ISSN: 0973-7510
E-ISSN: 2581-690X
Four oxalate degrading endophytic bacteria were isolated from oxalate rich Colocasia esculenta tubers. Based upon the oxalate oxidase (EC 1.2.3.4) activity produced in nutrient medium, one bacterium was selected and identified as Ochrobactrum intermedium by 16S rDNA sequencing. Studies on effect of nutritional and non-nutritional parameters showed that oxalate oxidase production is inducible, requires Manganese ions in the medium, and very low fill-up volume is beneficial. Shake flask fermentation carried out with medium comprising Sucrose, Ammonium chloride, Sodium oxalate along with basal salts gave 0.5 UmL-1 oxalate oxidase activity and 0.454 Umg-1specific activity after 65h of fermentation.
Colocasia esculenta, Endophyte, Ochrobactrum intermedium, Oxalate oxidase.
Oxalic acid and its salts occur as end products of metabolism in number of plant tissues. High oxalate levels are found in Taro (Colocasia esculenta), Sweet potato (Ipomoea batatas), and Yam (Dioscorea alata) tubers (275 – 574 mg/100 g fresh weight)1. Oxalate oxidase (EC 1.2.3.4), an enzyme found in nature degrades oxalate to hydrogen peroxide and carbon dioxide2. Oxalate oxidase activity has been detected in Barley3, Beet4, Maize5, sorghum6 and wheat7. Functionally diverse protein super family known as Cupins, concerned with cell wall, fungal defence, salt tolerance and floral induction are found to possess oxalate oxidase activity8. Oxalate oxidase activity has been reported in white rot basidiomycete, Ceriporiopsis subvermispora9 and a bacterium, Pseudomonas sp. OX-5310 as well. Oxalate oxidase is extensively used in kits to assay oxalate levels in blood and urine. Regular assessment of oxalate levels in urine helps to monitor and control hyperoxaluria and Urolithiasis 2. Removal of oxalic acid from bleaching filtrates using the enzyme oxalate oxidase is a possibility to prevent problems with scaling in the pulp and paper industry11.
Endophytes are endosymbionts that colonizes interior organs of plants, but does not have pathogenic effects on its host. Endophyte can be a bacterium or a fungus and in their symbiotic association, the host plant protects and feeds the endophyte, which in return produces bioactive metabolites to augment the growth and competitiveness of the host and protect it from being attacked by herbivores and plant pathogens12,13 and to date, all plant species studied have been found to harbour at least one endophyte14. Endophytic microorganisms are also known to produce wide range of industrial important enzymes like xylanases, hemicellulases, cellulases, protease15,16. The fact that substrate for these enzymes are available in excessive amount in plants17, could possibly have caused endophytes to evolve their genetic system to produce enzymes specific to the substrate available inside the host plant 18. The current study deals with the isolation and screening of novel endophytes from plant rich in oxalates for the purpose of production of oxalate oxidase. Further, shake flask studies were carried out to enhance the enzyme titre.
Isolation and identification of oxalate oxidase producing bacteria
Bacterial endophytes were isolated from the tuber of Colocasia esculenta grown wildly in Mangalore suburb of Karnataka state, India. The tuber of the plant was cut into small pieces of approximate 1.5 cm length. The explants were washed with distilled water and surface sterilized by dipping the explants in 70% (v/v) ethanol for 5 minutes, followed by 5%(w/v) sodium hypochlorite solution for 5 minutes, and 70% (v/v) ethanol for 30 s. The surface sterilized samples were washed in sterile distilled water three times to remove surface sterilization agents19. The explants were then cut into small pieces of 0.5 cm length with a flame sterilized scalpel and transferred to sterilized tryptic soy agar medium supplemented with antibiotic cyclohexamide of 10 ppm concentration for the isolation of bacteria. To check the efficacy of surface sterilization, the water from the final rinse was inoculated in tryptic soy agar plates. All the petriplates were then incubated at 30° C for 7 – 20 days. Isolates were screened for oxalate oxidase activity by streaking the isolates on solidified basal mineral medium having the composition (gL-1): KH2PO4– 3, Na2HPO4 – 6, NaCl – 5, NH4Cl – 2, MgSO4.7H2O – 0.1, Agar – 15 and 5 – calcium oxalate. Solubilisation of calcium oxalate was evidenced by the formation of clear zones around the colonies due to dissolution of calcium oxalate. Control plate was kept to avoid any false result. The positive isolates after the screening stages were maintained in tryptic soy agar slants and the organisms were preserved for long storage in 20% (w/v) glycerol stock supplemented with 1% (v/v) host plant extract. The host plant extract was prepared by crushing 1g of small pieces of tubers of Colocasia esculenta in a mortar and pestle with 10mL of distilled water. The homogenate was centrifuged for 10 min at 8,000 RPM in a refrigerated centrifuge and the recovered supernatant was filter sterilized using 0.2 µm filter membrane. The positive isolates were cultured in the common production medium and the most productive strain CL6 was chosen for further study. CL6 strain was sent to MTCC, IMTECH, Chandigarh, India for identification by 16 S rDNA sequencing.
Oxalate oxidase activity
Oxalate oxidase was assayed by modified 3-Methyl-2-Benzothiazolinone Hydrazone (MBTH) method20. The reaction mixture was comprised of 1.7 mL of sodium succinate buffer (50 mM, pH 3.8), 0.04 mL of 200 mM oxalic acid solution and 1 mL of enzyme solution. After incubation for 30 min at 55°C, the reaction was stopped by adding 0.1 mL of 100 mM EDTA solution. This was followed by the addition of 1 mL of colouring reagent comprising 0.1 mM MBTH (3-Methyl-2-Benzothiazolinone Hydrazone) and 0.72 mM DMA (N, N-Dimethylaniline) along with 3 U of 20 µL of peroxidase (Sigma Aldrich) solution. Hydrogen peroxide produced by the oxidation of oxalate by oxalate oxidase enzyme combines with MBTH and DMA (N, N-Dimethylaniline) in the presence of peroxidase to form purple colour indamine dye. The concentration of indamine dye is directly proportional to the concentration of hydrogen peroxide which in turn proportional to the activity of oxalate oxidase enzyme. The colour produced due to indamine dye formation was read at 600 nm. Standard curve was prepared using commercially available hydrogen peroxide. One unit of oxalate oxidase was defined as the amount of enzyme required to produce 1 µmol of H2O2 per minute at 55°C. For all the trials, highest enzyme activity and biomass content corresponding to highest enzyme activity was reported. The soluble protein in the culture supernatant was determined by Bradford method 21 with bovine serum albumin as standard.
Effect of nutritional and non-nutritional parameters on Oxalate oxidase production
Inoculum for all the shake flask studies was prepared in tryptic soy broth supplemented with 2.5 gL-1 sodium oxalate having an initial medium pH of 6.5. The medium was inoculated with a loop full of culture from slant, incubated at 30°C in an incubator shaker at 150 rpm. The culture was harvested after 18 h of growth, added as inoculum at the rate of 2 % (v/v). All the shake flask trials were conducted in 250 ml Erlenmeyer flasks, incubated at 30°C in an incubator shaker at 150 rpm, unless otherwise stated. The unoptimized medium had the following(gL-1): KH2PO4 – 3, Na2HPO4 – 6, NaCl – 5 , NH4Cl – 2 , MgSO4. 7H2O – 0.1, MnSO4 – 0.05, Disodium oxalate-5 and Biotin- 0.0015, with the initial pH of 6.5. Samples were taken at definite time intervals to analyze cell growth and oxalate oxidase production. Growth was monitored by measuring the optical density at 600 nm and the cell free supernatant obtained after centrifugation at 8,000 RPM for 15 min in a refrigerated centrifuge, was analysed for oxalate oxidase activity. The obtained optical density values were converted to dry cell weight (gL-1) using a calibration curve22.
The effect of different carbon sources on the production of oxalate oxidase and biomass were investigated in shake flasks containing above explained nutrient medium supplemented with 20 gL-1 carbon sources (sucrose, glucose, glycerol). Once the best carbon source was determined, the effect of supplemented carbon source concentration was determined by varying the selected carbon source (sucrose) in the range of 20-60 gL-1. The effect of nitrogen sources was determined using the nutrient medium explained above supplemented with 20 gL-1 sucrose and 2 gL-1 of various organic and inorganic nitrogen sources (Yeast extract, Peptone, Casein hydrolysate, Ammonium chloride, Sodium nitrate). Once the best nitrogen source was selected, the effect of concentration of selected nitrogen source (NH4Cl) on enzyme production was determined by varying its concentration in the range of 2-8 gL-1.
Several reports suggest presence of Mn2+ ions is important for functional oxalate oxidase enzyme production in plants23,24 and in fungus Ceriporiopsis subvermispora25. So the effect of Mn2+ was studied for its role in the production of functional oxalate oxidase by incorporating MnSO4 (0.025 – 0.1gL-1) in the media. Effect of fill up volume on the overall growth and enzyme production was studied. Fill up volume of one-half, one-fourth, one-fifth, one-tenth were maintained in different flasks and the biomass and enzyme production was estimated for each of the flasks. The effect of temperature was studied for growth and enzyme production by incubating the flasks at 25, 30 and 35°C. The effect of initial medium pH on oxalate oxidase enzyme production as studied in the pH range of 5 to 8. Medium pH was adjusted using either dilute HCl or dilute NaOH before autoclaving the medium. Effect of agitation speed for 150, 175, 200 rpm on the growth and enzyme production was determined. Effect of Inoculum from inoculum age of 12, 18 and 24 h was studied for enhanced production of enzyme.
After optimizing the media composition and physical parameters by one factor at a time method, time course study was conducted with optimized condition and growth requirements to obtain the product profile.
Isolation and identification of oxalate oxidase positive bacteria
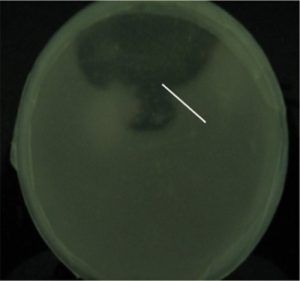
After primary screening on minimal mineral medium agar plates supplemented with 5 gL-1 calcium oxalate, 4 bacterial cultures were selected for shake flask studies based on their ability to form clear zones by dissolution of calcium oxalate during primary screening. One isolate, CL6 exhibited large clear zone on screening plate (Fig. 1) and good growth on sodium oxalate supplemented production medium in shake flask studies. The isolate was deposited and identified in the “Microbial Type Culture Collection and Gene bank” (MTCC), Institute of Microbial Technology (IMTECH Chandigarh, India). The organism was identified based on 16S rDNA sequence analysis as Ochrobactrum intermedium and the Gene-Bank accession number for the nucleotide sequence is KM658164. To the best of our knowledge, this is the first report on the presence of oxalate oxidase activity in the genus Ochrobactrum and in an endophyte from oxalate-rich Colocasia plant.
The taxonomy of Ochrobactrum reveals that it belongs to the order rhizobiales, which also comprises of bacteria of rhizosphere. The test organism Ochrobactrum intermedium CL6 has been isolated from the tubers of Colocasia as endophyte, so it can be stated that the test organism Ochrobactrum intermedium CL6 might have invaded the Colocasia plant tissue over a period of time from its rhizosphere to the internal tissues of Colocasia tuber. It had been previously believed that endophytic root bacterial communities comprise a subset of microbial population originating from the surrounding rhizosphere soil 26,27. Our results support this speculation.
The microbial community structure of any niche is influenced by surrounding soil and environmental parameters. Oxalate-degrading bacteria form a close association with plant roots that are rich in oxalic acid, where the oxalate exudates form a source of carbon for a variety of microorganisms in the rhizosphere of the yam (Dioscorea alata, Typhonium trilobatum and Amorpho phallus)28. Based on this it can be argued that endophytic microbial community of Colocasia esculenta might have also been influenced by high amount of oxalate in its living tissue. It appears that Ochrobactrum intermedium CL6 isolated from Colocasia tuber have evolved the enzyme system, oxalate oxidase in order to utilize oxalate available in host plant tissues.
Effect of nutritional parameters on oxalate oxidase production
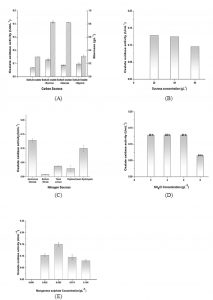
Initially the maximum biomass of the strain Ochrobactrum intermedium CL6 obtained with sodium oxalate as an only source of carbon was 0.3 gL-1 after 120 h of incubation but a comparative higher growth was obtained with sucrose as well as with glucose as additional carbon sources within 48 h of incubation (Fig. 2A). Sucrose as an additional carbon source exhibited maximum enzyme activity for oxalate oxidase. However, glucose as an additional carbon source was not very effective for enzyme production. There was no change in biomass content and enzyme activity with glycerol as additional carbon source. Sucrose was therefore selected for further study as it could reduce the fermentation time from 120 h to 48 h. The oxalate oxidase production with different concentrations of sucrose was also tested. Sucrose at 20 gL-1exhibited the maximum oxalate oxidase activity 0.128 U mL-1 (Fig. 2B). NH4Cl, found to be the best for the maximum production of oxalate oxidase. NH4Cl at 2gL-1 exhibited an oxalate oxidase activity of 0.128 UmL-1. Enhancement of enzyme activity was low in case of Yeast extract and Peptone. The only organic nitrogen source which showed some enzyme production was casein hydrolysate (Fig. 2C). NH4Cl was selected for further study. Varied concentration of NH4Cl didn’t contribute in further improving the oxalate oxidase activity. The enzyme activity remained same for NH4Cl concentration of 2-6 gL-1. However, as the nitrogen source concentration increased, peak enzyme activity got shifted from 48 to 84 h. At NH4Cl concentration of 8gL-1, reduction in peak enzyme activity was observed at 84h (Fig. 2D).
Mn2+ ions was found to be extremely important for active form of enzyme production. No oxalate oxidase activity was observed in the fermentation broth, when source of Mn2+ ions in the form of MnSO4 was not supplemented in the production media. The concentration of MnSO4 was varied from 0.025 gL-1 to 0.1 gL-1. Optimum concentration of MnSO4 was found to be 0.05 gL-1. Higher concentration of the MnSO4 supplementation in the media was leading to lower activity of the enzyme (Fig. 2E). Manganese sulphate concentration of 0.05 gL-1 was selected for further study. There are reports where presence of manganese at the active site of the enzyme oxalate oxidase has been documented. Ceriporiopsis subvermispora oxalate oxidase was the first bicupin enzyme identified that exhibit manganese dependent oxidation of oxalate 25. Barley oxalate oxidase, a homohexameric glycoprotein, contains one Mn atom which is ligated by three histidine residues, one glutamate residue and two water molecules 23,24,29. Koyama has reported the presence of 1.12 atoms of manganese and 0.36 atoms of zinc per subunit in oxalate oxidase produced from the soil bacterium Pseudomonas sp. OX- 53 10.
Fig. 2. Effect of nutritional parameters on biomass growth and enzyme production. (A) Effect of various sugars in medium on biomass growth () and enzyme activity (). Medium contains 20 gL-1carbon source, 5 gL-1 Sodium oxalate and 2 gL-1 NH4Cl along with basal salts. (B) Effect of sucrose concentration on the enzyme activity was studied by changing its concentration in the medium having 5 gL-1 Sodium oxalate and 2 gL-1 NH4Cl along with basal salts. (C) Effect of various nitrogen sources in medium on enzyme production was studied. The medium contains 20 gL-1 of sucrose as carbon source, 2 gL-1 nitrogen source and 5 gL-1 sodium oxalate as inducer along with basal salts. (D) Effect of NH4Cl Concentrations on oxalate oxidase activity was studied by changing its concentration in the medium having 20 gL-1 of sucrose and 5 gL-1sodium oxalate along with basal salts. (E) Effect of MnSO4 on enzyme production was studied by changing its concentration in medium having 20 gL-1 of sucrose, 5 gL-1sodium oxalate and 2 gL-1NH4Clalong with basal salts. Control had no MnSO4 in the medium.
Effect of non-nutritional parameters on oxalate oxidase production
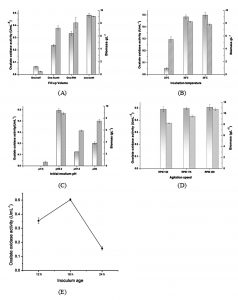
Fill up volume of one-half, one-fourth, one-fifth, one-tenth of shake flask was tried in four different experiments. The observations revealed that fill up volume of one-fifth and one- tenth gave higher biomass in short duration (Fig. 3A). There was a progressive increase in enzyme activity as the fill up volume is reduced from one- half to one-tenth. An oxalate oxidase activity of 0.5 UmL-1 was obtained at one-tenth of fill up volume. From this we can infer that oxygen requirement of Ochrobactrum intermedium CL6 is high and meeting the requirement is crucial. Higher oxygen requirement could be attributed to the fact that oxygen is a co-substrate utilized during oxalate catabolism30. Moreover it is the proved fact that in shake flasks, the oxygen transfer rate (OTR) is reversely proportional to the volume of culture 31.
Temperature of incubation during growth had a significant influence on the enzyme production. Observation suggests that the organism was growing very well at temperature of 35°C during the first few hours after inoculation. But after 24 h, the growth profile of the organism at 35°C and 30°C was almost same. Temperature of 25°C was suboptimal for enzyme production. The peak enzyme activity observed at 30°C and 35°C was almost similar (Fig. 3B).
It was observed that the growth pattern of the organism was almost similar in initial medium pH range of 6.5, 7.5 and 8. However, the activity of the enzyme was relatively higher in pH 6.5 (Fig. 3C). The result showed that the yield of the enzyme oxalate oxidase by strain Ochrobactrum intermedium CL6 was relatively less at initial medium pH 7.5. The enzyme activity profile also revealed that an activity of 0.2 UmL-1 was obtained for initial medium pH 8, which was marginally higher than that of initial medium pH 7.5, but was almost one – third of the activity obtained at pH 6.5. Pseudomonas sp.OX-53, which was the only bacterial species reported till date for oxalate oxidase production, could produce the enzyme at initial medium pH 8 10.
There was an increase in biomass content as the agitation speed was increased from of 150 to 200 rpm. However there was no significant increase in oxalate oxidase production with increase in agitation speed (Fig. 3D). Yield of oxalate oxidase was found to be dependent on inoculum age. Inoculum of age 24 h exhibited poor production in comparison to 18 h old inoculum (Fig. 3E) Time course study indicates that 24 h inoculum took more time to produce the enzyme and the maximum enzyme activity had gone down to 0.15 UmL-1 from 0.5 UmL-1 obtained with 18 h inoculum. Cells of younger inoculum were found to be more active in product formation whereas old inoculum could be partially induced for product formation 32, 33.
Fig. 3. Effect of non-nutritional parameters on biomass growth and enzyme production. (A) Effect of fill up volume on the biomass growth () and oxalate oxidase activity () was studied in a medium having 20 gL-1 of sucrose, 5 gL-1sodium oxalate and 2 gL-1NH4Cl along with basal salts. While finding the effect of fill up the volume, MnSO4 concentration of 0.05 gL-1 was taken in the medium. (B) Effect of fermentation temperature on biomass growth and enzyme production. Fill up the volume of one-tenth was maintained for all the experiments. Biomass content at peak enzyme activity was considered. (C) Effect of initial medium pH on enzyme production was studied with a medium having 20 gL-1 of sucrose, 5 gL-1sodium oxalate and 2 gL-1 NH4Cl, 0.05 gL-1 MnSO4 along with basal salts. Experiments were conducted at 30°C, fill up volume of one-tenth, agitation rate of 150 rpm and initial medium pH of 6.5. (D) Effect of agitation speed (E) Effect of inoculum age.
Time course study of oxalate oxidase production
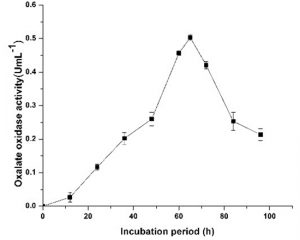
Enzyme activity with the present optimized condition of 20 gL-1 sucrose as additional carbon source, 5 gL-1 sodium oxalate as inducer, 2 gL-1 NH4Cl as nitrogen source, 0.05 gL-1 MnSO4, Fill up volume of one-tenth, pH of 6.5, fermented with inoculum age of 18 h, 30ºC at 150 rpm showed maximum activity of 0.5 UmL-1 after 65h of fermentation (Fig. 4). The 25 mL culture supernatant yielded 27.5 mg of protein with total oxalate oxidase activity of 12.5 U and specific activity of 0.454 Umg-1.
Fig. 4. Time course study of oxalate oxidase production in optimized medium and conditions. Medium having 20 gL-1 of sucrose, 5 gL-1 sodium oxalate and 2 gL-1 NH4Cl, 0.05 gL-1 MnSO4 along with basal salts was used. Experiments were conducted at 30°C, fill up volume of one-tenth, agitation rate of 200 rpm and initial medium pH of 6.5 with inoculum age of 18 h.
Comparison of specific activity of oxalate oxidase from Ochrobactrum intermedium CL6 and other sources suggests that specific activity of oxalate oxidase obtained in the present study is 0.454 Umg-1 (Table 1), which is much higher than that of other microbial sources 10 as well as plant sources34-36. The total enzyme activity from wheat seedling and barley roots is also lower than the total enzyme activity obtained from the culture supernatant of Ochrobactrum intermedium CL6. Higher specific activity in the cell free broth also indicates that the enzyme purification could be relatively easy.
Table (1):
Comparison of oxalate oxidase activity from Ochrobactrum intermedium CL6 and other sources.
Culture supernatant /Crude extract |
Specific activity (Umg-1) |
|---|---|
Ochrobactrum intermedium CL6 |
0.454 |
Pseudomonas sps. OX-53 |
0.29 |
Barley roots |
0.031 |
Wheat seedlings |
0.107 |
Costus pictus leaves |
0.0374 |
A growing body of evidence suggests that plant-associated microorganisms, especially endophytic and rhizosphere bacteria and fungi, represent a huge and largely untapped resource of natural products with chemical structures that have been optimized by evolution for biological and ecological relevance13,14. Since endophytes were first described in the Darnel (Lolium temulentum) 37, they have been isolated from various organs of different plant species, above ground tissues of liverworts, hornworts, mosses, lycophytes, equisetopsids, ferns, and spermatophytes from the tropics to the arctic, and from the wild to agricultural ecosystems 38 and to date, all plant species studied have been found to harbour at least one endophyte14. Endophytes usually produce the enzymes necessary for the colonization of plant tissues. It was demonstrated that most endophytes are able to utilize, at least in vitro, most plant cell components. As Colocasia esculenta tubers were found to contain large amount of oxalate crystals, it was hypothesized that at least few endophytes inhabiting tuber will have the ability to produce oxalate degrading enzyme, oxalate oxidase. Successful isolation of four bacterial strains supports our hypothesis. Presence of substantial amount of oxalic acid in the tubers of Colocasia might have put selective pressure on these endophytes to develop a means to use these energy rich molecules. This first report of isolation of Ochrobactrum intermedium, a novel oxalate oxidase producing bacterium from Colocasia esculenta also reiterate the fact that plants harbour endophytes that have the potential to produce many novel metabolites of human importance.
Four oxalate degrading bacteria were isolated as result of our effort. The bacterium which showed highest oxalate oxidase activity was identified as Ochrobactrum intermedium. The nutritional and non-nutritional factors were optimized in shake flasks. Sucrose at 20 gL-1, NH4Cl at 2 gL-1,MnSO4 at 0.05 gL-1and sodium oxalate as inducer at 5gL-1, fill up volume in shake flask at one – tenth, incubation temperature of 30°C and initial medium pH of 6.5 and inoculum age of 18 h was found to be optimum. Oxalate oxidase activity of 0.5UmL-1 and specific activity of 0.454 Umg-1was recorded in fermentation broth after 65h of fermentation under optimal conditions.
Acknowledgements
Mr. Kunal Kumar is supported by scholarship for doctoral course by the National Institute of Technology Karnataka (NITK) Surathkal, Karnataka, India.
Conflict Of Interest
The authors declares that there is no conflict of interest.
- Morrison S C, Savage G P. Oxalates, 2003, pp. 4282–4287. In Caballero B, Trugo LC, Finglas, PM (eds.), Encyclopedia of Food Sciences and Nutrition, Academic Press, London.
- Dunwell J M. Cupins: A New Superfamily of Functionally Diverse Proteins that Include Germins and Plant Storage Proteins. Biotechnol Genet Eng, 1998; 15: 1–32.
- Chiriboga J. Purification and properties of oxalic acid oxidase. Arch Biochem Biophys, 1966; 116: 516–523.
- Varalakshmi P, Richardson K E. Studies on oxalate oxidase from beet stems upon immobilization on concanavalin A. Biochem Int, 1992; 26: 153–162.
- Vuletic M, Sukalovic H T S. Characterization of cell wall oxalate oxidase from maize roots. Plant Sci, 2000; 157: 257–263.
- Satyapal, Pundir, C S. Purification and properties of an oxalate oxidase from leaves of grain sorghum hybrid CSH-5. Biochim Biophys Acta, 1993; 116: 1–5.
- Lane B G, Dunwell J M, Ray J A, Schmitt M R, Cuming A C. Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem, 1993; 268: 12239–12242.
- Dunwell J M, Khuri S, Gane P J. Microbial Relatives of the Seed Storage Proteins of Higher Plants: Conservation of Structure and Diversification of Function during Evolution of the Cupin Superfamily. Microbiol Mol Biol Rev, 2000; 64: 153–179.
- Aguilar C, Urzua U, Koenig C, Vicuna R. Oxalate oxidase from Ceriporiopsis subvermispora: biochemical and cytochemical studies. Arch Biochem Biophys, 1996; 366: 275–282.
- Koyama H. Purification and Characterization of Oxalate Oxidase from Pseudomonas sp. OX-53. Agric Biol Chem, 1988; 52: 743–748.
- Sjode A, Winestrand S, Nilvebrant N O, Jonsson L J. Enzyme-based control of oxalic acid in the pulp and paper industry. Enzyme Microb Technol, 2008; 43: 78–83.
- Dreyfuss M M, Chapela I H. Potential of fungi in the discovery of novel, low-molecular weight pharmaceuticals, 1994, pp. 49-80. In Gullo VP (ed.), The Discovery of Natural Products with Therapeutic Potential, Butterworth-Heinemann, Boston.
- Gunatilaka A A L. Natural Products from Plant-Associated Microorganisms: Distribution, Structural Diversity, Bioactivity, and Implications of Their Occurrence. J Nat Prod, 2006; 69: 509–526.
- Kusari S, Hertweck C, Spiteller M. Chemical Ecology of Endophytic Fungi: Origins of Secondary Metabolites. Chem Biol, 2012; 19: 792–798.
- Robl D, Delabona P, Mergel C M, Rojas J D, Costa P, Pimentel I C, Vicente V A, Pradella J G, Padilla G. The capability of endophytic fungi for production of hemicellulases and related enzymes. BMC Biotechnol, 2013; 13: 94.
- Zaferanloo B, Quang T D, Daumoo S, Ghorbani M M, Mahon P J, Palombo E A. Optimization of protease production by endophytic fungus, Alternaria alternata, isolated from an Australian native plant. World J Microbiol Biotechnol, 2014; 30: 1755–1762.
- Hadar Y. Sources for Lignocellulosic Raw Materials for the Production of Ethanol, 2013, pp. 21-38 In Faraco V (ed.), Lignocellulose Conversion, Springer, Berlin Heidelberg.
- Gurung N, Ray S, Bose S, Rai V. A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Research International, 2013; 1-18.
- Araujo W L, Marcon J, Maccheroni W, Van Elsas J D, Van Vuurde J W L, Azevedo J L. Diversity of Endophytic Bacterial Populations and Their Interaction with Xylella fastidiosa in Citrus Plants. Appl Environ Microbiol, 2002; 68: 4906–4914.
- Laker M F, Hofmann A F, Meeuse B J. Spectrophotometric determination of urinary oxalate with oxalate oxidase prepared from moss. Clin Chem, 1980; 26: 827–830.
- Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 1976; 72: 248–254.
- Prabha M S, Divakar K, Priya J D A, Selvam G P, Balasubramanian N, Gautam P. Statistical analysis of production of protease and esterase by a newly isolated Lysinibacillus fusiformis AU01: purification and application of protease in sub-culturing cell lines. Ann Microbiol, 2014; 65: 33–46.
- Requena L, Bornemann S. Barley (Hordeum vulgare) oxalate oxidase is a manganese-containing enzyme. Biochem J, 1999; 343: 185–190.
- Woo E J, Dunwell J M, Goodenough P W, Marvier A C, Pickersgill R W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat Struct Biol, 2000; 7: 1036–1040.
- Moomaw E W, Hoffer E, Moussatche P, Salerno J C, Grant M, Immelman B, Uberto R, Ozarowski A, Angerhofer A. Kinetic and Spectroscopic Studies of Bicupin Oxalate Oxidase and Putative Active Site Mutants. Plos One, 2013; 8: 57933.
- Cocking E C. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil, 2003; 252: 169–175.
- Hallmann J, Quadt-Hallmann A, Mahaffee W F, Kloepper J W. Bacterial endophytes in agricultural crops. Can J Microbiol, 1997; 43: 895–914.
- Sahin N. Isolation and characterization of a diazotrophic, oxalate-oxidizing bacterium from sour grass (Oxalis pes-caprae L.). Res Microbiol, 2005; 156: 452–456.
- Gane P J, Dunwell J M, Warwicker J. Modeling based on the structure of vicilins predicts a histidine cluster in the active site of oxalate oxidase. J Mol Evol, 1998; 46: 488–493.
- Opaleye O, Rose R S, Whittaker M M, Woo E J, Whittaker J W, Pickersgill R W. Structural and Spectroscopic Studies Shed Light on the Mechanism of Oxalate Oxidase. J Biol Chem, 2006; 281: 6428–6433.
- Maier U, Buchs J. Characterisation of the gas-liquid mass transfer in shaking bioreactors. Biochem Eng J, 2001; 7: 99–106.
- Neves A A, Vieira L M, Menezes J C. Effects of preculture variability on clavulanic acid fermentation. Biotechnol Bioeng, 2001; 72: 628–633.
- Wang J J, Shih J C H. Fermentation production of keratinase from Bacillus licheniformis PWD-1 and a recombinant B. subtilis FDB-29. J Ind Microbiol Biotechnol, 1999; 22: 608–616.
- Kotsira V P, Clonis Y D. Oxalate oxidase from barley roots: purification to homogeneity and study of some molecular, catalytic, and binding properties. Arch Biochem Biophys, 1997; 340: 239–249.
- Hu Y, Guo Z. Purification and characterization of oxalate oxidase from wheat seedlings. Acta Physiol Plant, 2009; 31: 229–235.
- Sathishraj R, Augustin A. Oxalic acid and oxalate oxidase enzyme in Costus pictus D. Don. Acta Physiol Plant, 2012; 34: 657–67.
- Freeman E M. The seed-fungus of Lolium temulentum, L., the Darnel. Philos Trans R Soc Lond B Biol Sci, 1904; 196: 1–27.
- Arnold A E. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev, 2007; 21: 51–66.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.