ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.35 | © The Author(s). 2019
Use of multifunctional plant growth promoting rhizobacteria (PGPR) for managing plant growth and health could not only facilitate higher positive effects on plants but also enable their predominant rhizospheric prevalence. While multi-functional PGPR are common, those harbouring both direct and indirect traits of growth promotion are relatively fewer. The present work aimed at isolating and characterizing the otherwise unusual multipotential PGPR with P-solubilizing ability in combination with broad-spectrum biocontrol abilities from diverse soils and analysing their relative prevalence. Primary screening yielded 50 isolates with varying P-solubilizing potential; of which only 8 showed In vitro antibiosis of E. coli. Selected 14 isolates with varying degree of P-solubilizing and antibacterial potential were evaluated for siderophore, HCN and indole-3-acetic acid (IAA) production. While all selected isolates produced HCN, 13 of them produced IAA and 10 showed siderophore production, at varying levels. Biochemical characterization of these isolates indicated that siderophore production was maximum with fluorescent Pseudomonas isolates while isolates of Enterobacteriaceae family were best IAA producers. However, molecular characterization of isolates capable of efficient P-solubilization along with strong ability to exhibit all the three biocontrol traits, identified them as Pseudomonas spp., typically P. aeruginosa. Overall, these results indicate that categorically P. aeruginosa species are likely to predominate as rhizobacteria with co-existence of discrete abilities to solubilize P as well as produce IAA, siderophore and HCN. The study also implies relatively higher metabolic versatility of P. aeruginosa species as compared to other members of fluorescent Pseudomonas family; thus, accounting for their rhizospheric abundance.
Multipotential PGPR; Rhizospheric Pseudomonas aeruginosa; P-solubilization; Biocontrol.
Incorporation of beneficial microorganisms, often known as plant growth promoting rhizobacteria (PGPR), into agricultural systems has been an approach well-explored to improve plant health and productivity in an eco-friendly manner. PGPR usually benefit the plant growth and health either by direct growth promotion or indirect biocontrol of disease-causing microbial population. Direct plant growth promoting mechanisms include enhancing availability of essential nutrients like phosphorus (P), nitrogen and iron as well as regulating the production of hormones like indole-3-acetic acid (IAA), gibberellic acid, cytokinin and ethylene. On the other hand, indirect mechanism of plant growth promotion mainly involves antibiosis of disease-causing microbes, inducing systemic resistance in plants and competing for the rhizosphere niches (Dutta and Podile 2010; Beneduzi et al. 2012) through production of lytic enzymes as well as siderophores, hydrogen cyanide (HCN), 2,4-diacetylphloroglucinol, IAA and other secondary metabolites. Besides, through secretion of these various metabolites, PGPR play a pivotal role in regulating the chemical and microbial composition of soil and its dynamics (Bergsma-Vlami et al., 2005; Vacheron et al., 2013; Igiehon, 2018). To be classified as an efficient PGPR, it should additionally possess high rhizospheric competence with aggressive root colonizing ability and tolerance to edaphic environment (Vejan et al., 2016). Majority of the PGPR isolated and characterized based on these criteria and subsequently commercialized, belong to genera Azotobacter, Azospirillium, Rhizobium, Bradyrhi-zobium, Burkholderia, Serratia, Arthrobacter, Enterobacter, Klebsiella, Bacillus, Pseudomonas and Agrobacterium (Glick, 2012; Ahemad and Kibret, 2014). Amongst these, Bacillus, Pseudomonas and Enterobacter form the most frequently encountered genera of root-associated beneficial rhizobacteria and hence are most explored (Beneduzi et al. 2012). However; performance of these PGPR in soil is subject to biotic and abiotic environment prevailing in the rhizosphere.
Two approaches have been employed in order to maximize the PGPR-mediated beneficial effect on the plant (i) combined use of different PGPR strains each with desired growth-promoting trait in the form of microbial consortia and (ii) use of multifunctional PGPR strains. Recent findings suggest that consortium-based inoculation may not necessarily produce sustainable cumulative beneficial effect; rather could evoke competitive process within the microbial species used as well as with the resident rhizobacteria. Moreover, additional efforts are required in identifying compatible strains to develop successful consortia. On the other hand, multipotential PGPR expressing both primary and secondary beneficial traits under a given rhizospheric environment could facilitate enhanced positive effects on the host plants while competitively prevailing in the rhizosphere. Multi-functional PGPR although are not unusual, those simultaneously harbouring both direct and indirect traits of growth promotion are relatively fewer yet capable as potential bioinoculants for sustainable agriculture (Avis et al. 2008).
In this context, considering that P as a nutrient is one of the most essential yet unavailable from most agricultural soils (Gyaneshwar et al., 2002), rhizobacteria with P-solubilizing ability along with efficient biocontrol abilities could make promising bio inoculants with greater rhizospheric competence. The present study aims at screening and characterization of multipotential PGPR possessing strong P-solubilizing ability co-existing with selected biocontrol abilities like production of HCN, IAA and siderophores; with subsequent analysis of relative prevalence of such dual ability within the known rhizobacteria.
Rhizospheric soil sampling and screening for P-solubilizing bacteria
About 5 g soil from rhizospheres of sorghum, paddy, tobacco, tea and maize crops was collected from local field in the vicinity of Changa village, Dist. Anand, Gujarat (India). Soil sample obtained from about 10 cm depth along the root system was considered as rhizospheric soil. Besides, soil samples were also collected from the rhizosphere in domestic garden as well as non-agricultural surfaces like river bank, lake-side and factory-side locations and stored at 4°C. Soils from these locations were collected across a period of one month and used for bacterial isolation within 3 days of sampling. For isolation of P-solubilizing bacteria, one gram of soil sample was suspended in 9ml sterile distilled water under aseptic condition and was subjected to vigorous shaking (100 rpm, 30°C) for one hour. This soil solution was used to prepare 10-2 10-3 10-4 10-5 and 10-6 dilutions serially (Sharma et al., 2011). For primary screening, an aliquot of 0.1 ml of each dilution was spread on Pikovskaya agar plate, and the colonies obtained after 24 to 48 h incubation at 30°C were examined to screen P-solubilizing bacteria. Presence of zone of clearance surrounding the colony was considered as qualitative measure of P-solubilizing ability. Selected P-solubilizing isolates were subsequently sub-cultured on nutrient agar medium and used for further characterization as discussed in following sections. All the isolates were maintained as pure cultures on nutrient agar medium at 4°C, throughout the study.
For quantitation of P-solubilizing ability, bacterial inocula for test isolates were prepared by inoculating single colony of each isolate aseptically in 3 ml nutrient broth and allowing the cultures to grow overnight under shaking conditions in a rotary shaker (Orbitek, Scigenics Biotech Pvt. Ltd., India) maintained at 150 rpm and 30°C. One millilitre of freshly grown culture of each isolate was harvested by centrifuging at 10,000 rpm for 2 minutes. Resulting pellets were washed with 1 ml of sterile normal saline and finally re-suspended in 1 ml of normal saline. A three-microliter aliquot of inoculum for each isolate was aseptically spotted on pre-solidified Pikovskaya agar. The petri plates were incubated at 30°C for 24 to 48 h to allow bacterial growth and formation of clear halo. Normal saline similarly spotted was used as control. P-solubilizing ability was quantified as phosphate solubilising index (PSI), calculated as ratio of diameter of clear halo (mm) to diameter of zone of growth (mm) (Nosrati et al. 2014).
Antibacterial activity
Selected isolates were tested for ability to antagonise E. coli using soft agar overlay technique with slight modification (Hockett and Baltrus 2017). Bacterial inocula of test isolates were prepared as described above, with final re-suspension of cultures in 3 ml of normal saline for further use. To pour the overlay, 24h freshly grown E. coli culture (50µl) was aseptically mixed with sterile warm molten agar (0.6%) and poured onto the already solidified nutrient agar. Plates were carefully tilted in all directions so as to ensure even distribution of overlay, allowed to solidify and used to create 3 mm bores using sterile cork borer. Thereafter, 20µl of fresh test inoculum was transferred into the bore and the plates were incubated at 30°C for 3 days. Clear zone around the zone of growth of test isolate was taken as a measure of antibacterial ability. Antibacterial ability of all the test isolates was recorded qualitatively as positive or negative.
IAA production
Phytohormone IAA was quantitated and detected by method given by Ahmad et al. (2008). Briefly, test cultures were grown in 3 ml Luria broth supplemented with 500µg/ml tryptophan to induce IAA production. Each culture similarly inoculated on Luria broth in absence of tryptophan was used as corresponding control. Cultures grown for 48 h under shaking conditions (150 rpm, 30°C) were harvested and centrifuged 10,000 rpm for 2 minutes at room temperature. Resulting cell free supernatants were used for spectrophotometric determination of IAA at 535 nm using Salkowski reagent (Ahmad et al. 2008). IAA was quantified using a standard curve prepared by similar spectrophotometric measurement of pure IAA in range of 10 to 100µg/ml. IAA quantification for each culture was carried out in triplicates.
Siderophore Production
Siderophore production by test isolates was measured using CAS solution assay (Christina Jenifer et al. 2015). Briefly, the isolates were grown in King’s B medium under shaking conditions (150 rpm, 30°C) for 24 h and centrifuged to derive cell free supernatants. To 0.5 ml of these supernatants, 0.5 ml CAS solution was added and colour developed after 10 minutes of incubation at room temperature was measured spectrophotometrically at 630nm. Un-inoculated medium similarly treated was used as the control. For quantitation, % siderophore units were calculated by following formula [{Ar-As}/Ar]*100; where Ar = absorbance of reference and As = absorbance of sample. Siderophore quantitation for each culture was carried out in triplicates.
HCN production
HCN production by test isolates was measured qualitatively as per method given by Reetha et al. (2014). Test bacteria were streaked on Luria agar plate amended with 4.4 g/L of glycine. Sterile filter paper soaked in 2% sodium carbonate and 0.5% picric acid solution was placed in lid of the petri dishes which were subsequently sealed with parafilm and incubated at 30°C for 48 h to allow bacterial growth. Change in colour of the filter paper from yellow to orange/light brown, brown or reddish-brown indicated weak, moderate or strong production of HCN respectively.
Microscopic and biochemical characterization of bacterial isolates
Test isolates were characterized on preliminary basis using selected microscopic and biochemical tests including Gram staining, catalase, indole production test, nitrate reduction test, Vogues-Proskauer test, Methyl red test and oxidase test, performed according to Bergey’s Manual of Determinative Bacteriology. Besides, fluorescent pigment production on King’s B agar after 24 h of growth at °C was also checked to identify fluorescent Pseudomonas species.
Molecular characterization of bacterial isolates
Identification of selected bacterial isolates was carried out on the basis of 16s rRNA gene sequencing. Genomic DNA of test isolates was isolated using standard procedure as described by Sambrook and Russel (2001) while 16S rRNA gene was amplified using universal primers 8F(5’-AGAGTTTGATCCTGGC-TCAG-3’) and 1492R(5’-ACGGCTACCTTGTTAC-GACTT-3’). The PCR reaction mixture in a total volume of 20µl contained, 20 picomoles of each primer, 0.2mM dNTPs, ~80ng of template DNA, 1X PCR buffer and 1U of Taq DNA polymerase (procured from Takara Bio USA, Inc). The thermal cycler (MJ Mini, Bio-Rad) was programmed as follows: 94°C for 4 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds and primer extension at 72°C for 1 minute; followed by final extension at 72°C for 10 minutes. The PCR amplicons were analysed using 1% agarose gel, stained with ethidium bromide post electrophoresis. PCR products of appropriate sizes were then partially sequenced in single pass reaction using 8F primer (services out sourced from 1st Base, Singapore). The sequences obtained were subjected to BLAST analysis (http://www.ncbi.nlm.nih.gov) to identify the isolates on the basis of sequence similarities. Moreover, these partial nucleotide sequences were deposited at GenBank and the accession numbers were obtained.
Statistical analysis
Data for quantitative experiments is represented as Mean ± S.D as indicated in the respective figure legends. Correlation analysis between P-solubilization and biocontrol traits was performed using Microsoft Excel, for which absolute values of PSI values were retained while siderophore, IAA and HCN producing abilities were rated on a relative scale of 0-3, based on quantitative and qualitative determinations as described earlier.
Isolation and screening of potential P-solubilizing bacteria from diverse soil types and their antibacterial activity
Soil samples from randomly selected diverse crop rhizospheres, domestic garden, river banks as well as non-agricultural factory-side location were collected and used for bacterial isolation. Primary screening on the basis of P-solubilizing ability yielded fifty different isolates capable of solubilizing calcium phosphate in Pikovskya agar up to varying efficacies, as evident from PSIs ranging between 0.96-3.4 (Fig. 1). Twelve of these isolates showed PSI ≥ 2, indicating strong P-solubilizing capacity, while the remaining isolates showed moderate P-solubilization. Additionally, these isolates were also tested for their antibacterial capacity as an additional trait. Eight of these fifty isolates viz PR3, PR4, TR4, TR7, MR3, MP1, MP2 and FS2 showed antibacterial activity against E. coli as measured qualitatively after 24h of growth.
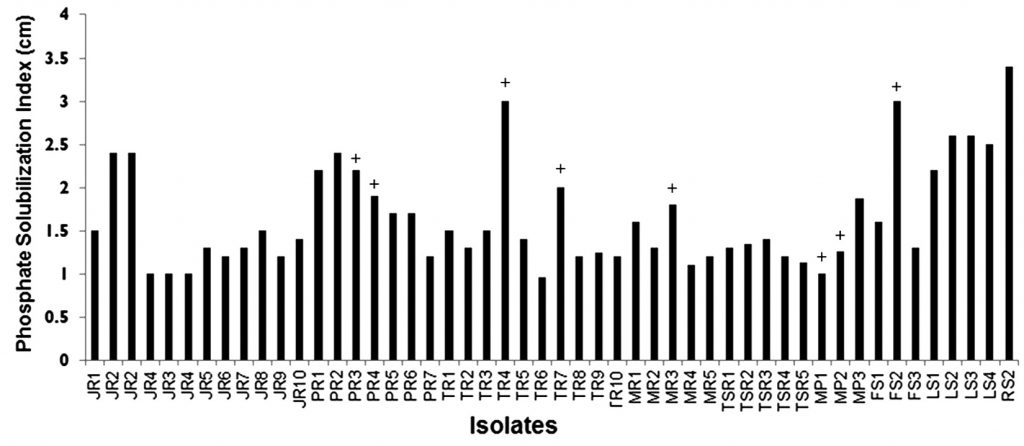
Fig. 1. Phosphate solubilisation indices of rhizospheric isolates Isolate codes depict the soil source as follows:MR-maize;JR-Jowar (sorghum); PR-Paddy; TR- Tobacco; MP-Money plant; TSR- Tea soil; LS-Lake side, RS- River side, FS-Factory side rhizospheres. + depicted above the bar for several isolate indicates presence of primary antibacterial ability; with that being absent in remaining unmarked isolates.
Characterisation of biocontrol traits in selected P-solubilizing isolates
Fourteen out of 50 isolates were selected with various combinations of P-solubilizing and antibacterial abilities for further characterization on the following basis: (i) strong P-solubilizer (PSI>2) with positive antibacterial ability – 4 isolates viz. PR3, TR4, TR7, FS2 (ii) moderate P-solubilizer (PSI between 1-2) with positive antibacterial ability – 4 isolates viz. PR4, MR3, MP1, MP2(iii) strong P-solubilizer without antibacterial ability – 5 isolates viz. LS1, LS2, LS3, LS4, RS2 and (iv) non-P-solubilizer without antibacterial ability -1 isolate PC, which served as negative control. These selected isolates were then subjected to determining the presence of important biocontrol traits like production of HCN, siderophores and IAA.
Quantitative determination of IAA production revealed that most selected isolates, except FS2 showed varied indigenous IAA producing ability ranging from 2-75 µg/ml/OD600nm, when grown in standard Luria broth upon induction with L-tryptophan (Fig. 2a). IAA production by eight of thirteen isolates was induced with the presence of L-tryptophan in the medium; of which LS1 and LS2 were highest producers of IAA. Incidentally, for remaining five isolates, IAA production was relatively low and independent of L-tryptophan. Similarly, 10 out of 14 isolates demonstrated varied capacity to produce siderophores (Fig. 2b), with five isolates viz. MR3, PR3, PR4, TR4 and TR7 showing siderophore production more than 70% units. Four isolates viz. LS1, LS3, LS4 and FS2 failed to show siderophore production. Furthermore, all 14 isolates showed varying ability to produce HCN as qualitatively monitored (Fig. 2C).
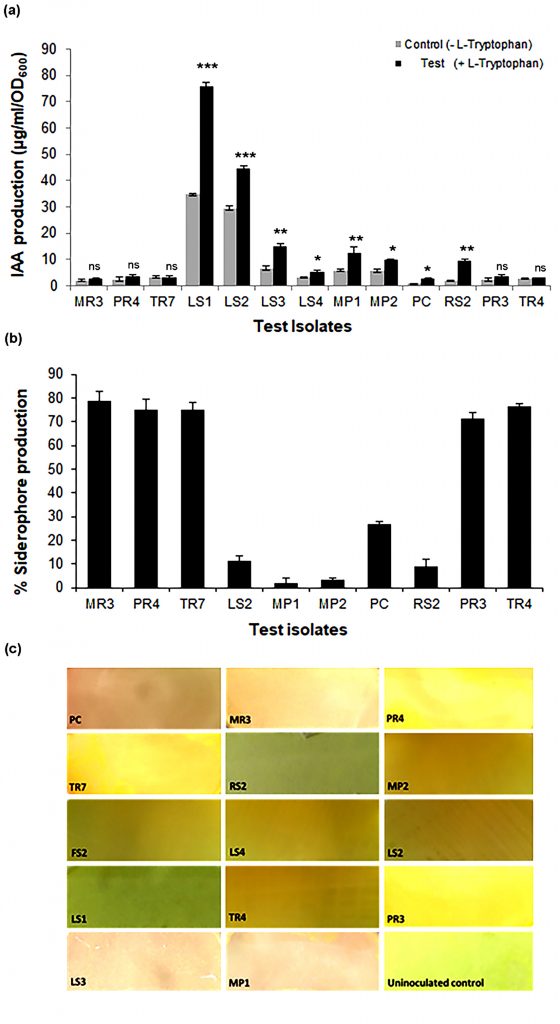
Fig. 2. IAA, siderophore and HCN producing abilities of selected isolates
(A) IAA production in presence and absence of L-Tryptophan supplemented in Luria broth. Results are expressed as Mean ± SD of 3 independent observations; *p<0.05, **p<0.01, ***p<0.001 and ns non-significant as compared to the corresponding uninduced control (B) Siderophore production as measured by CAS solution assay, Results are expressed as % siderophore production and values are represented Mean ± SD of 3 independent observations (C) Qualitative determination of HCN producing ability as monitored after 72 h of incubation.
Statistical correlation derived for coexistence of P-solubilizing ability as well as ability to produce biocontrol metabolites like IAA, HCN and siderophores across 14 selected isolates, showed about 53% positive correlation between siderophore and HCN producing traits, while siderophore production negatively correlated IAA production and P-solubilization. P-solubilizing ability showed no positive correlation with any of the biocontrol traits (Table 1).
Table (1):
Correlation analysis to indicate co-existence of P-solubilizing and multiple biocontrol traits in selected isolates
| Isolate | Siderophore | IAA | HCN | PSI | ||
|---|---|---|---|---|---|---|
| MR3 | 3 | 1 | 2 | 1.8 | ||
| PR4 | 3 | 1 | 3 | 1.91 | ||
| PR3 | 3 | 1 | 2 | 2.2 | ||
| TR4 | 3 | 1 | 3 | 3 | ||
| TR7 | 3 | 1 | 3 | 2 | ||
| LS1 | 0 | 3 | 2 | 2.2 | ||
| LS2 | 2 | 3 | 2 | 2.6 | ||
| LS3 | 0 | 2 | 1 | 2.6 | ||
| LS4 | 0 | 1 | 2 | 2.5 | ||
| MP1 | 1 | 2 | 1 | 1 | ||
| MP2 | 1 | 2 | 3 | 1.26 | ||
| RS2 | 1 | 1 | 1 | 3.4 | ||
| PC | 2 | 1 | 2 | 0 | ||
| FS2 | 0 | 0 | 2 | 3 | ||
| Correlation analysis | ||||||
| Siderophore | IAA | HCN | PSI | |||
| Siderophore | 1.00 | |||||
| IAA | -0.24 | 1.00 | ||||
| HCN | 0.53 | -0.18 | 1.00 | |||
| PSI | -0.18 | -0.12 | -0.11 | 1.00 | ||
Siderophore, IAA and HCN producing abilities were rated on a relative scale of 0-3 based of quantitative and qualitative determinations. Absolute PSI values were used to derive correlation matrix.
Biochemical and molecular characterization of selected isolates
To understand the distribution of dual ability of P-solubilization and multiple biocontrol across bacterial genera, biochemical characteristics of selected 14 isolates were deciphered (Table 2). Five of total isolates were found to belong to fluorescent Pseudomonas species, five isolates were identified as members of Enterobacteriaceae family while one isolate was characterized as Bacillus-like. Three isolates could not be clearly classified based on the set of biochemical tests performed in the present study and hence were discontinued from further study. Based on this preliminary analysis and considering the varying degree of coexistence of P-solubilizing ability with biocontrol traits, five isolates were short listed for molecular identification. Isolates TR7, MR3 and PR4 were selected as those possessing significant P-solubilizing as well as all three biocontrol abilities, each representing a different rhizosphere. Conversely, RS2 represented a strong P-solubilizer with all the three biocontrol traits at moderate levels while FS2 was selected as a strong P-solubilizer with relatively poor spectrum of biocontrol traits. Partial 16S rDNA sequence-based BLAST analysis identified TR7, MR3 and PR4 as P. aeruginosa spp. while RS2 and FS2 were identified as Rosenbergiella spp. and Cronobacter sakazakii, both known to be members of Enterobacteriaceae family (Table 3; Supplementary Material 1).
Table (2):
Microscopic and biochemical characterization of selected isolates
Isolates |
Gram staining |
Catalase Test |
Indole Test |
Nitrate reductase Test |
Vogues Proskauer Test |
Methyl red Test |
Oxidase Test |
Pigment
(fluorescent) |
Predicted family/genus |
|---|---|---|---|---|---|---|---|---|---|
MR3 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
positive |
positive |
Pseudomonas |
PR4 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
Positive |
positive |
Pseudomonas |
PR3 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
Positive |
positive |
Pseudomonas |
TR4 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
Positive |
positive |
Pseudomonas |
TR7 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
Positive |
positive |
Pseudomonas |
LS1 |
Negative |
Positive |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative |
Enterobacteriaceae |
LS2 |
Positive |
Positive |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative |
Identification unclear |
LS3 |
Positive |
Positive |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative |
Identification unclear |
LS4 |
Negative |
Positive |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative |
Enterobacteriaceae |
MP1 |
Negative |
Positive |
Negative |
Positive |
Negative |
Negative |
Positive |
Negative |
Identification unclear |
MP2 |
Negative |
Positive |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative |
Enterobacteriaceae |
RS2 |
Negative |
Positive |
Negative |
Negative |
Positive |
Negative |
Negative |
Negative |
Enterobacteriaceae |
PC |
Positive |
Positive |
Negative |
Negative |
Positive |
Negative |
Negative |
Negative |
Bacillus related |
FS2 |
Negative |
Positive |
Negative |
Negative |
Positive |
Negative |
Negative |
Negative |
Enterobacteriaceae |
Table (3):
Molecular characterization of five selected isolates/span>
Sr. No |
Isolate |
Query Length
(bp) |
Query Coverage
(%) |
Homology
(%) |
Identified species |
GenBank Accession Numbers |
|---|---|---|---|---|---|---|
1 |
TR7 |
1004 |
98 |
98 |
Pseudomonas aeruginosa |
MK372993 |
2 |
MR3 |
1086 |
95 |
95 |
Pseudomonas aeruginosa |
MK372994 |
3 |
PR4 |
970 |
99 |
98 |
Pseudomonas aeruginosa |
MK372995 |
4 |
FS2 |
1337 |
99 |
97 |
Cronobacter sakazakii |
MK372996 |
5 |
RS2 |
1270 |
97 |
99 |
Rosenbergiella spp. |
MK372997 |
Rhizo-microbiome constitutes biologically diverse community of soil microorganisms that inhabit the rhizosphere influencing the growth and productivity of their host plants by various beneficial biochemical and microbial interactions as well as exchange of several important signal molecules (Mendes et al. 2013; Venturi and Keel 2016). Microbial species with efficient multiple beneficial traits are likely to prevail abundantly in rhizosphere and at the same time benefit the plant health while successfully modulating the overall microbial population in the rhizosphere. Such bacterial species offer strong potential to be developed as bio-inoculant for sustainable agriculture. A structured study about prevalence of these multiple coexisting plant beneficial traits across microbial species may also help in targeted screening of efficient beneficial rhizobacteria with better probabilities of performance in fields. Present study aimed at isolation, characterization and identification of potent P-solubilizing soil bacteria simultaneously capable of exhibiting efficient biocontrol traits specifically production of metabolites like IAA, siderophores and HCN.
Presence of P-solubilizing ability was considered as primary screening criterion since P is the most important element in plant nutrition and P-solubilizing microbes are known to enhance the bioavailability of inorganic P by facilitating mineralization of insoluble P forms into soluble ones, mostly by secretion of organic acids (Gyaneshwar et al. 2002; Buch et al. 2008). P-solubilizing bacteria are ubiquitous and may constitute upto 1-50% of total bacterial population in rhizospheric as well as non-rhizospheric soils (Sharma et al. 2013). Successful isolation of about 50 isolates in the present study irrespective of soil types was justified by the known abundance of these P-solubilizing bacteria. On the other hand, biocontrol traits like IAA, HCN and siderophore production were categorically selected in the study owing to implications of these metabolites in antibiosis, nutrient competition and induction of systemic resistance. IAA being a phytohormone stimulates plant growth and development. PGPR capable of producing IAA are known to facilitate development of ordered and systematic root system (Vacheron et al. 2013). PGPR secreting siderophores, the iron chelating molecules, improve status of Fe nutrition in plants (Sharma and Johri, 2003; Ahemad and Kibret, 2014). Siderophores are also known to elicit defence mechanisms in plants and suppress soil-borne fungal pathogens by competing for iron acquisition (Beneduzi et al., 2012; Aznar and Dellagi, 2015). HCN on the other hand is well-known antifungal agent, production of which facilitates antibiosis of phytopathogens thus benefiting plant health and growth (Ahemad and Kibret 2014). While all isolates in the present study could produce HCN irrespective of its classification, siderophore production was drastically higher in isolates characterized as fluorescent Pseudomonas species. On the contrary isolates characterized as members of Enterobacteriaceae family showed maximum IAA production, inducible in presence of L-tryptophan. Interestingly, IAA production by fluorescent Pseudomonas isolates was although significantly low relatively, was found to be independent of L-tryptophan.
Correlation analysis between the existing plant beneficial traits measured across selected isolates also revealed a clear pattern of co-occurrence of these traits within diverse rhizobacterial species. Within biocontrol traits, occurrence of HCN production and siderophore production traits positively correlated suggesting their frequent co-existence; with no clear correlation being derived for occurrence of IAA production trait. Co-occurrence of all the three biocontrol traits with strong expression was prominent in isolates belonging to fluorescent Pseudomonas species unlike across the isolates of Enterobacteriaceae family. This was supported by an earlier report describing common co-occurrence of HCN production with production of hydroxamate-typesiderophores observed in cyanogenic Pseudomonas strains (Rijavec and Lapanje 2016). An elaborate analysis of genes contributing to plant-beneficial traits and their accumulation across PGPR genomes demonstrated preferential association of genes hcnABC involved in HCN production with phlACBD genes involved in production of potent antifungal metabolite 2,4-diacetylphloroglucinol (Bruto et al. 2014). However, any genetic association between HCN production and siderophore production across PGPR genomes is not yet demonstrated. The same study also identified ipdC gene implicated in IAA production to be clustered with budC gene involved in 2,3-butanediol production while being completely independent of the genes responsible for other plant beneficial traits; thus, substantiating our experimental observation. Incidentally, our results showed no obvious correlation between co-occurrence of P-solubilizing ability with any of the three biocontrol traits tested; which was in accordance with earlier report where pqqBCDE genes contributing to P-solubilization phenotype were found clustered separately from genes involved in IAA and HCN production (Bruto et al. 2014). Therefore, these results also could explain relatively low prevalence of rhizobacteria with co-existing P-solubilizing and biocontrol traits.
In this regard, eventually strains possessing moderate to strong P-solubilizing ability co-existing with all the three biocontrol traits were characterized as fluorescent Pseudomonas strains, typically P. aeruginosa spp.These results not only demonstrated selective predominance of fluorescent Pseudomonas spp. as multipotential PGPR across diverse rhizospheres but also suggested that within fluorescent pseudomonads, P. aeruginosa spp. may possess relatively better genetic and metabolic capacity to enable co-existence of P-solubilization and broad-spectrum biocontrol traits; thereby accounting for their abundance as root-associated Pseudomonas spp. As far as Pseudomonas spp. as potent bionoculants are considered, P. fluorescens and related species within the complex including P. protegens, P. chlororaphis and P. corrugata as well as species like P. putida and P. cepacia are largely recognized for their biocontrol potential and beneficial associations with diverse plant hosts (Mercado-Blanco and Bakker, 2007; Avis et al., 2008; Couillerot et al., 2009; Dorjey et al., 2017). The biocontrol ability of these species has been attributed to secretion of HCN, 2,4-Diacetyl-phloroglucinol, pyoluteorin, pyrrolnitrin, salicylate, ACC deaminase and siderophores while their growth promoting effect has been often attributed to IAA production. Most of these studies have exclusively explored either their plant growth promoting ability or biocontrol potential (Yasmin et al. 2017). However, broad spectrum biocontrol ability along with biofertlizer ability as in P-solubilization has been demonstrated mostly in P. aeruginosa (Islam et al., 2014; Radhapriya et al., 2015; Yasmin et al., 2017; Uzair et al., 2018).
To conclude, P. aeruginosa spp. apparently predominate as multipotential PGPR with co-occurring P-solubilization and wide-spectrum biocontrol traits irrespective of the rhizosphere; a combination rarely explored in known beneficial rhizobacteria. Investigating the robust metabolic and genetic framework underlying these abilities may reveal interesting regulatory networks which may further be useful in identifying and developing efficient PGPR with sustained performance. Furthermore, it may also provide insights into differentiating the beneficial P. aeruginosa strains from their pathogenic counterparts; thus, enabling development of various biotechnological interventions to optimize their use as model bioinoculants.
The work has been partially funded by Charotar University of Science and Technology while the publication has been supported by Department of Science and Technology, Government of India.
On behalf of all authors, the corresponding author states that there is no conflict of interest.
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizo-bacteria: Current perspective. J King Saud Univ – Sci, 2014; 26: 1–20.
- Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res, 2008; 163: 173–181.
- Avis TJ, Gravel V, Antoun H, Tweddell RJ. Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol Biochem, 2008; 40:1733–1740.
- Aznar A, Dellagi A. New insights into the role of siderophores as triggers of plant immunity/ : what can we learn from animals/J Exp Bot,2015;66: 3001–3010.
- Beneduzi A, Ambrosini A, Passaglia LMP. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Gen Mol Biol, 2012; 35: 1044–1051.
- Bergsma-Vlami M, Prins ME, Raaijmakers JM. Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol., 2005; 52: 59–69.
- Bruto M, Prigent-combaret C, Muller D, Moe Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizo-bacteria and related Proteobacteria Sci. Rep. 2014; 4: 6261.
- Buch A, Archana G, Naresh Kumar, G. Metabolic channeling of glucose towards gluconate in phosphate-solubilizing Pseudomonas aeruginosa P4 under phosphorus deficiency. Res. Microbiol., 2008; 159: 635–642.
- Christina Jenifer A, Aruna Sharmili S, Anbumalar-mathi J, Umamaheswari K, Shyamala K. Studies on siderophore production by microbial isolates obtained from aquatic environment.Eur. J Exp Biol, 2015; 5: 41–45
- Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moכnne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol., 2009; 48: 505–512.
- Dorjey S, Dolkar D, Sharma R. Plant growth promoting rhizobacteria Pseudomonas: A Review. Int J Curr Microbiol App Sci,2017; 6: 1335–1344.
- Dutta S, Podile AR. Plant growth promoting rhizobacteria (PGPR): The bugs to debug the root zone. Crit. Rev. Microbiol., 2010; 36: 232–244.
- Glick BR.Plant growth-promoting bacteria: Mechanisms and Applications. Scientifica (Cairo), 2012; 2012: 1–15.
- Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS. Role of soil microorganisms in improving P nutrition of plants. Plant and Soil, 2002; 245: 83–84.
- Hockett KL, Baltrus DA. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp, 2017; 119: e55064.
- Igiehon NO. Rhizosphere microbiome modulators: Contributions of nitrogen fixing bacteria towards sustainable agriculture.Int J Environ Res. Public Health, 2018; 15: 574.
- Islam F, Yasmeen T, Ali Q, Ali S, Arif MS, Hussain S, Rizvi H. Influence of Pseudomonas aeruginosa as PGPR on oxidative stress tolerance in wheat under Zn stress. Ecotoxicol Environ. Saf., 2014; 104: 285–293.
- Mendes R, Garbeva P, Raaijmakers JM.The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev.,2013; 37: 634–663.
- Mercado-Blanco J, Bakker PAHM. Interactions between plants and beneficial Pseudomonas spp.: Exploiting bacterial traits for crop protection. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol., 2007; 92: 367–389.
- Nosrati R, Owlia P, Saderi H, Rasooli I, Malboobi MA.Phosphate solubilization characteristics of efficient nitrogen fixing soil Azotobacter strains. Iran J. Microbiol., 2014; 6: 285–295
- Radhapriya P, Ramachandran A, Anandham R. Pseudomonas aeruginosa RRALC3 enhances the biomass, nutrient and carbon contents of Pongamia pinnata seedlings in degraded forest soil. PLoS One, 2015; 10: e0139881.
- Reetha AK, Pavani SL, Mohan S. Original research article hydrogen cyanide production ability by bacterial antagonist and their antibiotics inhibition potential on Macrophomina phaseolina (Tassi.) Goid Int. J. Curr. Microbiol. App. Sci., 2014; 3:172–178.
- Rijavec T, Lapanje A. Hydrogen cyanide in the rhizosphere/ : Not suppressing plant pathogens , but rather regulating availability of phosphate. Front Microbiol., 2016; 7: 1–14.
- Sambrook JF, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, USA, 2001.
- Sharma A, Johri BN. Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS 9 in maize (Zea mays L.) under iron limiting conditions. Microbiol. Res., 2003;158: 243–248.
- Sharma S, Kumar V, Tripathi RB. Isolation of phosphatesolubilizing microorganism (PSMs ) from soil isolation of phosphate solubilizing microorganism (PSMs) from soil. J. Microbiol. Biotechnol. Res., 2011; 1: 90–95
- Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springer plus, 2013; 2: 587.
- Uzair B, Kausar R, Bano SA, Fatima S, Badshah M, Habiba U, Fasim F. Isolation and molecular characterization of a model antagonistic Pseudomonas aeruginosa divulging in vitro plant growth promoting characteristics. Bio. Med. Res. Int., 2018; 2018: 6147380.
- Vacheron J, Desbrosses G, Bouffaud M-L, Touraine B, Moכnne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyי F, Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci., 2013; 4: 356.
- Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq Boyce A. Role of Plant growth promoting rhizobacteria in agricultural sustainability-A review. Molecules, 2016; 21: 573.
- Venturi V, Keel C. Signaling in the Rhizosphere. Trends Plant Sci., 2016; 21: 187–198.
- Yasmin S, Hafeez FY, Mirza MS, Rasul M, Arshad HMI, Zubair M, Iqbal M. Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front Microbiol., 2017; 8: 01895.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


