcus aureus in food. J Food Agric Environ. 2012;10(3):252-256.
- Zadernowska A, Chajęcka W. Detection of Salmonella spp. presence in food. Salmonella-A Dangerous Foodborne Pathogen, 2012;20:21.
- Yeh HY, Shoemaker CA, Klesius PH. Evaluation of a loop-mediated isothermal amplification method for rapid detection of channel catfish Ictalurus punctatus important bacterial pathogen Edwardsiella ictaluri. J Microbiol Methods. 2005;63(1):36-44.
Crossref - Itano T, Kawakami H, Kono T, Sakai M. Detection of fish nocardiosis by loop-mediated isothermal amplification J Appl Microbiol. 2006;100(6):1381-1387.
Crossref - Ye Y, Wang B, Huang F, et al. Application of in situ loop-mediated isothermal amplification method for detection of Salmonella in foods. Food Control. 2011;22(3):438-444.
Crossref - Kono T, Savan R, Sakai M, Itami T. Detection of white spot syndrome virus in shrimp by loop-mediated isothermal amplification. J Virol Methods. 2004;115(1):59-65.
Crossref - Mekata T, Kono T, Savan R, et al. Detection of yellow head virus in shrimp by loop-mediated isothermal amplification (LAMP). J Virol Methods. 2006;135(2):151-156.
Crossref - Savan R, Kono T, Itami T, Sakai M. Loop-mediated isothermal amplification: an emerging technology for detection of fish and shellfish pathogens. J Fish Dis. 2005;28(10):573-581.
Crossref - Jenkins DM, Kubota R, Dong J, Li Y, Higashiguchi D. Handheld device for real-time, quantitative, LAMP-based detection of Salmonella enterica using assimilating probes. Biosens Bioelectron. 2011;30(1):255-260.
Crossref - Ravan H, Yazdanparast R. Development of a new loop-mediated isothermal amplification assay for prt (rfbS) gene to improve the identification of Salmonella serogroup D. World J Microbiol Biotechnol. 2012;28(5):2101-2106.
Crossref - Srisawat M, Panbangred W. Efficient and specific detection of Salmonella in food samples using a stn-based loop-mediated isothermal amplification method. Biomed Res Int. 2015;2015:356401.
Crossref - Zhang YQ, Shan XX, Shi L, et al. Development of a fimY-based loop-mediated isothermal amplification assay for detection of Salmonella in food. Food Res Int. 2012b;45(2):1011-1015.
Crossref - Garrido-Maestu A, Azinheiro S, Carvalho J, et al. . Combination of microfluidic loop-mediated isothermal amplification with gold nanoparticles for rapid detection of Salmonella spp. in food samples. Front Microbiol. 2017a;8:2159.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.