ISSN: 0973-7510
E-ISSN: 2581-690X
The human cytomegalovirus (HCMV) is a global opportunistic β-herpes virus causing severe diseases in immune-compromised patients, such as malignant tumor patients, especially those undergoing chemotherapeutic treatment. This study aimed to determine the prevalence of HCMV-DNA in chemotherapeutic treatment naive cancer patients, and after chemotherapy, to compare between conventional nested PCR and ELISA techniques for the detection of HCMV, and to detect glycoprotein B genotypes. Plasma and serum samples before and after three chemotherapy cycles were collected from 49 chemotherapy-naive cancer patients. DNA was extracted from plasma samples using QIAamp® DNA Mini kit. HCMV-DNA was detected using a nested PCR technique. Multiplex nested PCR was used for HCMV-glycoprotein B (gB) genotyping. HCMV-IgG and -IgM were detected using ELISA technique. Thirty one (63.3 %) of the 49 plasma samples of the chemotherapy-naïve cancer patients were positive for HCMV-DNA; 21 of which remained positive after chemotherapy. However, 18 samples were negative of which 16 became positive after chemotherapy. gB-5 was the most common glycoprotein genotype detected (80.6 %), followed by gB-1, gB-3, gB-4, and gB-2. HCMV IgG was detected in the 49 serum samples of chemotherapy-naïve patients, and after exposure to chemotherapy. HCMV-DNA is commonly identified in cancer patients. Its detection after chemotherapy exposure may suggest HCMV reactivation. The most common genotype detected in cancer patients in Egypt is gB-5 in contrast to earlier research. IgG was detected in all patients. This indicates that HCMV is endemic in Egypt, necessitating the development of public awareness campaigns about HCMV infection and preventive strategies.
Human Cytomegalovirus, Genotyping, Cancer, ELISA, Nested PCR
Human cytomegalovirus (HCMV) is a global β-herpes virus that is highly pervasive. It is also known as human herpes virus 5 and belongs to the Herpesviridae family. The genus cytomegalovirus consists of a genome of approximately 240 kb encoding 165 genes, which is thought to be the biggest herpes virus infecting humans.1 HCMV is a well-known opportunistic pathogen that can cause severe diseases in immune-compromised patients. HCMV prevalence varies geographically and socioeconomically. Individuals from lower socioeconomic levels have a higher HCMV prevalence rate.2 Most primary infections caused by HCMV are either asymptomatic or subclinical, which mainly occur in childhood. Cytomegalovirus then enters into a latent state in both monocytes and macrophages.3,4 HCMV abandons latency and reactivates when the immune system of patients is compromised. HCMV reactivation prevalence in severely ill patients is very high, maybe up to 71%.4,5 Immune-compromised individuals, such as AIDS patients, recipients of organ transplantation or blood transfusion, newborns, and neonates often contract severe diseases caused by HCMV.6,7 Individuals experiencing HCMV reactivation may experience generalized symptoms, such as malaise, fever, leukopenia, as well as HCMV-associated syndromes, such as hepatitis, meningitis, pneumonitis, encephalitis, enterocolitis, gastroenteritis, nephritis, or retinitis.8-10 It may also cause severe fatal systemic infections in immune-compromised patients. Malignant tumor patients, especially those undergoing chemotherapeutic treatment, are an important sector in immune-compromised individuals who are vulnerable to HCMV infection and reactivation.11-14 It was demonstrated that HCMV has oncogenic transforming potential in vitro.15,16 HCMV diseases incidence and prognosis are frequently related to the immunosuppression level, host susceptibility factors, and the virulence of different HCMV strains.17 Genetic diversity is found among genes involved in tissue tropism and cell penetration or replication affecting HCMV strains virulence.18 HCMV pathogenicity varies according to the viral genetic variation within viral genes.19 Gene coding for the viral envelope glycoproteins express great genetic diversity among HCMV genes, e.g., UL55 which encodes glycoprotein B (gB).20 gB genotyping is composed of five genotypes, numbered 1–5.21 The main component of viral envelopes are glycoproteins. They are important for viral penetration into the cell and viral transmission from cell to another.22 A person can be infected by several HCMV strains during their lifetime.23
People of all ages can acquire HCMV infection according to the Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO). It was reported in the United States that more than half the adults infected with HCMV aged 40 years, and nearly 33% of children infected with HCMV aged five years. The most common infectious cause of birth defects in the United States is HCMV. The CDC made June as “National CMV Awareness Month” to increase awareness of congenital cytomegalovirus. In Africa, HCMV is neglected due to a widespread belief that HCMV is endemic from childhood, there is less possibility of maternal reactivation or reinfection during pregnancy, and as a consequence, there is a lower incidence for severe congenital infection to occur compared to primary infection.24-26 Maternal reactivation or reinfection causes majority of congenital HCMV infections even in high-income populations.27 In addition, African patients might not receive immunosuppressive therapy that may cause HCMV diseases, which is becoming increasingly obsolete as several advanced therapies for non-transmissible diseases become more widely available.28,29 Finally, diagnosis and treatment of HCMV-related diseases are largely ignored.30 In literature, much of the recent research in Egypt addresses infections in those undergoing transplantation, AIDS/HIV, and pregnancy. However, there is limited data on HCMV DNAemia in cancer patients undergoing chemotherapeutic treatment.
Aim of the Study
The current study aimed to determine the prevalence of HCMV DNAemia in chemotherapeutic treatment naïve cancer patients. Moreover, it aimed to investigate the effect of chemotherapeutic treatment on HCMV DNAemia and to compare between conventional nested PCR and ELISA techniques for HCMV reactivation detection. In addition, it aimed to detect glycoprotein B genotypes and the prevalence of HCMV-IgG antibodies in these patients.
Patients and Sample Collection
Plasma and serum samples were collected from 49 chemotherapy-naïve cancer patients before and after three chemotherapy cycles (each cycle = 21 days) from December 2018 to August 2019 at the Damanhur Oncology Center. Plasma and serum samples were kept in a −80°C freezer until the DNA was extracted for PCR processing and ELISA, respectively.
DNA Extraction and Nested PCR for HCMV Detection
The QIAamp® DNA Mini kit (QIAGEN, Hilden, Germany) was used according to the manufacturer’s instructions to extract DNA from plasma samples and then were kept at −80°C until PCR processing. HCMV-DNA was identified using a nested PCR technique in both chemotherapy-naïve and post-three-cycle-chemotherapy samples.31 Two sets of primers were designed to target the fourth exon of the HCMV Immediate Early gene.31,32 The primer sequences were as follows: the external primers: 1a: 5ʹ-GGTCACTAGTGACGCTTGTATGATGA-3ʹ and 1b: 5ʹ-GATAGTCGCGGGTACA GGGGACTCT-3ʹ; the internal primers: 2a: 5ʹ-AAGTGAGTT CTGTCGGGTGCT-3ʹ and 2b: 5ʹ- GTGACACCAGAGAATCAGAGGA-3ʹ. A thermal cycler (BOECO, Hamburg. Germany) was used for PCR amplification. Briefly, the first step of PCR was performed in a 25 μL total volume comprising 1 μL of forward and reverse primers (10 pmol/μL), 3 μL of plasma DNA extract, 7.5 μL of free RNase water, and 12.5 mL of MyTaqTM HS Red Mix. The thermal cycling conditions were an initial denaturation at 95°C for 1 min, followed by 35 cycles [denaturation: 95°C for 15 s, annealing: 55°C for 15 s, and extension: 72°C for 10 s], and followed by final extension at 72°C for 10 min. The second step of PCR was similar to the first, but with the following differences: an initial denaturation step of 95°C for 3 min, 3 μL of the first step amplicon as a template, an annealing temperature of 53°C for 15 s 10 μL of PCR products were visualized via ultraviolet illumination (Analytik Jena, Germany) of 2% agarose gel (GeneDireX, USA) stained with ethidium bromide (Sigma-Aldrich, US).
Glycoprotein B Genotyping
HCMV DNAemia positive samples of chemotherapy-naïve cancer patients were subjected to the multiplex nested PCR (M-nPCR) assay for gB genotyping.33 The M-nPCR for gB genotype used primers illustrated in (Table 1). The PCR technique was carried out as reported by Pignatelli et al. and Tarrago et al.34,35 A PCR master mix [One PCR, Master mix (GENEDIREX)] was used for the amplification. Concisely, the first PCR step was performed in a 25 μL total volume containing 1 μL of each forward and reverse primers (10 pmol/μL), 3 μL of the DNA extract, 7.5 μL of free RNase water and 12.5 μL of master mix. The thermal cycling condition for the first PCR step was as follows: denaturation: 94°C for 4 min, followed by 35 cycles of [denaturation: 94°C for 30 s, annealing: 60°C for 1 min, extension: 72°C for 1 min], and final extension at 72°C for 10 min. The second PCR step was performed in 20 μL containing 10 μL of master mix, 5 μL of the first step amplicon as a template, 0.5 μL of the five forward primers, and 2.5 μL reverse primer (10 pmol/μL). The second step was performed at the same thermal cycling conditions. However, the annealing temperature was at 58°C. Ultraviolet illumination (Analytik Jena, Germany) of 2.5% agarose gel (GeneDireX, USA) stained with ethidium bromide (Sigma-Aldrich, US) were used to visualize PCR products.
Table (1):
Primer’s sequences used in HCMV glycoprotein b genotyping by multiplex nested PCR.
| Gene | primer sequence | product size (bp) | |
|---|---|---|---|
| PCR for HCMV detection | UL55 | UL55-F: 5`-TTTGGAGAAAACGCCGAC- 3` | 751 |
| UL55-R: 5`- CGCGCGGCAATCGGTTTGTTGTA- 3 | |||
| PCR for Glycoprotein B genotyping | gB1 | gB1-F: 5`-ATGACCGCCACTTTCTTATC- 3` gB1-R: 5`- GTTGATCCACRCACCAGGC- 3` |
420 |
| gB2 | gB2-F: 5`- TTCCGACTTTGGAAGACCCAACG- 3` gB2-R: 5`- GTTGATCCACRCACCAGGC- 3` |
613 | |
| gB3 | gB3-F: 5`- TAGCTCCGGTGTGAACTCC- 3` gB3-R: 5`- GTTGATCCACRCACCAGGC- 3` |
190 | |
| gB4 | gB4-F: 5`- ACCATTCGTTCCGAAGCCGAGGAGTCA -3` gB4-R: 5`- GTTGATCCACRCACCAGGC- 3` |
465 | |
| gB5 | gB5-F: 5`- TACCCTATCGCTGGAGAAC-3` gB5-R: 5`- GTTGATCCACRCACCAGGC- 3` |
139 |
Serology
Serum samples were collected from each chemotherapy-naïve patient and after three cycles of chemotherapy for serological analysis. Samples were subsequently kept at −80°C until ELISA processing. Prechek® (HCMV IgG EIA test kit, Prechek Inc, USA) and a Microtitre plate ELISA reader (BioTek Instruments, Inc, USA) were used to identify IgG antibodies against HCMV. Absorbance was measured at 450 nm. IgM antibodies to HCMV were identified in serum samples after chemotherapy treatment using a commercially available HCMV-IgM ELISA kit (EIA KIT, Prechek, Inc, USA) according to the manufacturer’s instructions, and Microtitre plate ELISA reader (BioTek Instruments, Inc, USA). Absorbance was measured at 450 nm.
Statistical Analysis
The IBM SPSS software program version 20.0 was used for all statistical analyses (IBM Corporation, Armonk, New York). A p-value below 0.05 was considered statistically significant.
Patients and Sample Collection
Plasma and serum samples were collected from 49 chemotherapy-naïve cancer patients aged 25–65 years old. A total of 44 patients were females and 5 were males having different types of cancers (Table 2).
Table (2):
Characteristics of patients enrolled in this study.
| Demographic Data | No. (%) | |
|---|---|---|
| Gender | Male | 5(10.2%) |
| Female | 44(89.8) | |
| Age (yrs) | 25 – <45 | 16 (32.7%) |
| 45 – <65 | 29(59.2%) | |
| ≥65 | 4(8.2%) | |
| Cancer types | Breast | 27(55.1%) |
| NHL | 13(26.5%) | |
| Ovary | 4(8.2%) | |
| Colorectal | 4(8.2%) | |
| Uterus | 1(2.0%) | |
Detection of HCMV DNAemia using Nested PCR Before and After Chemotherapy
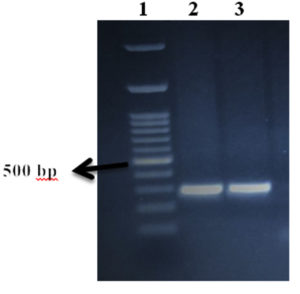
A total of 31 (63.3 %) of the 49 plasma samples taken from the chemotherapy-naïve cancer patients involved in this research were positive for HCMV DNAemia by conventional nested PCR, and 18 (36.7%) were negative. The bands were detected at 293 bp, as anticipated (Figure 1). A total of 17 (55%) out of the 31 HCMV-DNA-positive patients, had breast cancer, 10 (32%) had NHL, 2 (6.5%) had colorectal cancer, and 2 (6.5%) had ovarian cancer.
Figure 1. Detection of HCMV DNAemia using nested PCR. PCR products were visualized in 2% agarose gel with ethidium bromide staining. Lane 1: a 100 bp DNA ladder, Lanes 2 and 3: positive samples detected at 293 bp
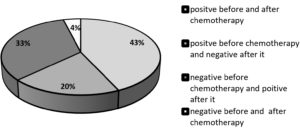
Following three cycles of chemotherapy, samples were obtained from the same patients for HCMV DNAemia detection. A total of 10 samples out of the 31 positive HCMV DNAemia were negative (became latent), 21 samples remained positive after chemotherapy exposure. On the other hand, out of the 18 negative HCMV DNAemia chemotherapy-naive, 16 samples were positive (reactivation or reinfection), and two remained negative after chemotherapy exposure. Therefore, 16/49 (32.7%) represented reactivation or reinfection, and 10/49 (20.4%) entered into latency stage (Figure 2).
Glycoprotein B (gB) Genotyping
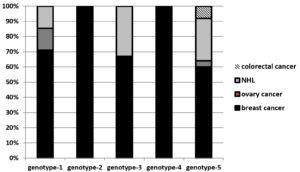
Multiplex nested PCR was used to detect gB genotypes in the 31 HCMV positive samples collected from the chemotherapy-naïve cancer patients. Various HCMV genotypes were detected as follows: gB-5 was the most common glycoprotein genotype accounting for 25/31 (80.6 %), followed by gB-1: 7/31 (22.6%), gB-3 6/31 (19.4), gB-4 2/31 (6.5%), and gB-2 1/31 (3.2%) (Figure 3). Out of the 31 samples, seven (22.5 %) samples harbored a mixture of two or three genotypes. All specimens of mixed genotypes surprisingly belonged to breast cancer patients (Table 3).
Table (3):
Distribution of mixed glycoprotein B genotypes among 7 breast cancer chemotherapy naive patients.
Isolate no. |
Glycoprotein B genotypes |
|---|---|
1 |
gB-2, gB-4, gB-5 |
10 |
gB-1, gB-5 |
14 |
gB-4, gB-5 |
15 |
gB-1, gB-5 |
19 |
gB-1, gB-3, gB-5 |
31 |
gB-1, gB-3 |
40 |
gB-1, gB-3, gB-5 |
Figure 3. Distribution of HCMV glycoprotein B genotypes among the 31 PCR positive chemotherapy naïve patients with different cancer types
ELISA
Remarkably, all the 49 (100 %) chemotherapy-naïve patients’ serum samples were positive for HCMV-IgG antibodies using the ELISA technique. All samples results also came out positive for HCMV-IgG antibodies after exposure to chemotherapy. HCMV-IgM was also detected in three (6.1%) of the 49 samples after exposure to chemotherapy. Unfortunately, there was no significant relationship (X2 = 0.135, p = 1.000) between the results of IgM detection by ELISA and HCMV detection by PCR (Table 4).
Table (4):
Comparison between IgM detection by ELISA and HCMV detection by PCR after chemotherapy exposure.
| ELISA | Total | |||
|---|---|---|---|---|
| ELISA negative | ELISA positive | |||
| PCR | PCR negative | 11 | 1 | 12 |
| PCR positive | 35 | 2 | 37 | |
| Total | 46 | 3 | 49 | |
HCMV is a prominent human infection that affects the vast majority of the world’s population. Examples of instances where HCMV reactivation is intentionally sought include pregnancy, hematopoietic stem cell and solid organ transplantation, HIV infection, and recipients on immunosuppressive medications.35,36 Immunosuppression-induced HCMV reactivation results in a severe clinical presentation. HCMV has been shown to have oncogenic transforming potential in vitro.16,37 In addition, HCMV has been studied as an oncomodulatory virus and is an important factor in prognosis and survival in immune compromised patients. Preventive/prophylactic methods have resulted in a considerable decrease in HCMV-related mortality and morbidity. It was reported that chemotherapy causes immunosuppression in cancer patients.38 Unfortunately, very few studies regarding HCMV infections and related morbidities due to reactivation were reported in the literature. There is no documented report regarding HCMV reactivation among cancer patients undergoing chemotherapeutic treatment in Egypt. This study aimed to determine the incidence HCMV reactivation after chemotherapy, glycoprotein (gB) genotypes among cancer patients, and serological prevalence of HCMV antibodies.
In this study, we collected 49 samples from chemotherapy-naïve cancer patients and after chemotherapy exposure in Egypt. Our results revealed that HCMV DNAemia was detected in 31/49 (63.3%) chemotherapy-naïve patients using conventional nested PCR. On the other hand, El Shazly et al. reported that 20% of their chemoradiotherapy naive breast cancer patients had HCMV-DNA in their blood samples.36 In 2020, Lv et al. reported that 35.66% of the gastrointestinal cancer patients and 9.74% of the control individuals were HCMV-DNA positive. Therefore, they supposed that HCMV infection is related with an increased risk of gastrointestinal risk.37 In addition, Handous et al. reported that 19.35% of patients with lymphoma revealed presence of HCMV-DNA with a significant higher frequency than the control group.38 Torres et al. reported that HCMV-DNA was significantly associated with non-Hodgkin’s lymphoma especially at active and late-stage lymphoma in another study on lymphoma patients.39 Several studies had detected HCMV in malignant glioma, glioblastoma, colon cancer, salivary gland cancer, and prostate carcinoma.40-44 In contrast, El-Shinawi et al. were unable to detect HCMV-DNA in the peripheral blood of their 77 breast cancer patients. However, they detected it in the breast cancer tissue of 62.3% tested females with a higher percent in inflammatory breast cancer revealing HCMV contribution in cancer pathogenesis and prognosis.29 A possible explanation for the oncogenic potential of HCMV is that the virus has the power to manage apoptosis and evade immune surveillance, providing an advantage of survival to the infected cells. Viral proteins were identified in several cells, such as smooth muscle cells, inflammatory cells, tumor cells, epithelial cells, and blood vessel walls.45 HCMV can infect different cell types and enter the latency phase in myeloid progenitor cells, specifically CD34+ cells.46-47
HCMV DNAemia and HCMV-IgM seroprevalence among cancer patients after exposure to chemotherapy in Egypt in this study were detected using both PCR and ELISA, respectively. Conventional nested PCR analysis revealed that 37/49 (75.5%) cancer patients were HCMV DNAemia positive after chemotherapy exposure. In terms of prognosis, 16/49 (32.7%) of cancer patients turned from negative HCMV DNAemia before chemotherapy exposure to positive after exposure which may be due to either reactivation or reinfection. On the other hand, 10/49 (20.4%) cancer patients turned from HCMV DNAemia positive before chemotherapy exposure to negative after exposure which may be due to the viral latency. In addition, 21/49 (42.9%) of cancer patients were positive for HCMV DNAemia before and after chemotherapy exposure, and 2/49 (4.1%) cancer patients were negative for HCMV DNAemia before and after chemotherapy exposure. HCMV-DNA presence in plasma indicates human cell disturbance due to viral replication. Besides, it is thought that the presence of DNA in serum is closely linked to symptomatic infection.48,49 Similar to our results, HCMV reactivation upon chemotherapy exposure was reported in a previous pilot study on 15 cancer patients.50 HCMV reactivation was observed in all their patients except for one throughout the chemotherapy treatment course with viral load peaking during the third treatment session. In addition, Schlick et al. reported that HCMV reactivation was observed in 12% of participants in another retrospective analysis of hematological and oncological patients.51 Besides, 41% of adult patients with solid tumors showed positive HCMV-DNA after chemotherapy exposure.52 It was reported that HCMV reactivation was not only present in terminally advanced cancer patients, but with immune-competent ones as well making it necessary to suspect HCMV with pyrexia of unknown origin.39,53-56 HCMV reactivation could not be detected in any of the 93 cancer patients with mild to moderate level of immunosuppression after chemotherapy exposure, which is contrary to our results.57 Schlick et al. reported the case study of a patient with fever, pancreatic adenocarcinoma, and metastasis to the liver, lungs, and peritoneal cavity. He succumbed with sepsis after chemotherapy, and HCMV was the only pathogen detected in post-mortem blood cultures.51 Previous findings showed that cancer patients are at an increased HCMV reactivation risk and its associated mortality.14,58
In this study, HCMV-specific IgM was detected in 3/49 (6.1%) of cancer patients after chemotherapy exposure. Notably, all these positive HCMV-IgM patients were NHL patients. Similarly, Salman et al. reported that 8.45% of the breast cancer patients under their study were HCMV-IgM positive.59 In addition, El Shazly reported that none of their breast cancer patients were HCMV-IgM positive.36 Moreover, IgM was not detected in any of the patients in a study of 130 patients with benign and malignant breast tumors.60 In addition, Rådestad et al. reported that 12% of ovarian cancer patients were HCMV-IgM positive in comparison to 3% of benign tumor patients.61 About 76.6% of the HCMV-IgM positive patients had breast cancer with invasive ductal carcinoma, which is in contrast to our findings.62
In our study, one of the three positive HCMV-IgM samples (33.3%) was considered false positive as they were HCMV-IgM positive and HCMV-DNA negative. The difficulty in detecting HCMV-DNA in IgM positive patients could be related to the persistence of IgM antibodies for a long time after primary infection.63,64 HCMV-specific IgM can be detected for an average of 6 to 9 months after the initial infection has resolved.65 The presence of HCMV-IgM in serum samples may be due to reinfection or reactivation, and not restricted to primary infection. Therefore, a follow-up test as molecular tests should be performed to ascertain the time of HCMV infection when IgM is detected.66 In contrast, conventional nested PCR revealed positive HCMV DNAemia in 35/46 of the negative HCMV-IgM patients, indicating that IgM tests may be negative or under-detectable in immune-compromised patients who are actively infected67 or re-infected and peaks within few weeks after infection.68 Therefore, serological techniques alone are not reliable in detecting active cytomegalovirus infection. Molecular approaches should be performed for accurate diagnosis.69
Previous research in literature revealed that HCMV strains differ genetically, which greatly affects disease pathogenesis and progression. Furthermore, the HCMV potential to infect numerous organs and cells have been linked to diversity in gene sequence between strains.23,70,71 Certain areas exhibit higher rates of mutation, although the genomes of different HCMV strains are 95% identical.72,73 One of the most well-studied polymorphic gene is UL55, encoding the viral gB which is required for viral entry, cell fusion, and the key motive for neutralization.73,74 Our results showed that the genotype gb-5 is the most predominant in contrast to previous literature; 80.6% of HCMV-DNA positive patients in this study. In contrast to our findings, Mohamed et al. reported the prevalent distribution of gB-1 in their breast cancer patients.73 In addition, Nogueira et al. also reported the prevalence of gB-1 in kidney transplant recipients, and solid organ transplant recipients with accelerated development of invasive diseases in Brazil.75 On the other hand, the prevalence of gB2 among AIDS patients was reported by other studies in Canada, Brazil, United States, and Austria.21,76-79 Moreover, Dieamant et al. reported that hematopoietic stem cell transplantation patients with gB-3 genotype had higher morbidity and mortality than patients with the gB-1, gB-2, and gB-4 genotypes.80 The existence of HCMV-gB-3 aggressive genotype in IBC patients may alter the disease’s poor prognosis and morbidity.73 The aggressive activity of HCMV-gB-3 could be attributed to its unique biological pathways involved in host-virus interactions.81
Mixed HCMV infection with more than one genotype of HCMV-gB was detected in seven samples of HCMV-DNA positive chemotherapy-naïve cancer patients in this present study. Remarkably, all detected mixed genotypes were breast cancer patients. In fact, it was reported that mixed infections of different HCMV genotypes were detected in both healthy and immune-compromised individuals, including pulmonary transplanted patients,82 AIDS patients,73,74 and inflammatory breast cancer patients.73 The mixture of strains was related with higher morbidity and mortality in solid organ transplant patients,83,84 higher disease progression in inflammatory breast cancer.73, fetal death during pregnancy,85 higher viral load, postponed viral eradication, and a higher rate of viral recurrence following treatment in both transplant recipients and AIDS patients.86 The increased pathogenesis of HCMV caused by mixed infection of different strains may cause the secretion of chemokines, cytokines, and growth factors as a result of distinct viral strain replication.87 In this study, out of 31 samples, 7 (22.5%) had mixed genotypes with the following distribution: 2 samples had (gB-1+gB-5), 2 had (gB-1+gB-3+gB-5), 1 had (gB-1+gB-3), 1 had (gB-4+gB-5), and 1 had (gB-2+gB-4+gB-5). Notably, six of these seven samples with mixed genotypes contained the gB-5. Wu et al. reported that a mixture of genotypes gB-1 and gB-3 was more commonly detected in hematopoietic stem cell transplant patients, and gB-3 was linked to the incidence of pneumonia.88
Surprisingly, serological detection of IgG antibodies for HCMV in chemotherapy-naïve cancer patients’ samples in this study revealed 100% HCMV-IgG prevalence. All patients except one were HCMV-IgG positive after exposure to chemotherapy. Similarly, both Mohammed et al. and el Shazly et al. reported 100% HCMV-IgG prevalence among invasive ductal carcinoma patients.36,62 There were significant changes when they compared ELISA readings of IgG optical densities between breast cancer patients and healthy women (p = 0.05). Similarly,32 Fagundes et al. reported that the HCMV antibody titer was considered to be an important objective indicator of fatigue in their study.89 HCMV-IgG antibodies were detected in 49 (70%) of their 70 serology samples in another study reported by Richardson et al.; however, HCMV-DNA was not detected in any of the tumor samples by QPCR.90 Furthermore, IgG levels were raised to 81.69% in breast cancer patients, compared to only 40% of the control group in another study reported by Salman et al.59 Radestad et al. reported that a higher HCMV-IgG level was linked to a higher disease stage as HCMV-IgG level was greater in ovarian cancer patients (p = 0.002) or benign cystadenoma patients than control groups (P.0001).61
In contrast, Pandey et al. demonstrated that HCMV-IgG level was much greater in cancer-free people than in breast cancer patients, supporting the idea that HCMV host immunity may play a role in keeping people cancer-free.91 Moreover, 96.6 % of the subjects tested positive for HCMV-IgG in a study of voluntary blood donors from the Alexandria Regional Blood Transfusion Centre.92 In addition, in a study of 130 patients that had breast swelling and either malignant or benign surgical excision, Surendran & Chisthi reported that HCMV-IgG antibodies were detected in all investigated patients. Therefore, they inferred that there was no correlation between HCMV-IgG seropositivity and breast cancer.60 Moreover, HCMV-IgG antibodies were detected in 67/71 (94.36 %) Iraqi breast cancer patients, and in 19/20 (95%) of the control group in the immunological phase of a study assessing HCMV seroprevalence. On the other hand, in a study of 37 patients with colon adenocarcinoma, Avni et al. reported that an increased HCMV-IgG antibody titer only in patients treated with chemotherapy. This was often due to secondary infections due to the chemotherapy immunosuppression and is not associated with the colorectal cancer. Numerous detection methods, such as immunohistochemistry and DNA hybridization revealed that HCMV was not discovered at a significantly greater incidence in cancer tissue than normal tissue in subsequent studies.93
HCMV-DNA is commonly identified in cancer patients. Its detection after chemotherapy exposure may suggest HCMV reactivation or reinfection. The most common genotype detected in cancer patients in Egypt is gB-5 in contrast to earlier research, indicating that the prevalence of this genotype requires additional investigations. IgG was detected in all patients indicating that HCMV is endemic in Egypt, necessitating the development of public awareness campaigns about HCMV infection and preventive strategies. HCMV-specific IgM was detected in 6.1% of patients after chemotherapy. Therefore, we recommend HCMV screening for cancer patients suffering from a fever of unknown etiology. Larger investigations are needed to determine the risk factors for HCMV infection. The rising number of older people getting chemotherapy, along with the fact that HCMV prevalence rises with age, predicts that HCMV response and disease will become more common in the future.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ISS, RA, KB, AAS and SMA contributed to the study concept and design. ISS and KB prepared material, collected data and performed analysis. ISS drafted the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Research Ethics Committee of the Faculty of Pharmacy, Damanhur University, Egypt with approval reference number 109PM12.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Nikolich-Zugich J, van Lier RAW. Cytomegalovirus (CMV) research in immune senescence comes of age: overview of the 6th International Workshop on CMV and Immunosenescence. GeroScience. 2017;39(3):245-249.
Crossref - Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20(4):202-213.
Crossref - Kondo K, Kaneshimat H, Mocarski ES. Human cytomegalovirus latent infection of granulocyt macrophage progenitors (herpvs/gene exprdon/myelold/persstent infection). Proc Nati Acad Sci USA. 1994;91(December):11879-11883.
Crossref - Lachance P, Chen J, Featherstone R, Sligl WI. Association between cytomegalovirus reactivation and clinical outcomes in immunocompetent critically ill patients: A systematic review and meta-analysis. Open Forum Infectious Diseases. 2017;4(2):1-9.
Crossref - Papazian L, Hraiech S, Lehingue S, et al. Cytomegalovirus reactivation in ICU patients. Intensive Care Med. 2016;42(1):28-37.
Crossref - H Al Mana, Yassine HM, Younes NN, et al. The current status of cytomegalovirus (CMV) prevalence in the MENA region: A systematic review. Pathogens. 2019;8(4):213.
Crossref - Sylwester AW, Mitchell BL, Edgar JB, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202(5):673-685.
Crossref - Hebart H, Einsele H. Clinical aspects of CMV infection after stem cell transplantation. Hum Immunol. 2004;65(5):432-436.
Crossref - Hsu JW, Hiemenz JW, Wingard JR, Leather H. Viral infections in patients with hematological malignancies. Neoplastic Diseases of the Blood. 2018;1079-127.
Crossref - Wang YC, Lee HS, Lin TY, Wang NC. Cytomegalovirus colitis mimics amebic colitis in a man with AIDS. Am J Med Sci. 2008;336(4):362-364.
Crossref - Breathnach OS, Donnellan P, Collins D, McNicholas W, Crown J. Cytomegalovirus pneumonia in a patient with breast cancer on chemotherapy: Case report and review of the literature. Ann Oncol. 1999;10(4):461-465.
Crossref - Chen IH, Lai YL, Wu CL, et al. Immune impairment in patients with terminal cancers: Influence of cancer treatments and cytomegalovirus infection. Cancer Immunol Immunother. 2010;59(2):323-334.
Crossref - van den Brande J, Schrijvers D, Colpaert C, Vermorken JB. Cytomegalovirus colitis after administration of docetaxel-5- fluorouracil-cisplatin chemotherapy for locally advanced hypopharyngeal cancer. Ann Oncol. 1999;10(11):1369-1372.
Crossref - Wang YC, Wang NC, Lin JC, et al. Risk factors and outcomes of cytomegalovirus viremia in cancer patients: A study from a medical center in northern Taiwan. J Microbiol Immunol Infect. 2011;44(6):442-448.
Crossref - Heybar H, Alavi SM, Nejad MF, Latifi M. Cytomegalovirus infection and atherosclerosis in candidate of coronary artery bypass graft. Jundishapur J Microbiol. 2015;8(3).
Crossref - Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G. HCMV Activates the IL-6-JAK-STAT3 Axis in HepG2 Cells and Primary Human Hepatocytes. PLoS ONE. 2013;8(3):59591.
Crossref - Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis. 2004;39(2):155-161.
Crossref - Halary F, Amara A, Lortat-Jacob H, et al. Human Cytomegalovirus Binding to DC-SIGN Is Required for Dendritic Cell Infection and Target Cell trans-Infection. Immunity. 2002;17(5):653-664.
Crossref - Dolan A, Cunningham C, Hector RD, et al. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85(5):1301-1312.
Crossref - Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol. 2004;14(6):383-410.
Crossref - Shepp DH, Match ME, Lipson SM, Pergolizzi RG. A fifth human cytomegalovirus glycoprotein B genotype. Res Virol. 1998;149(2):109-114.
Crossref - Feire AL, Roy RM, Manley K, Compton T. The Glycoprotein B Disintegrin-Like Domain Binds Beta 1 Integrin To Mediate Cytomegalovirus Entry. J Virol. 2010;84(19):10026-10037.
Crossref - Meyer-Konig U, Ebert K, Schrage B, Pollak S, Hufert FT. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet. 1998;352(9136):1280-1281.
Crossref - Foulon I, Naessens A, Foulon W, Casteels A, Gordts F. A 10-Year Prospective Study of Sensorineural Hearing Loss in Children with Congenital Cytomegalovirus Infection. J Pediatr. 2008;153(1):84-88.
Crossref - Ross SA, Fowler KB, Ashrith G, et al. Hearing loss in children with congenital cytomegalovirus infection born to mothers with preexisting immunity. J Pediatr. 2006;148(3):332-336
Crossref - Fowler K, Stagno S, Pass R, Britt W, Boll T, Alford C. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. Int J Gynaecol Obstet. 1992;39(2):153.
Crossref - de Vries JJC, van Zwet EW, Dekker FW, Kroes ACM, Verkerk PH, Vossen ACTM. The apparent paradox of maternal seropositivity as a risk factor for congenital cytomegalovirus infection: a population-based prediction model. Rev Med Virol. 2013;23(4):241-249.
Crossref - Ajayi SO, Raji Y, Salako BL. Renal Data from Asia-Africa Ethical and Legal Issues in Renal Transplantation in Nigeria. Saudi J Kidney Dis Transpl. 2016;27(1):125-128.
Crossref - Spearman CWN, McCulloch MI. Challenges for paediatric transplantation in Africa. Pediatr Transplant. 2014;18(7):668-674.
Crossref - Bates M, Brantsaeter AB. Human cytomegalovirus (CMV) in Africa: a neglected but important pathogen. J Virus Erad. 2016;2(3):136-142.
Crossref - Zhang S, Zhou YH, Li L, Hu Y. Monitoring human cytomegalovirus infection with nested PCR: Comparison of positive rates in plasma and leukocytes and with quantitative PCR. Virol J. 2010;7:73.
Crossref - El-Shinawi M, Mohamed HT, El-Ghonaimy EA, et al. Human Cytomegalovirus Infection Enhances NF-κB/p65 Signaling in Inflammatory Breast Cancer Patients. PLoS ONE. 2013;8(2):e55755.
Crossref - Tarrago D, Quereda C, Tenorio A. Different cytomegalovirus glycoprotein B genotype distribution in serum and cerebrospinal fluid specimens determined by a novel multiplex nested PCR. J Clin Microbiol. 2003;41(7):2872-2877.
Crossref - Pignatelli S, Maurizio D, Ladini MP, Monte PD. Development of a multiplex PCR for the simultaneous amplification and genotyping of glycoprotein N among human cytomegalovirus strains. New Microbiologica. 2010;33(3):257-262.
- Ramanan P, Razonable RR. Cytomegalovirus infections in solid organ transplantation: A review. Infect Chemother. 2013;45(3):260-271.
Crossref - Mocarski ES. Cytomegaloviruses and their replication. Fields Virology. 1996;2:2447-2492.
- Clanton DJ, Jariwalla RJ, Kress C, Rosenthal LJ. Neoplastic transformation by a cloned human cytomegalovirus DNA fragment uniquely homologous to one of the transforming regions of herpes simplex virus type 2. Proce Natl Acad Sci USA. 1983;80(12):3826-3830.
Crossref - Brower V. Accidental passengers or perpetrators? Current virus-cancer research. J Natl Cancer Inst. 2004;96(4):257-258.
Crossref - El Shazly DF, Bahnassey AA, Omar OS, et al. Detection of Human Cytomegalovirus in Malignant and Benign Breast Tumors in Egyptian Women. Clin Breast Cancer. 2018;18(4):629-642.
Crossref - Lv YL, Han FF, An ZL, et al. Cytomegalovirus Infection Is a Risk Factor in Gastrointestinal Cancer: A Cross-Sectional and Meta-Analysis Study. Intervirology. 2020;63(1-6):10-16.
Crossref - Handous I, Hannachi N, Achour B, et al. Presence of cytomegalovirus and human herpesvirus 6 in-patients with lymphomas. Int J Infect Dis. 2020;101:298.
Crossref - Torres HA, Kontoyiannis DP, Aguilera EA, et al. Cytomegalovirus infection in patients with lymphoma: An important cause of morbidity and mortality. Clinical Lymphoma and Myeloma. 2006;6(5):393-398.
Crossref - Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170(3):998-1002.
Crossref - Cobbs CS, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62(12):3347-3350. PMID: 12067971
- Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: Increasing evidence and open questions. Neoplasia. 2009;11(1):1-9.
Crossref - Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157(2):193-203.
Crossref - Taher C, de Boniface J, Mohammad AA, et al. High Prevalence of Human Cytomegalovirus Proteins and Nucleic Acids in Primary Breast Cancer and Metastatic Sentinel Lymph Nodes. PLoS ONE. 2013;8(2):e56795.
Crossref - Straat K, Liu C, Rahbar A, et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst. 2009;101(7):488-497.
Crossref - Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87(7):1763-1779.
Crossref - Soderberg-Naucler C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91(1):119-126.
Crossref - Tang W, Elmore SH, Fan H, Thorne LB, Gulley ML. Cytomegalovirus DNA Measurement in Blood and Plasma Using Roche LightCycler CMV Quantification Reagents. Diagn Mol Pathol. 2008;17(3):166-73.
Crossref - Kanaan A, Cour I, Alvarez-Lafuente R, et al. Significance of nested PCR and quantitative real time PCR for cytomegalovirus detection in renal transplant recipients. Int J Antimicrob Agents. 2004;24(5):455-462.
Crossref - Kuo C-P, Wu C-L, Ho H-T, Chen CG, Liu S-I, Lu Y-T. Detection of cytomegalovirus reactivation in cancer patients receiving chemotherapy. Clin Microbiol Infect. 2008;14(1):211-27.
Crossref - Schlick K, Grundbichler M, Auberger J, et al. Cytomegalovirus reactivation and its clinical impact in patients with solid tumors Clinical oncology. Infect Agents Cancer. 2015;10(1):10-45.
Crossref - Agrawal AK, Rajendra A, Noronha V, et al. Cytomegalovirus infection in solid malignancies. Cancer Research, Statistics, and Treatment. 2020. 3(1);19-24.
Crossref - Wang HW, Kuo CJ, Lin WR, et al. The clinical characteristics and manifestations of cytomegalovirus esophagitis. Dis Esophagus. 2016;29(4):392-399.
Crossref - Matsuda Y, Kishida S, Miyamoto H, et al. Cytomegalovirus-associated ulceration of gastric conduit after chemoradiotherapy following esophagectomy for cancer. Esophagus. 2015;12(3):300-303.
Crossref - Ohnuma H, Sato Y, Takayama T, et al. Esophageal cancer complicated by cytomegalovirus esophagitis during chemoradiotherapy: Case report. Gastrointestinal Endoscopy. 2003;57(4):622-626.
Crossref - Murakami D, Harada H, Yamato M, Amano Y. Cytomegalovirus-associated esophagitis on early esophageal cancer in immunocompetent host: a case report. Gut Pathogens. 2021;13(1).
Crossref - Demi̇rel A, Nur Pi̇lanci K, Inan N, et al. Investigation of the effect of chemotherapy on cytomegalovirus reactivity in patients with solid organ tumors. The European Research Journal. 2021;7(1):38-43.
Crossref - Torres HA, Kontoyiannis DP, Bodey GP, et al. Gastrointestinal cytomegalovirus disease in patients with cancer: A two decade experience in a tertiary care cancer center. Eur J Cancer. 2005;41(15):2268-2279.
Crossref - Salman OH, Al-Azzawi RH, Al-Azzawi RH. Seroprevalence of human cytomegalovirus in iraqi breast cancer patients association of human cytomegalovirus with her2 proto-oncogene overexpression in iraqi breast cancer patients View project immunogenetic View project seroprevalence of human cytomegalovirus in iraqi breast cancer patients. Plant Archives. 2020;20(Suppl 2):729-731. Available from: https://www.researchgate.net/publication/343351059
- Surendran A, Chisthi MM. Breast Cancer Association with Cytomegalo Virus-A Tertiary Center Case-Control Study. J Invest Surg. 2019;32(2):172-177.
Crossref - Radestad AF, Estekizadeh A, Cui HL, et al. Impact of Human Cytomegalovirus Infection and its Immune Response on Survival of Patients with Ovarian Cancer. Translational Oncology. 2018;11(6):1292-300.
Crossref - Mohammed A, Kadhim H, Ghani A. Investigation the role of human cytomegalovirus in the invasive ductal breast carcinoma. Clin Cancer Investig J. 2015;4(2):199.
Crossref - Enan KA, Rennert H, El-Eragi AM, El Hussein ARM, Elkhidir IM. Comparison of Real-time PCR to ELISA for the detection of human cytomegalovirus infection in renal transplant patients in the Sudan. Virol J. 2011;8:222.
Crossref - Drew WL. Diagnosis of Cytomegalovirus Infection. Rev Infect Dis. 1988;10(Suppl 3):S468-S476.
Crossref - Udeze A, Odebisi-Omokanye M, Ajileye T. Cytomegalovirus infection among Human Immunodeficiency Virus (HIV) infected individuals on highly active anti-retroviral therapy in North-Central Nigeria. Afr Health Sci. 2018;18(4):1057-1065.
Crossref - Gutierrez J, Piedrola G, Maroto MDC. Value of cytomegalovirus (CMV) IgG avidity index for the diagnosis of primary CMV infection. J Infect Dis. 1998;178(2):599-600.
Crossref - Ross SA, Novak Z, Pati S, Boppana SB. Overview of the Diagnosis of Cytomegalovirus Infection. Infectious Disorders – Drug Targets. 2011;11(5):466-474.
Crossref - Kangro H 0, Griffiths PD, Huber TJ, Heath R 6. Specific IgM Class Antibody Production Following Infection With Cytomegalovirus. J Med Virol. 1982;10(3):203-211.
Crossref - Stojcevic-Maletic J, Baculov K, Bogdanovic-Vasic S, Milanovic B, Vucinic N, Milutinovic A. Comparison of serological and molecular methods in the diagnosis of cytomegalovirus infections in dialysis patients. Medicinski Pregled. 2020;73(1-2):43-48.
Crossref - Chou S. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology. 1992;188(1):388-390.
Crossref - Fries BC, Chon S, Boeckh M, Torok-Storb B. Frequency distribution of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J Infect Dis. 1994;169(4):769-774.
Crossref - Chandler SH, McDougall JK. Comparison of restriction site polymorphisms among clinical isolates and laboratory strains of human cytomegalovirus. J Gen Virol. 1986;67(10):2179-2192.
Crossref - Mohamed HT, El-Shinawi M, Nouh MA, et al. Inflammatory breast cancer: High incidence of detection of mixed human cytomegalovirus genotypes associated with disease pathogenesis. Front Oncol. 2014;4:246.
Crossref - Navarro D, Paz P, Tugizov S, Topp K, Vail J la, Pereira L. Glycoprotein B of Human Cytomegalovirus Promotes Virion Penetration into Cells, Transmission of Infection from Cell to Cell, and Fusion of Infected Cells. Virology. 1993;197(1):143-158.
Crossref - Nogueira E, Ozaki KS, Tomiyama H, Camara NOS, Granato CFH. Clinical correlations of human cytomegalovirus strains and viral load in kidney transplant recipients. Int Immunopharmacol. 2009;9(1):26-31
Crossref - Chern KC, Chandler DB, Martin DF, Kuppermann BD, Wolitz RA, Margolis TP. Glycoprotein B subtyping of cytomegalovirus (CMV) in the vitreous of patients with AIDS and CMV retinitis. J Infect Dis. 1998 Oct;178(4):1149-53.
Crossref - Correa C, Kouri V, Perez L, Soto Y, Limia C. Diagnosis, gB genotype distribution and viral load of symptomatic congenitally infected CMV patients in Cuba. J Perinatol. 2016;36(10):837-842.
Crossref - Cunha AA, Aquino VH, Mariguela V, Nogueira ML, Figueiredo LTM. Evaluation of glycoprotein B genotypes and load of CMV infecting blood leukocytes on prognosis of AIDS patients. Rev Inst Med Trop Sao Paulo. 2011;53(2):83-88.
Crossref - Gilbert C, Handfield J, Toma E, Lalonde R, Bergeron MG, Boivin G. Human cytomegalovirus glycoprotein B genotypes in blood of AIDS patients: Lack of association with either the viral DNA load in leukocytes or presence of retinitis. J Med Virol. 1999;59(1):98-103.
Crossref - Dieamant DC, Bonon SHA, Peres RMB, et al. Cytomegalovirus (CMV) genotype in allogeneic hematopoietic stem cell transplantation. BMC Infect Dis. 2013;13:310.
Crossref - Torok-Storb B, Boeckh M, Hoy C, Leisenring W, Myerson D, Gooley T. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood. 1997;90(5):2097-102.
Crossref - Gorzer I, Kerschner H, Redlberger-Fritz M, Puchhammer-Stockl E. Human cytomegalovirus (HCMV) genotype populations in immunocompetent individuals during primary HCMV infection. J Clin Virol. 2010;48(2):100-103.
Crossref - Lisboa LF, Kumar D, Wilson LE, Humar A. Clinical utility of cytomegalovirus cell-mediated immunity in transplant recipients with cytomegalovirus viremia. Transplantation. 2012;93(2):195-200.
Crossref - Pang X, Humar A, Preiksaitis JK. Concurrent genotyping and quantitation of cytomegalovirus gB genotypes in solid-organ-transplant recipients by use of a real-time PCR assay. J Clin Microbiol. 2008;46(12):4004-4010.
Crossref - Arav-Boger R, Willoughby RE, Pass RF, et al. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-a and β-chemokine receptors in congenital CMV disease. J Infect Dis. 2002;186(8):1057-1064.
Crossref - Drew WL, Sweet ES, Miner RC, Mocarski ES. Multiple Infections by Cytomegalovirus in Patients with Acquired Immunodeficiency Syndrome: Documentation by Southern Blot Hybridization. J Infect Dis. 1984;150(6):952-923.
Crossref - Cicin-Sain L, Podlech J, Messerle M, Reddehase MJ, Koszinowski UH. Frequent Coinfection of Cells Explains Functional In Vivo Complementation between Cytomegalovirus Variants in the Multiply Infected Host. J Virol. 2005;79(15):9492-502.
Crossref - Wu X, Wang Y, Xu Y, et al. Cytomegalovirus Glycoprotein B Genotype in Hematopoietic Stem Cell Transplant Patients from China. Biol Blood Marrow Transplant. 2010;16(5):647-652.
Crossref - Fagundes CP, Glaser R, Alfano CM, et al. Fatigue and herpesvirus latency in women newly diagnosed with breast cancer. Brain Behav Immun. 2012;26(3):394-400.
Crossref - Richardson AK, Currie MJ, Robinson BA, et al. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS ONE. 2015;10(2):1-14.
Crossref - Pandey JP, Gao G, Namboodiri AM, et al. Humoral immunity to cytomegalovirus Glycoprotein B in patients with breast cancer and matched controls: Contribution of immunoglobulin g, k, and Fcg receptor genes. J Infect Dis. 2016;213(4):611-617.
Crossref - Gawad AA, Hashish M, Abaza AF, El-Kayal A. Cytomegalovirus immunoglobulin G avidity index among blood donors in Alexandria, Egypt. Central Eur J Public Health. 2016;24(4):314-20.
Crossref - Avni A, Haikin H, Feuchtwanger MM, et al. Antibody Pattern to Human Cytomegalovirus in Patients with Adenocarcinoma of the Colon. Intervirology. 1981;16.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.