ISSN: 0973-7510
E-ISSN: 2581-690X
Enzymes which would be active in cold conditions can be used in a wide range of fields from molecular biology to detergent industry due to their low processing capacity and high activity. In this study, sixty cold-adapted bacteria were isolated from water and sludge samples collected from Erzurum and Van provinces. Identification of eight isolates by molecular [(GTG)5-PCR and 16S rRNA sequencing] techniques and tests for temperature (4-35°C), pH (3-11) and salt (2-15% (w/v) requirements were performed. These bacteria were belonging to Pseudomonas chlororaphis subsp. aureofaciens (SM011A), Psychrobacter faecalis (SM012D), Rahnella aquatilis (SM015A), Shewanella putrefaciens (SM018A), Pseudomonas lactis (SM0110A), Flavobacterium chryseum (SM0112E), Exiguobacterium mexicanum (SM0117A) and Glutamicibacter arilaitensis (SM0118A). The physicochemical requirements for all isolates ranged between 4-25°C, pH 5-7 and 2-15% salt (NaCl) concentration. However, E. mexicanum did not require salt in growth medium. All bacteria were evaluated for protease, lipase and amylase enzymes and all were found to be multiple enzyme producers. The eight isolates were identified from the resources of Turkey, for the first time and enzyme production abilities of some isolates to produce enzymes were declared. The originating of the producers of these enzymes from Turkey shows that Turkey has a remarkable reservoir for cold-adaptive microorganisms and these microorganisms will make important contributions to the detergent industry worldwide.
Cold-adaptive, Industrial potential, Molecular characterization, Multiple enzyme.
Microorganisms and their components in physical, chemical, biological environments are called ecosystems. Aquatic (rivers, lakes, seas, etc.) and terrestrial (soil, sand dunes, subsurface depths) environments are important microbial ecosystems1.
Psychrophilic microorganisms generally grow and reproduce in the range of (-20°C) and (+15°C)2. These microorganisms can survive at permanently cold environments such as polar regions and deep-sea regions, and pockets in deep-sea glaciers which have high salinity rate3. Even, a certain amount of microbial activity has been measured in frozen soil below -39°C4. The taxonomic diversity of psychrophilic can be quite high. Psychrobacter sp. and some species of the genera Halomonas, Pseudomonas, Hyphomonas, Sphingomonas and Arthrobacter are taxonomic branches of psychrophilic bacteria5.
It is known that cold environments generally stop the metabolic activity of microorganism or reduce to very low levels. In this context, there is a differentiation within psychrophilic microorganisms. Some facultative bacterial species may exhibit activity at low temperatures, in spite of not as much as the original psychrophilic. Those organisms with low living temperatures may also survive at higher temperatures and are called psychrotolerant. Psychrotolerant bacteria are called vital bacteria, which are also able to function even when temperatures are much lower than their normal life temperature of psychrophilic bacteria, and are essentially spread over a wider area than the obligate ones. One of the best known examples of this group is, Trichococcus patagoniensis PmagG1 which has been isolated from Guano Penguins in Chile6. This bacterium, which has an optimal life temperature of 28-30°C, can also show vital activity at lower temperatures like -7°C or -5°C6.
Enzymes are suitable for use in industrial fields today because of their ability to work in vitro and exhibit the same catalytic activity. The enzymes play a small unlocking role in synthesizing a desired product in suitable environments or removing the desired region within a synthesized product. Today’s use of enzymes is a catalytic process that is not achieved by conventional chemistry, often requiring no cofactors. In some types of reactions that require the use of cofactors, wild or recombinant organisms producing this enzyme can be used. Therefore, in cases where conventional chemistry is inadequate, enzymes or organisms that produce the enzyme become a subject of the application7.
Many benefits of cold-adapted enzymes can be counted when compared to mesophilic and thermophilic homologs. Under optimum conditions, less amounts of psychrophilic enzymes have the same work capacity and higher catalytic activities than mesophilic and thermophilic ones. In addition, reaction systems using psychrophilic enzymes allow simple control operations by increasing the temperature. All of these features provide profits on cost and the operation time in certain reactions. It has been observed that psychrophilic enzymes reduce the cost of the operation systems in which they are used, hence psychrophilic enzymes are the most remarkable enzyme group in the detergent industry due to their special capabilities such as cold work. The most popular class of enzymes for energy-efficient washing and dishwashing machines is psychrophilic enzymes, which are considered as a solution for dirtiness at low temperatures8.
The aim of this study was the isolation and identification of cold-adaptive or psychrophilic organisms in Turkey which had potential to produce commercially important enzymes. Thus, Turkey, microbial enzyme reservoir of Turkey will be determined in order to be used especially in detergent industry.
Microorganisms
Water and sludge samples were taken from Erzurum and Van provinces, by using sterilized glass bottles and pipette tips9. Samples with the presence of nutrient broth (NB) were incubated at 4°C for 3-10 days to isolate psychrophilic bacteria10. Bacterial suspension was diluted (10-1-10-9 dilution factors) and spread onto nutrient agar (NA) media and incubated 4°C for five days. The growing colonies were spread on NA to obtain pure cultures and were eliminated by morphology differences in plates, as a first step. The pure, single and different colonies were stored in the Nutrient Broth (NB) with 15% glycerol content at -86°C for further studies.
Molecular identification
A single colony from each bacterial isolates grown in NA media were selected and transferred to NB media. After incubation at 15°C for 3-5 days, genomic DNA isolation was carried out for each isolate according to the procedure of Promega™ Wizard® Genomic DNA Purification. (GTG)5-PCR was preferred due to the reliability in the amplification of the universally conserved gene regions and 16S rRNA sequencing was used to examine the evolutionary gene of the prokaryotic ribosomes of each bacterial isolates.
To obtain the amplified products, 50 ng of purified DNA was used as the template in 30µL reaction mixture. 27µL of the reaction cocktail was prepared as follows: Gitschier Buffer 5µL, dimethyl sulfoxide 2.5µL (100%, 20X), dNTPs (10mM) 1.25µL, bovine serum albumin 1.25µL (20 mg/mL), primer (5 mM) [5’-GTGGTGGTGGTGGTG-3’ for (GTG)5] 3.0µL, Taq polymerase (250U) 0.3µL and water 13.7µL. A negative control (no DNA) was included in each PCR assay10.
The PCR products (30µL) were mixed with 3µL gel loading buffer (6X) and subjected to agarose (1.5% w/v) gel electrophoresis in Tris–Acetate–EDTA (TAE) buffer at 90 V, 120 min for (GTG)5-PCR reaction. After separation of the amplification products by the gel, the fragments were stained with ethidium bromide solution (2µL Etbr/100 mL 1X TAE buffer) and monitored using the Quantum Vilber Lourmat Gel Documentation System (Australia).
The 16S rRNA of the test isolates was amplified by Polymerase Chain Reaction (PCR) with the special primers [27-F: 5’AGAGTTTGATYMTGGCTCAG3′ and 1492-R: 5′ GGTTACCTTGTTACGACTT 3′]11. The amplified fragments for 16S rRNA sequencing were cloned into E. coli JM101 with a vector system (pGEM-T, Promega, the UK) and the clones were sequenced (Macrogen, Amsterdam, the Netherlands). The results of 16S rRNA gene sequencing were analyzed using the GenBank and EzTaxon (http://blast.ncbi.nlm.nih.gov/blast.cgi and http://www.eztaxon.org) server11. Considering the results of the study, a phylogenetic tree was formed via the neighbor joining method using the software package MEGA 4.012.
Conventional identification
The test isolates were subjected to conventional tests for pH, temperature, and salt (NaCl) requirements in growth media. The pH, temperature and NaCl requirements for bacterial growth were measured in Nutrient Broth (NB) media at 600 nm wavelength in spectrophotometer13. Firstly, NB media were prepared and, then a full loop of each test isolate on NA was transferred to broth media and incubated at different temperatures (4-35°C) for 48 hours. To measure the response to pH changes during growth, NB media were prepared and pHs of broth media were adjusted to different points in pH 3-11 range, before autoclaving. Then, a full loop of each test isolate on NA was transferred to broth media and incubated at 15°C for 48 hours. The salt (NaCl) requirement for growth was also tested in NB media containing 2-15 (w/v). To this aim, NB media were prepared and salt at predetermined concentrations was added to broth media for each isolate, before autoclaving. Then, a full loop of each test isolate on NA was transferred to broth media and incubated at 15°C for 48 hours. For each test, microbial growth was measured at 600 nm wavelength at the end of the incubation period.
Cell and colony morphology (Leica ICC50 HD light microscope), Gram and endospore staining, motility and the presence of catalase and oxidase reactions were also investigated13-15.
Screening for enzyme production
Protease
Each bacterial isolate in NB were propagated in Skimmed Milk Agar (20 g/L nutrient agar, 10 g/L skim milk powder and 15 g/L agar) and incubated at 15°C for 3-5 days to assay proteolytic activities. The plates were evaluated according to the zone formations, and the observation of a halo zone indicated positive protease activity9.
Lipase
The bacterial isolates were inoculated on tributyrin agar (23 g/L) medium which contained 1% tributyrin (glycerol tributyrate) and incubated at 15°C for 3-5 days. The strains with transparent and the highest zone formation (lipolytic activity) were determined as lipase producers16.
Amylase
Amylase assay was performed at 15°C for 3-5 days in a medium which contained nutrient agar (20 g/L), soluble starch (5 g/L) and agar (15 g/L). The amylase activity was screened by Lugol’s solution. Clear and large zones were evaluated as positive, or as amylase production; however, no zone formation was evaluated as negative, or as no amylase production17.
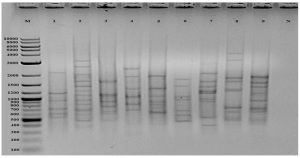
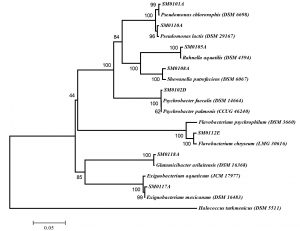
Water and sludge samples from Erzurum and Van provinces were collected and the microbial flora was induced to grow by incubating in fresh nutrient broth and sixty bacterial isolates were determined due to the colony formation differences in agar plate. To determine molecular differences of these cold-adaptive isolates, (GTG)5-PCR was carried out and eight different isolates were detected (Fig. 1). These isolates were identified by 16S rRNA sequencing and the results of 16S rRNA as phylogenetic tree were given in Fig. 2. The isolates, SM011A, SM012D, SM015A, SM018A, SM0110A, SM0112E, SM0117A and SM0118A were similar to Pseudomonas chlororaphis subsp. aureofaciens (MN192404), Psychrobacter faecalis (MN192405), Rahnella aquatilis (MN192429), Shewanella putrefaciens (MN192428), Pseudomonas lactis (MN192431), Flavobacterium chryseum (MN192434), Exiguobacterium mexicanum (MN192433) and Glutamicibacter arilaitensis (MN192913) at a rate of approximately 99%, respectively (Table 1).
Table (1):
16S rRNA sequence similarity ratios and GenBank accession numbers.
Isolate code |
Closest phylogenetic relative |
Similarity (%) |
Accession number |
|---|---|---|---|
SM011A |
Pseudomonas chlororaphis subsp. aureofaciens |
99.9 |
MN192404 |
SM012D |
Psychrobacter faecalis |
99.4 |
MN192405 |
SM015A |
Rahnella aquatilis |
99.2 |
MN192429 |
SM018A |
Shewanella putrefaciens |
99.9 |
MN192428 |
SM0110A |
Pseudomonas lactis |
99.3 |
MN192431 |
SM0112E |
Flavobacterium chryseum |
98.3 |
MN192434 |
SM0117A |
Exiguobacterium mexicanum |
99.9 |
MN192433 |
SM0118A |
Glutamicibacter arilaitensis |
98.9 |
MN192913 |
Fig. 1. (GTG)5 profile of cold-adaptive isolates (M: marker, 1:SM011A, 2:SM012D, 3:SM015A, 4:SM018A, 5:SM018F, 6:SM0110A, 7:SM0112E, 8:SM0117A, 9:SM0118A, N: negative control).
Fig. 2. Neighbor joining phylogenetic tree on the basis of 16S rRNA gene sequence data of the cold-adaptive isolates in Turkey. Halococcus turkmenicus was used as out-group. Bootstrap values based on 1000 replications are listed as percentages at branching points. The accession numbers are given in parentheses. The scale bar represented 5% divergence.
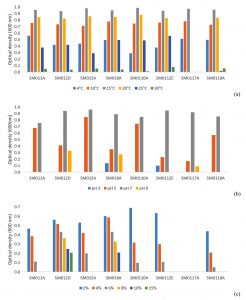
As conventional analysis, all the isolates were bacilli, Gram (except SM0117A and SM0118A) and endospore negative, catalase and oxidase (except SM0118A) positive and motile (except SM012D and SM0118A). The conventional methods were needed to determine especially the growth conditions of cold-adaptive microorganisms. The identified isolates were subjected to growth at different mediums with varying temperature, pH and salt concentrations. In order to determine the optimal temperature conditions, the isolated strains were inoculated into the NB at different temperatures and it was observed that cardinal growth temperatures ranged from 4°C to 30°C (Fig. 3a) while the growth of each isolate after 25°C and 30°C was weak. However, SM0117A could not grow higher than 15°C.
The pH ranges in which the cold-adaptive isolates could develop, were determined by screening the growths in NB medium with pH 3-11. All isolates showed growth in the range of pH 5-7 however, optimum value was pH 7. No growth was observed at pH 11, although weak growth was observed rarely in pH 3 and pH 9 (Fig. 3b).
Salt tolerances of the isolated strains were determined in nutrient broth containing 2-15%(w/v) NaCl and the growth of the strains was monitored (Fig. 3c). Although all of the isolates needed salt to grow at any concentration, the isolate SM0117A didn’t required NaCl.
Fig. 3. Growth of cold-adaptive isolates according to different (a) temperature (b) pH and (c) salt (NaCl) concentrations.
All of the eight isolates were able to produce at least two of protease, lipase and amylase enzymes so that all the isolates were multienzyme producers (Table 2). SM011A, SM015A, SM018A, SM0110A, SM0112E and SM0118A were also strong producers for some of the related enzymes.
Table (2):
Industrial enzyme production abilities of cold-adaptive isolates.
| Isolate | Protease | Lipase | Amylase | |
|---|---|---|---|---|
| SM011A | Pseudomonas chlororaphis subsp. aureofaciens | +* | +* | +* |
| SM012D | Psychrobacter faecalis | – | + | + |
| SM015A | Rahnella aquatilis | + | + | +* |
| SM018A | Shewanella putrefaciens | + | + | +* |
| SM0110A | Pseudomonas lactis | +* | +* | + |
| SM0112E | Flavobacterium chryseum | + | – | +* |
| SM0117A | Exiguobacterium mexicanum | + | + | + |
| SM0118A | Glutamicibacter arilaitensis | + | +* | + |
(+/+*: producer / strong producer; -: non-producer)
The isolate SM011A was identified as Pseudomonas chlororaphis subsp. aureofaciens (MN192404). It was gram and endospore negative, catalase and oxidase positive with growth ranges of 4-25°C, pH 5-7 and 2-6% salt concentration. It was also remarkable producer of protease, lipase and amylase. Wang et al.18 were identified P. chlororaphis subsp. aureofaciens as plant growth stimulating bacteria and determined presence of cellulase and absence of protease. P. chlororaphis subsp. aureofaciens was reported as producer of natural antibiotics named phenazines which could control fungi, however it was not able to produce protease19. The isolate in this study was moderate psychrophilic with proteolytic activity18-22 however, this is the first report of amylase and lipase production from P. chlororaphis subsp. aureofaciens.
The non-motile isolate, SM012D, was similar to Psychrobacter faecalis (MN192405) optimum growth at 4-25°C, pH 7 and 2-15% salt concentration. It was gram and endospore negative, catalase and oxidase positive and had lipase and amylase production. The identification of P. faecalis was carried out from pigeon faeces by Kämpfer et al.23 and it was halotolerant and rarely opportunistic pathogen24. They were defined non-sporulating P. faecalis as gram negative, oxidase and catalase positive and non-motile. It had tolerance at 10% salt condition but intolerance at 15%. The growth temperature was varied between 4-36°C, however the isolate in this study could not survive at higher than 30°C25. P. faecalis in this study, was able to produce amylase and lipase however, there is no report about any enzyme production, until now.
The isolate SM015A, was identified as Rahnella aquatilis (MN192429), a member of Enterobacteriaceae family. It was motile, gram and endospore negative, catalase and oxidase positive with optimum growth at 4-25°C, pH 5-7 and 2-6% salt concentration. R. aquatilis was first isolated by Izard, PA26. It was gram negative and enteric bacteria but Harrell et al.27 were isolated from bronchial contaminant. The Rahnella species in the study of Demirci et al.28 were motile, gram and endospore negative, had antimicrobial activity against a forest pest. They could grow at 10-37°C, pH 4-9 and 3-10% NaCl concentration and produce amylase and protease enzymes. R. aquatilis could survive at washing room of a minimally proceed plant with a temperature of 5°C and was not able to produce protease29. Also R. aquatilis, which had antagonistic activity against olive knot disease was not able to produce protease30, amylase and lipase. However, R. aquatilis in this study, was determined with protease, lipase and strong amylase activities, so until now, this is the first report of lipase production from cold-adaptive R. aquatilis.
The motile isolate SM018A was similar to Shewanella putrefaciens (MN192428) with growth ranges of 4-25°C, pH 3-9 and 2-10% salt concentration. It was gram and endospore negative, catalase and oxidase positive. Braun, Sutherland31 incubated S. putrefaciens within a cocktail at 2-20°C, pH 4-7.5 and they could produce lipase and protease from this consortium. S. putrefaciens in this study was able to also produce amylase however, there is no available report about amylase production.
SM0110A was identified as Pseudomonas lactis (MN192431). It was gram and endospore negative, motile, catalase and oxidase positive. Optimum ranges for growth were 4-25°C, pH 5-7 and 2-6% NaCl concentration. P. lactis WS 4992T which had proteolytic and lipolytic activity was identified from raw bovine milk by von Neubeck et al.32 with optimum growth at 35°C, pH 5-8 and 0-6% NaCl concentration. Pseudomonas genus were known with proteolytic and lipolytic activity and psychrotolerant nature33,34 however, this is the first report on amylase production from P. lactis.
The motile isolate SM0112E was identified as Flavobacterium chryseum (MN192434). It was gram and endospore negative, catalase and oxidase positive, could survive at 4-25°C, pH 3-7 and 2-6% NaCl. The initial report of F. chryseum was arranged by Králová et al.35 and the isolate (CCM 8826T) was gram and endospore negative, catalase and oxidase positive. Optimum growth ranges were 1-25°C, pH 6-9 and only 1% NaCl concentration. The lipase production was declared35 but, our study is the first report of protease and amylase production from F. chryseum.
The isolate SM0117A was motile and identified as Exiguobacterium mexicanum (MN192433). It was gram, catalase and oxidase positive, endospore negative and could survive at 4-15°C, pH 5-9 and in the absence of salt. E. mexicanum was firstly described by López-Cortés et al.36 as a mesophilic bacteria which could grow at 20-41°C. It was gram, catalase and oxidase positive, motile and endospore negative. Orozco-Medina et al.37 were studied on protease production of E. mexicanum and Venkatachalam et al.38 were able to produce cold-adaptive protease, amylase and lipase form E. mexicanum.
The non-motile isolate SM0118A which was gram and catalase positive, endospore and oxidase negative, was identified as Glutamicibacter arilaitensis (MN192913). It could optimally grow at 4-20°C, pH 5-7 and 2-6% NaCl. The genus Glutamicibacter, which was belonging to the class of Actinobacteria was firstly proposed by Busse39. G. arilaitensis Re117T was able to grow optimum at 28°C, pH 5-10 and 0-10% NaCl40. Mesophilic G. arilaitensis ALA4 was able to produce protease and higher amounts of amylase41. However, there is no report of lipase production from G. arilaitensis.
A wide range of microorganisms from diverse habitats, permanently cold as well as those exposed to cold during a part of the year, are known to produce cold-active enzymes. In recent years, many studies have been conducted to find new cold-adapted enzymes or to extend the use of existing enzymes. Cold adaptive enzymes are preferred because of their low processing capacity and high catalytic activity. This is why psychrophilic or psychrotolerant enzymes replace most mesophilic enzymes today. There are studies indicating that psychrophilic or cold-adaptive enzymes are used to increase the effect of detergents at low temperatures and that some psychrophilic or psychrotolerant enzymes are commercially available as brands. Considering all these results, it is planned to investigate the potential of adaptability and usability in industrial applications since the bacteria identified in this study are adaptable to cold and whose industrial enzyme profiles are not known.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AM collected the water and sludge samples, MS carried out the study, GB and AS participated in design to draft, AA wrote the manuscript. All the authors read and approved the final manuscript
FUNDING
None.
AVAILABILITY OF DATA
All datasets obtained or studied during this study are incorporated in the manuscript.
ETHICS STATEMENT
This article does not contain any studies about human or animal objects.
- Madigan M.T., Martinko J.M., Brock T.D. Brock biology of microorganisms. Pearson Prentice Hall, Upper Saddle River, NJ, 2006.
Crossref - Arabaci N., Arikan B. Isolation and characterization of a cold-active, alkaline, detergent stable α-amylase from a novel bacterium Bacillus subtilis N8. Prep. Biochem. Biotechnol., 2018; 48(5): 419-426.
Crossref - D’Amico S., Collins T., Marx J.C., Feller G., Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Reports, 2006; 7(4): 385-389.
Crossref - Panikov N., Flanagan P., Oechel W., Mastepanov M., Christensen T. Microbial activity in soils frozen to below− 39 C. Soil Biol Biochem., 2006; 38(4): 785-794.
Crossref - Siddiqui K.S., Williams T.J., Wilkins D., Yau S., Allen M.A., Brown M.V., Lauro F.M., Cavicchioli R. Psychrophiles. Annual Review of Earth and Planetary Sciences, 2013; 41: 87-115.
Crossref - Pikuta E.V., Hoover R.B., Bej A.K., Marsic D., Whitman W.B., Krader P.E., Tang J. Trichococcus patagoniensis sp. nov., a facultative anaerobe that grows at− 5°C, isolated from penguin guano in Chilean Patagonia. International Journal of Systematic and Evolutionary Microbiology, 2006; 56(9): 2055-2062.
Crossref - Schmid A., Dordick J., Hauer B., Kiener A., Wubbolts M., Witholt B. Industrial biocatalysis today and tomorrow. Nature, 2001; 409(6817): 258.
Crossref - Sarmiento F., Peralta R., Blamey J.M. Cold and hot extremozymes: industrial relevance and current trends. Frontiers in Bioengineering and Biotechnology, 2015; 3: 148.
- Baltaci M.O., Genc B., Arslan S., Adiguzel G., Adiguzel A. Isolation and Characterization of Thermophilic Bacteria from Geothermal Areas in Turkey and Preliminary Research on Biotechnologically Important Enzyme Production. Geomicrobiol. J., 2017; 34(1): 53-62.
Crossref - Bowman J. Methods for psychrophilic bacteria. Methods in Microbiology, 2001; 30: 591-614.
Crossref - Altschul S.F., Gish W. Local alignment statistics, In: Methods in Enzymology, 1996; 266(27). pp 460-480. Academic Press.
Crossref - Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 1999; 27(2): 573.
Crossref - Prescott L.M., Harley J.P., Klein D.A. Laboratory exercises in microbiology. McGraw-Hill Companies, 2002.
- Sari B., Faiz O., Genc B., Sisecioglu M., Adiguzel A., Adiguzel G. New xylanolytic enzyme from Geobacillus galactosidasius BS61 from a geothermal resource in Turkey. International Journal of Biological Macromolecules, 2018; 119: 1017-1026.
Crossref - Buck J.D. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl. Environ. Microbiol., 1982; 44(4): 992-993.
Crossref - Albayrak S., Genc B., Ozkan H., Taskin M., Adiguzel A. Presence of Different Bacterial Species in Thermal Sources and Novelty in Their Industrial Enzyme Productions. Journal of Pure and Applied Microbiology, 2019; 13 (3): 1375-1387.
Crossref - Yanmis D., Baltaci M.O., Gulluce M., Adiguzel A. Identification of thermophilic strains from geothermal areas in Turkey by using conventional and molecular techniques. Res. J. Biotechnol., 2015; 10(1): 39-45.
- Wang X., Mavrodi D.V., Ke L., Mavrodi O.V., Yang M., Thomashow L.S., Zheng N., Weller D.M., Zhang J. Biocontrol and plant growth-promoting activity of rhizobacteria from Chinese fields with contaminated soils. Microbial Biotechnology, 2015; 8(3): 404-418.
Crossref - Raio A., Reveglia P., Puopolo G., Cimmino A., Danti R., Evidente A. Involvement of phenazine-1-carboxylic acid in the interaction between Pseudomonas chlororaphis subsp. aureofaciens strain M71 and Seiridium cardinale in vivo. Microbiol. Res., 2017; 199: 49-56.
Crossref - Haslun J.A., Ostrom N.E., Hegg E.L., Ostrom P.H. Estimation of isotope variation of N2O during denitrification by Pseudomonas aureofaciens and Pseudomonas chlororaphis: implications for N2O source apportionment. Biogeosciences (Online), 2018; 15(12).
Crossref - He F., Zhang Z., Cui M., Liu L., Xue Q. Soft rot disease alters soil characteristics and root-associated, culturable microbial community of Amorphophallus konjac. Journal of General Plant Pathology, 2018; 84(1): 44-57.
Crossref - Brilli F., Pollastri S., Raio A., Baraldi R., Neri L., Bartolini P., Podda A., Loreto F., Maserti B.E., Balestrini R. Root colonization by Pseudomonas chlororaphis primes tomato (Lycopersicum esculentum) plants for enhanced tolerance to water stress. J. Plant Physiol., 2019; 232: 82-93.
Crossref - Kampfer P., Albrecht A., Buczolits S., Busse H.-J. Psychrobacter faecalis sp. nov., a new species from a bioaerosol originating from pigeon faeces. Syst. Appl. Microbiol., 2002; 25(1): 31-36.
Crossref - Maria O.-A.J., Miguel S.-C.J., Fabiola G.-A., Elizabeth G.-D., Araceli R.-C., Patricia A.-P., Claudia W.-A., Maribel G.-V., Gloria L.-Á., Jeanette G.-C.A. Fatal Psychrobacter sp. infection in a pediatric patient with meningitis identified by metagenomic next-generation sequencing in cerebrospinal fluid. Archives of Microbiology, 2016; 198(2): 129-135.
Crossref - Deschaght P., Janssens M., Vaneechoutte M., Wauters G. Psychrobacter isolates of human origin, other than Psychrobacter phenylpyruvicus, are predominantly Psychrobacter faecalis and Psychrobacter pulmonis, with emended description of P. faecalis. International Journal of Systematic and Evolutionary Microbiology, 2012; 62(3): 671-674.
Crossref - Izard D., PA T. Rahnella aquatilis, nouveau membre de la famille des Enterobacteriaceae. 1979.
- Harrell L., Cameron M., O’hara C. Rahnella aquatilis, an unusual gram-negative rod isolated from the bronchial washing of a patient with acquired immunodeficiency syndrome. J. Clin. Microbiol., 1989; 27(7): 1671-1672.
Crossref - Demirci M., Sevim E., Demir I., Sevim A. Culturable bacterial microbiota of Plagiodera versicolora (L.)(Coleoptera: Chrysomelidae) and virulence of the isolated strains. Folia Microbiol., 2013; 58(3): 201-210.
Crossref - Meireles A., Fulgrncio R., Machado I., MergulhNo F., Melo L., Sim M. Characterization of the heterotrophic bacteria from a minimally processed vegetables plant. LWT-Food Science and Technology, 2017; 85: 293-300.
Crossref - Bouaichi A., Benkirane R., El-kinany S., Habbadi K., Lougraimzi H., Sadik S., Benbouazza A., Achbani E.H. Potantial effect of antagonistic bacteria in the management of olive knot disease caused by Pseudomonas savastanoi pv. savastanoi. The Journal of Microbiology, Biotechnology and Food Sciences, 2019; 8(4): 1035.
Crossref - Braun P., Sutherland J. Predictive modelling of growth and enzyme production and activity by a cocktail of Pseudomonas spp., Shewanella putrefaciens and Acinetobacter sp. International Journal of Food Microbiology, 2003; 86(3): 271-282.
Crossref - von Neubeck M., Huptas C., Gluck C., Krewinkel M., Stoeckel M., Stressler T., Fischer L., Hinrichs J., Scherer S., Wenning M. Pseudomonas lactis sp. nov. and Pseudomonas paralactis sp. nov., isolated from bovine raw milk. International Journal of Systematic and Evolutionary Microbiology, 2017; 67(6): 1656-1664.
Crossref - Tanaka C., Yamada K., Takeuchi H., Inokuchi Y., Kashiwagi A., Toba T. A lytic bacteriophage for controlling Pseudomonas lactis in raw cow’s milk. Appl. Environ. Microbiol., 2018; 84(18): e00111-00118.
Crossref - Hantsis-Zacharov E., Halpern M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol., 2007; 73(22): 7162-7168.
Crossref - Kralova S., Švec P., Busse H.-J., Stankova E., Vaczi P., Sedlacek I. Flavobacterium chryseum sp. nov. and Flavobacterium psychroterrae sp. nov., novel environmental bacteria isolated from Antarctica. International Journal of Systematic and Evolutionary Microbiology, 2018 68 (10): 3132-3139.
Crossref - Lopez-Cortes A., Schumann P., Pukall R., Stackebrandt E. Exiguobacterium mexicanum sp. nov. and Exiguobacterium artemiae sp. nov., isolated from the brine shrimp Artemia franciscana. Syst. Appl. Microbiol., 2006; 29(3): 183-190.
Crossref - Orozco-Medina C., Maeda-Martinez A.M., Cordova-Murueta J.H., Lopez-Cortes A., Navarrete-del-Toro M.d.l.A. Activity, molecular mass and hydrolysis on baker’s yeast protein of extracellular proteases from the putative probiotic bacteria Microbacterium sp. strain 8L and Exiguobacterium mexicanum strain 8N. Aquacult Res., 2009; 41(1): 45-52.
Crossref - Venkatachalam S., Gowdaman V., Prabagaran S.R. Culturable and culture-independent bacterial diversity and the prevalence of cold-adapted enzymes from the Himalayan mountain ranges of India and Nepal. Microb. Ecol., 2015; 69(3): 472-491.
Crossref - Busse H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. International Journal of Systematic and Evolutionary Microbiology, 2016; 66(1): 9-37.
Crossref - Feng W.-W., Wang T.-T., Bai J.-L., Ding P., Xing K., Jiang J.-H., Peng X., Qin S. Glutamicibacter halophytocola sp. nov., an endophytic actinomycete isolated from the roots of a coastal halophyte, Limonium sinense. International Journal of Systematic and Evolutionary Microbiology, 2017; 67(5): 1120-1125.
Crossref - Aarti C., Khusro A., Agastian P. Goat dung as a feedstock for hyper-production of amylase from Glutamicibacter arilaitensis strain ALA4. Bioresources and Bioprocessing, 2017; 4(1): 43.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.