ISSN: 0973-7510
E-ISSN: 2581-690X
Pyrostegia venusta is known as an ornamental plant with its source of antioxidants, cytotoxic, anti-inflammatory, and anti-HIV compounds. Ephypitic molds are potentially co-existed on the surface of this flower since it contains essential nutrients which support their growth. On the other hand, molds produce several enzymes that might involve flower growth. The presence of ephypitic molds on this flower provides information about its ability to produce amylase. This study successfully isolated molds from August flower (P. venusta) originating from Taman Nasional Bedugul, Bali, Indonesia. The study aimed to isolate potential amylase producer strains and optimize the enzyme production using Solid-State Fermentation (SSF) method. Ten mold isolates belonging to Universitas Negeri Jakarta Culture Collection (UNJCC) were selected according to their amylolytic index (IA) values, morphological identification, and colony count number. Selected strains were optimized for its growth to produce amylase using the SSF method under different temperatures (30, 40, 50°C) and pH (6, 7, 8) with a wheat brain fermentation medium. Results showed that UNJCC F100 (6.53 × 108 CFU/ml) and UNJCC F106 (9.83 x 108 CFU/ml) are the two isolates with the highest IA values of 1.34 ± 0.1 and 1.08 ± 0.12 among all isolates. Based on molecular identification using ITS region, UNJCC F100 and UNJCC F106 were identified as A. subflavus (97% homology) and A. fumigatus (99.52% homology), respectively. This study exhibited that both isolate UNJCC F100 and isolate UNJCC F106 have optimal amylase production conditions at 30°C and pH 6. The enzyme produced was 19.99 U/ml at 30°C and 34.33 U/ml at pH 6 for isolate UNJCC F100, and for isolate UNJCC F106 is 28.55±3.80 U/ml. The two isolates are potentially used for amylase production, referring to the specific environmental condition. However, to generate a higher amount with amylase activity, other external variables such as medium used, inoculum concentration, and fermentation method are important to consider further for a larger application.
Amylase, Molds, Pyrostegia venusta, Taman Nasional Bedugul, Solid-State Fermentation (SSF)
Starch serves as a carbon and energy source in the metabolic process of flower growth and development.1,2 Instead of a natural phenomenon, starch hydrolysis can be carried out by amylolytic enzymes. Amylases are a group of enzymes that can hydrolyze starch by breaking the glycoside bonds into simple sugars3 Amylases are significant in biotechnological applications, including food, fermentation, detergent, pharmaceutical, brewing and textile, and paper industries,4 According to,5 this enzyme has been used massively in many industrial applications and contributes around 30% of the total world enzyme production. Amylases can be obtained from plants, animals, and microorganisms. Nowadays, for industrial production, several types of enzymes can be produced by microorganisms such as amylase, pectinase, phytase, and xylanase.6-8 However, amylase production by molds is considered more effective in terms of cost, consistency, time, and space needed for production, as well as optimization in the production process.5 Molds that have the potential ability to produce amylases are called amylolytic molds.9,10 Several studies have obtained amylolytic fungi, including Aspergillus, Penicillium, Mucor, Rhizopus, Streptomyces, Curvularia, Hansfordia, Biporali, Rhizomucor, Rhizoctonia, Schizophyllum, Chalara, Thermoactinomyces, Thermomucorus, Curcularia, Hansfordia, Biporali, Rhizomucor, Rhizoctonia, Schizophyllum, Chalara. Nodilusporium.11
Molds can be associated with plants, animals, and other microorganisms. Several studies have successfully found various types of mold from various substrates such as animal feed,10 orange leaves (Citrus nobilis),12 and apples. Amylolytic molds are generally spread on substrates that contain carbohydrates. August flower (P. venusta (Ker Gawl.) Miers) is a dicotyledonous plant belonging to the group of Bignoniaceae,13 which is widely distributed in tropical and subtropical regions. In addition, the use of these flowers of their active compounds is also extensive. Many research on the botanical, pharmalogical and phytochemical characteristics of August flower (P. venusta) plant have been carried out.1 There are simple carbohydrate contents in the flower section of plants, such as galactose, glucose, fructose, sucrose, melibiosa.2,14 Carbohydrate content on the flower surface allows the presence of amylolytic molds that might involve co-metabolisms with the hosts.
Increasing the activity of amylase production from the mold can be done with optimization techniques. Optimization is done by regulating the environmental conditions of microorganism growth so that the amylase produced can be applied on an industrial scale.15 Growth conditions such as pH and temperature of the medium can affect the amylase activity.11 In this study, molecular identification of molds isolated from August flower (P. venusta) were performed, followed by culture-dependent analysis. The isolates were then tested for their ability to produce amylases and obtained the optimal temperature and pH of growth using the Solid State Fermentation (SSF) method.
Sampling
Ten stalks of August flower Pyrostegia Venusta (Ker Gawl.) Miers (Figure 1) were collected from Taman Nasional Bedugul, Taman Nasional Bedugul, Bedugul, Candikuning Village, Baturiti District, Tabanan Regency, Bali, Indonesia (8°07’42.9″S 114°28’28.6″E).
Mold Isolation
Mold isolation was carried out by direct method according to10 using Potato
Dextrose Agar (PDA) media. The flower sample is cut into small pieces until it reaches a weight of 1 g of each flower. The flower chunks were washed in 30 ml saline solution, followed by a vortex at 200 rpm for 1 hour. Each piece of flower petals was added with 0.05% tetracycline (after sterilization) and 0.1 ml of suspension was inoculated into the PDA medium with three replications and incubated for 72 hours at 27°C. After cultivation, all single colonies were picked up and placed into new plates to create colony libraries. The cultures from this study were deposited in Universitas Negeri Jakarta Culture Collection (UNJCC), Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Jakarta, Indonesia.
Culture-dependent Analysis and Molecular Identification
Culture-dependent analysis was performed by morphological identification after growing the isolates on a PDA medium for seven days.10 Macroscopic characteristics, including colony surface, texture, zoning, colony reverse color, and exudate drops, were observed. Microscopic characteristics such as conidial head shape, vesicle shape, hyphae bulkhead, and spore form16 were also recorded.
Two representatives of mold isolates were selected for molecular identification based on initial screening test values of amylolytic indexes. Mold isolates were grown on Potato Dextrose Broth (PDB) medium for three days and then put into a shaker at 200 rpm until a mycelium pellet was produced. For DNA extraction, two loopful of cells were added to a 1 ml Eppendorf tube. The DNA was extracted using the Genomic DNA Mini Kit Plant (Geneaid Lot.No. FC24112). PCR was performed under Go Taq Green Master Mix (Promega) protocol to amplify the ITS region of ribosomal DNA using primers ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′). The total reactions were 25 μL which contains 0.5 μL of each primer ITS4 and ITS5 (conc. 10 pmol), 3 μL of 100 ng DNA templates, GoTaq master mix 12.5 μL, and nuclease-free water 8.5 μL. PCR condition was set as follows: post denaturation 95°C for 2 minutes; denaturation 95°C for 30 seconds; annealing 58oC for 30 seconds; elongation 72°C for 1 minute; and 72°C for 10 minutes.17 The nucleotide sequences of ITS regions were determined using the First Base service. Sequence analysis was performed using the Basic Local Algorithm Search Tools (BLAST) program. Sequences of the ITS regions were manually edited and assembled using MEGA version 4 software.18 The distance matrix for the aligned sequences was calculated using Kimura’s two-parameter method. The neighbor-joining (NJ) method was used to construct all phylogenetic trees. The robustness for individual branches was estimated by bootstrapping with 1000 replicates.
Amylase Production and Optimization using Solid-State Fermentation (SSF) Method
Amylase production is carried out based on19 with four steps: formulation of wheat brain fermentation media; formulation of Mcilvein buffer with the addition of nutrients; inoculation of inoculum; incubation and enzyme extraction. Optimization of amylase activity was carried out by Solid-State Fermentation (SSF) with different pH conditions (6, 7, and 8) and temperatures (30, 40, 50°C).
In 250 ml of Erlenmeyer, 3 ml of Mcilvein buffer solution with nutrients (1% starch, 1% peptone, 0.2% MgSO4, 0.02% CaCl2) was added into 5 g of wheat brain until got moistened 60%. The Mcllvein buffer was prepared based on Mcilvein (1921) by mixing 0.2 M Na2HPO4 stock solution (3.56g /100ml) and 0.1 M citric acid (0.64/100 ml) until reaching the desired pH 6.0 (12.63 ml 0.2 M Na2HPO4 added 7.37 ml 0.1 M citric acid), pH 7 (16.47 ml 0.2 M Na2HPO4 added 3.53 ml 0.1 M citric acid), pH 8 (19.45 ml 0.2 M Na2HPO4 added 0.55 ml 0.1 M citric acid) as a solvent used for distilled water. Subsequently, the media was sterilized by autoclave at 121°C for 15 minutes. After sterilization, the media was cooled and inoculated with a 7-day-old mold suspension of 500 µL. Crude enzyme extraction was carried out after 5-days fermentation based on9,20 at different temperatures (30, 40, 50°C). A total volume of 50 ml cold sterile distilled water containing 0.1% tween 80 was added to the fermented media. The mixture is then put into a shaker for 60 minutes at 200 rpm. The mixture was then filtered using two muslin clothes, followed by filtration using Whatman No.1. The filtrate was then centrifuged at 8000 rpm at 4°C for 15 minutes. The remaining filtrate was then used for further analyses of amylase activity.
Amylase Enzyme Assay using DNSA Method
Estimating amylase activity was carried out according to the DNSA (3,5-dinitro salicylic acid) method and using starch as a substrate based on.21 0.5 ml of 1% (w/v) starch was incubated with 0.5 ml of the enzyme extract and 0.05M citrate buffer (pH 4.8). The reaction mixture was incubated at 50°C for 5 min. The reaction was stopped by adding 2ml of DNS and kept in a boiling water bath for 15 minutes. Furthermore, 6 ml of sterile distilled water is added to the sample. The absorbance was read at 540 nm using a Spectrophotometer (Shimadzu, Thermoelectric cell holder, S-1700) to measure the enzyme activity. One unit of enzyme activity is defined as the amount of enzyme which releases 1μmole of reducing sugar as glucose per minute under the assay conditions (U/ml/min). The experiments were carried out in triplicates, and standard error was calculated. The calculation of enzyme activity can be done using the following formula21:
EA= [Glucose] x 1000 x[Df] / Mw Glucose xVx t
Note:
EA = Enzyme Activity (U/ml)
[Glucose] = Glucose concentration produced by starch hydrolysis
Df = Dilution factor
Mw Glucose = Molecular weight of Glucose (Mw = 180)
t = Incubation time (15 min)
V = Volume of enzyme (0,5 ml)
Fungal Isolation and Qualitative Screening of Amylolytic Molds
A total of 130 isolates were obtained from P. venusta fruit samples using the washing and direct method. Isolation was carried out using PDA medium. The inoculated plates were incubated at 30°C for 72-hour. In this study, isolation using the direct method resulted in more molds obtained than using the washing method. Based on morphological characteristics, 10 isolates of ephyphitic molds were selected for further analysis.
Qualitative Screening of Amylolytic Molds
Based on the amylolytic index, 10 mold isolates were obtained. The isolates were then observed by the presence of a clear zone around the mold colonies. Amylolytic qualitative screening results showed that all mold isolates (UNJCC F100, UNJCC F106, UNJCC F101, UNJCC F97, UNJCC F103, UNJCC F104, UNJCC F105, UNJCC F99, UNJCC F102, and UNJCC F98) produced clear zones after staining with 0.1% Lugol iodine solution (Figure 2). Based on the amylolytic index values, two mold isolates, UNJCC F100 and UNJCC F106, have the highest IA values compared to 8 other isolates. IA values of UNJCC F100 and UNJCC F106 isolates were 1.34±0.10 and 1.08±0.12, respectively (Table 1). Other isolates ranged from 0.67±0.20 to 0.14±0.01 with the lowest IA value is UNJCC F98. Further to the analysis, the difference values between the isolates were analyzed using one-way ANOVA. The results showed differences in the amylolytic index in each mold isolate is Sig. 0.00 <a (0.05). This shows that all mold isolates have significantly different amylolytic index values, which continues to perform the Duncan Multiple Range Test (DMRT) to classify fungi based on the value of the amylolytic index.
Table (1):
Amylolytic index value of molds from August flower on YPSA medium, incubation temperature of 30°C for 72 hours.
Isolate codes |
Amylolitic Index (AI) (Mean ± SE) |
|---|---|
UNJCC F100 |
1.34e ± 0.10 |
UNJCC F106 |
1.08d ± 0.12 |
UNJCC F101 |
0.67c ± 0.20 |
UNJCC F97 |
0.59c ± 0.01 |
UNJCC F103 |
0.35b ± 0.01 |
UNJCC F104 |
0.31ab ± 0.05 |
UNJCC F105 |
0.30ab ± 0.01 |
UNJCC F99 |
0.19ab ± 0.10 |
UNJCC F102 |
0.18ab± 0.02 |
UNJCC F98 |
0.14a ± 0.01 |
Notes: Duncan Multiple Range Test (DMRT) shows that the number followed by the same letter is not significantly different at α = 0.05.
Figure 2. Screening of mold isolates based on amylolytic index by the well method. (A) Negative control; (B) UNJCC F100; and (C) UNJCC F106. On the YPSA medium, incubated at 30°C for 72 hours.
Amylolytic index values of UNJCC F100 and UNJCC F106 mold isolates have moderate enzymatic reactions. The extracellular enzyme ratio value of UNJCC F100 and UNJCC F106 mold isolates is less than 2 but more than 1. In contrast, the amylolytic index values of isolates UNJCC F97, UNJCC F98, UNJCC F99, UNJCC F101, UNJCC F102, UNJCC F103, UNJCC F104, and UNJCC F10 have weak enzymatic reactions. The extracellular enzyme ratio values of UNJCC F97 isolates, UNJCC F98, UNJCC F99, UNJCC F101, UNJCC F102, UNJCC F103, UNJCC F104, and UNJCC F105 are equal to or less than 1.
Culture-dependent Analysis and Molecular Identification
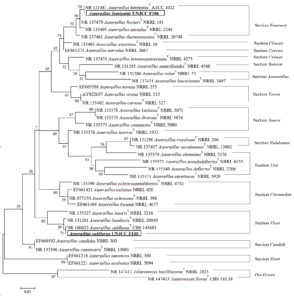
Isolates of UNJCC F100 and UNJCC F106 were identified by molecular approach to understand their phylogeny. ITS amplification using ITS 4 and ITS 5 primers generated nucleotide base sequences of 635 bp and 622 bp in length for UNJCC F100 and UNJCC F106, respectively. The area of ITS has a nucleotide base length ranging from 380-900 bp. The ITS sequences of UNJCC F100 and UNJCC F106 were then compared with the sequence of nucleotide bases stored in the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST) program to determine species closely related to UNJCC F100 and UNJCC F106 (Figure 3). In addition, the BLAST results of the ITS sequence of UNJCC F100 indicated one clade with type strain. This sequence belongs to the Aspergillus fumigatus ATCC 1022 with a 70% bootstrap value. Therefore, strain UNJCC F106 is located at the same clade with Aspergillus subflavus CBS 143683 with the bootstrap value of 75% (Figure 3).
Figure 3. Phylogenetic tree based on ITS rDNA region. Neighbor-joining method integrating Kimura-2distance was used. Data are bootstrap values issued from 1000 repetitions
Colony Measurements of the Two Potential Amylolytic Molds
The calculation results of the average colony density of mold colonies at the age of seven days showed that the A. subflavus UNJCC F100 has 6.53 × 108 CFU/ml while the A. fumigatus UNJCC F106 has of 9.83 × 108 CFU/ml (Table 3), showing that the isolates were all having high density of cells (Table 2).
Table (2):
Colony measurement of two isolates (cultivation on PDA medium, at 30°C for 48 hours).
| Isolate Codes | Dilution Factor | Colony numbers | AKK CFU/ml |
|||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Average | |||
| A. subflavus UNJCC F100 | 10-6 | 59 | 76 | 61 | 65.3 | 6.53 x 108 |
| 10-7 | 12 | 14 | 10 | 12 | ||
| A. fumigatus UNJCC F106 | 10-6 | 92 | 116 | 83 | 98.3 | 9.83 x 108 |
| 10-7 | 12 | 17 | 16 | 15 | ||
Table (3):
Effect of temperature and pH on the amylase activity (U/ml) of A. subflavus UNJCC F100, incubation on the fermentation media of wheat bran for 5 days.
| Isolates | Temperature (ºC) | pH | ||||
|---|---|---|---|---|---|---|
| 30 | 40 | 50 | 6 | 7 | 8 | |
| A. subflavus UNJCC F100 | 19.99±5.43b | 16.34±4.45a | 13.28±3.83a | 34.33±2.62b | 7.93±1.47a | 7.34±1.80a |
Note: The enzyme activity value followed by the same number is not significantly different in the temperature factor with α = 0.05 Duncan Multiple Range Test (DMRT).
Amylase Production and Optimization using Solid-State Fermentation (SSF)
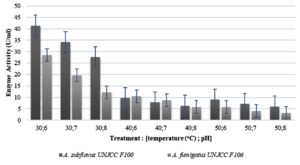
Overall, the data showed that amylase activity produced by both isolates at all temperatures decreased when the pH was higher (more alkaline condition). At 30°C the activity was higher than at any other temperature, and a significant decrease was shown when the pH condition was higher. Based on the optimization of enzyme activity results, A. subflavus UNJCC F100 and A. fumigatus UNJCC F106 molds have an optimum condition for amylase enzyme activity at 30°C and pH 6 (Figure 4). Under this optimum condition, the enzyme activity values of the two molds reached the highest values of 41.30 U/ml and 28.55 U/ml, respectively (Figure 4).
Indonesia is a tropical country with high biodiversity resources, including microorganisms.22 Indonesia has a variety of fungal and mold organisms, about 180.000-240.000 species (6-8% of the total 1.5 million species estimated) which are widely distributed in various types of sources such as soil, air, comestibles,22 plants,12 and fruits.23 Pyrostegia venusta (Ker.) (Miers). Bign, is a dicotyledon belonging to the group of Bignoniaceae.13 As it contains oleanolic acid compounds, Pyrostegia venusta (Ker.) (Miers). Bign. is often used as a chemical ingredient in traditional herb medicines in Brazil which are very important as cytotoxic, anti-consumer, antioxidant, anti-inflammatory, anti-HIV, acetylcholinesterase, alfa-beta glucosidase, antimicrobial, hepatoprotective, antipruritic, spasmolytic activity, antiangiogenic, anti-allergic, antiviral and immunomodulatory activities available on the aerial and flower parts of the plant.24
Research on epiphytic fungi on flowers has not been widely obtained. However, there have been several studies on epiphytic mold. Epiphytic microfungi in living tissue, such as young leaves, generally play a role in protection and anti-microbial activity for the host organism. Based on a review by25 Pyrostegia venusta has significant pharmacological potential and promising activities, especially in tropical diseases, skin problems, and respiratory diseases. It may serve as a vital natural bioactive medicinal source and promote a high interest in further studies.
The presence of 130 isolates from P. venusta in this study has been a proven that microorganisms did exist on the surface of flower. Pyrostegia venusta (Ker.) (Miers). Bign with an orange flower are characterized by a wide floral tube, and it contains a higher sucrose content and a higher amino acid compared to the yellow ones.3,25 The sugar content (consisting of sucrose, glucose and fructose) of nectar from Pyrostegia venusta flower was 22%, making it a good carbon source for molds to grow. Fungi can also be found on the surface of orange phylloplane.12
As a result of the amylase assay, 11 mold isolates have been confirmed to produce amylase, promising to be scaled up on an industrial scale. The use of the method allows the diffusion well because this method is an effective method to see the clear zone formed. The method for well diffusion is based on the ability of mold isolates to hydrolyze compounds in a medium after the iodine reagent has been dropped and produce a clear zone around the well testing.
In this research, starch with a concentration of 1% is used as the substrate because molds require the presence of starch as the source of energy for metabolic activity. It is found that the highest amylase enzyme activity is at 1% starch concentration.26 The addition of starch 2.5% and 5% showed a low rate of mold growth and lower amylase enzyme activity. This is due to the limitation of amylase enzyme produced by molds and is possibly caused by a feedback inhibition where the enzyme is used as an input to control the behavior of the process itself, frequently limiting the production of more enzymes.
The use of 1% yeast extract is intended as a source of nitrogen for mold to increase the growth rate and amylase enzyme activity. According to27 the highest enzyme activity was found in the addition of nitrogen in the form of 1% yeast extract. There are yeast cells, minerals, vitamins, coenzymes, and nitrogen components in the yeast extract that can support mold growth and enzyme production. The addition of MgSO4.7H2O and CaCl2.7H2O to the growth medium aimed to increase the amylase enzyme activity.
The test results showed that 10 amylolytic molds could grow well on YPSA selective media. The clear zone produced indicated that the mold can produce amylase. According to,28 clear zones are formed because the amylase enzyme has hydrolyzed starch. Active hydrolysis of starch by the amylase enzyme will cause the starch-iodine complex to decompose to form a clear zone. The iodine-starch reaction is caused by the presence of the amylose and iodine helices in forming I3- which fills the helix core.15 The absence of a clear zone is caused by starch is not hydrolyzed by the amylase enzyme so that the starch reacts with iodine and forms a deep blue color.12
The two isolates (UNJCC F100 and UNJCC F106) have the highest IA values compared to 8 other isolates. The difference in the amylolytic index value of each isolate is due to having different abilities in producing the amylase enzyme, according to the environmental conditions. Types and characteristics of molds can also affect the production of amylase enzymes. This is in line with the characteristics of each UNJCC mold isolate from August flowers (P. venusta).
Molecular identification of the two isolates based on the ITS region is commonly used to determine the closely related species. The ITS region has a high sequential variability that can be used to identify up to species level,29 and can identify molds to species level.30 Based on the phylogenetic analysis of the two isolates, it is shown that the isolate UNJCC F100 is in one monophyletic clade with A. subflavus NR160622 and isolate UNJCC F106 is located in the same clade with A. fumigatus NR121481. The length of the branches in the clade is not significantly different, indicating that there was no genetic change between the isolates. Branch length describes the number of base substitutions that can be in the form of DNA polymorphisms.31 This result is also supported by the UNJCC F100 isolate bootstrap against A. subflavus 143683, which has a high bootstrap value of 75%, and the UNJCC F106 isolate against A. fumigatus ATCC 1022 has a high bootstrap value of 70%. Bootstrap values between 70-100% indicate a high confidence level in the phylogeny tree topology formed.32 Talaromyces flavus and Talaromyces bacillisporus were used as outgroups during the reconstruction of the phylogenetic tree.33 Outgroup selection is often chosen randomly or based on clear relationships between ingroup and outgroup. Outgroups are taxa that have a close kinship with the species to be studied but are not part of the group.34
To optimize amylase production, the process is carried out using a Solid-State Fermentation (SSF) method based on.19 The use of Solid-State Fermentation (SSF) method has advantages in high productivity, low cost, and sufficient technology.35 The SSF was run using solid media containing wheat husk in this study. In several studies, wheat bran is used as the best substrate in the amylase enzyme fermentation process to increase the activity of the amylase enzyme.36,37 According to,38 the content of wheat bran consists of 60 – 75% carbohydrates, 9.6 – 18.6 protein, 9.1 – 38.9% starch, and 33.4 – 63% starch which is suitable for mold to grow.
Moisture medium is also essential in enzyme production using the SSF method. Humidity will affect the gas exchange process in the medium. According to39 too low humidity will cause negative microbial growth and decreased levels of nutrient acquisition, while high humidity in the medium will cause the gas phase to decrease and gas exchange to be inhibited so that the substrate conditions become anaerobic. Aspergillus group reached the highest enzyme activity in medium humidity around 60%.36,37,40 Research conducted by19,37 showed that the highest enzyme activity was obtained when using 10% of inoculum for the cultivation of A. flavus, Trichothecium roseum, and Thermomyces lanuginosus.
Amylase activity was measured using the DNS method to determine reducing sugar levels using dinitrosalicylate reagent. If there is reducing sugar in the sample, the DNS solution which is initially yellow, will react with the reducing sugar giving rise to a reddish-orange color. DNS is an aromatic compound that can react (redox) with reducing sugars in aldehyde groups to carboxyl groups to form 3-amino-5-nitrosalicylic acid.41
Optimization of amylase production by A. subflavus UNJCC F100 resulted in the optimal condition at 30°C of 19.99 U/ml and at pH 6 of 34.33 U/ml. The lowest enzyme activity value of A. subflavus UNJCC F100 was at a temperature of 50oC which is 13.28 U/ml and at pH 8 of 7.38 U/ml.27,37 stated that A. flavus has the optimum fermentation conditions for amylase enzyme activity at 30°C and pH 6 (680 – 685 U/mL).
Enzyme activity values were analyzed using two-way ANOVA to see the difference between temperature, pH, and their interaction with the enzyme activity value in the A. subflavus UNJCC F100. The results of univariate two-way ANOVA analysis (Table 3) showed significant differences in enzyme activity on temperature and pH factors, namely the Sig. 0.00 <a (0.05). However, there was no significant difference in enzyme activity in the interaction factors of temperature and pH (Sig. 0.110> a (0.05)). This shows that the A. subflavus UNJCC F100 is influenced by a single factor without considering the interaction between the two factors.
The results of the optimization of amylase enzyme activity in A. fumigatus UNJCC F106 obtained the highest value of enzyme activity at a temperature of 30°C and pH 6 of 28.55 U/ml and has the lowest activity value at a temperature of 50°C and pH 8 of 3.22 U/ml (Table 4). According to,42 the optimum growth conditions of A. fumigatus for amylase activity is at 30°C and pH 6. According to,24,43 the optimum temperature for A. fumigatus growth and amylase enzyme activity lies between 35°C – 37°C.
Table (4):
Amylase activity value (U/ml) of A. fumigatus UNJCC F106 (Incubation on the wheat husk fermentation media, 5 days).
| Isolate | Temperature (ºC) | pH | ||
|---|---|---|---|---|
| 6 | 7 | 8 | ||
| A. fumigatus UNJCC F106 | 30 | 28.55±3.80d | 19.57±2.67c | 12.19±3.90b |
| 40 | 10.37±0.57ab | 8.71±1.20ab | 5.67±1.01 ab | |
| 50 | 5.69±1.73ab | 3.98±0.49a | 3.22±0.66a | |
Note: The value of enzyme activity followed by the same letter is not significantly different in the temperature factor with α = 0.05 Duncan Multiple Range Test (DMRT).
The enzyme activity value was then analyzed using univariate two-way ANOVA to see the difference between the temperature and pH of the fermentation. The results of univariate two-way ANOVA analysis (Table 4) showed significant differences in enzyme activity in the temperature, pH, and interaction of both factors with the Sig. 0.00 <a (0.05). This shows that the amylase produced by A. fumigatus UNJCC F106 can be influenced by a single factor, and the interaction between environmental conditions should be considered.
Temperature and pH conditions outside the optimum conditions will cause a decrease in the value of the amylase enzyme activity. This is because the temperature in the SSF fermentation process will affect the growth rate of biomass and enzyme production. The effective temperature to increase growth and enzyme production depends on the type of mold.44 Temperature 25-37°C is the optimum incubation temperature for the activity of amylase enzymes from mesophilic molds.37 Meanwhile, A. fumigatus is a thermophilic with a growth temperature of 12-55°C. A decrease in enzyme yields at lower or higher temperatures is caused by reduced metabolic activity and disruption of fungal cell membranes.27 An increase in temperature can also cause a denatured enzyme that will change the structure of the enzyme so that the amount of substrate that can be bound by the active side of the enzyme is reduced, and the enzyme activity will definitely decrease.
In addition to physical parameters, the pH of growth media is also crucial in morphological changes in microbes and enzyme secretion.45,46 Microorganisms are sensitive to the concentration of hydrogen ions present in the medium; pH is considered an important factor in determining growth, morphology, and product formation. Most molds can grow well at a wide pH, for instance, in the pH range of 3.0-8.5, and have an optimum pH of 5.0-7.0.46,47 Therefore, pH of the substrate is crucial for the growth of fungi because certain enzymes will only break down a substrate according to its activity at a certain pH level.
Amylase production in solid-state fermentation by molds isolated from August flower (Pyrostegia Venusta (Ker.) (Miers). Bign) has been confirmed. 10 mold isolates have the potential ability to produce amylase which is promising to be used as amylase resources for industrial needs. The two isolates of UNJCC F100 and UNJCC F106 have the highest IA values of 1.34 ± 0.1 and 1.08 ± 0.12, respectively. Based on phylogenetic analysis, isolate UNJCC F100 was identified as A. subflavus, while isolate UNJCC F106 was identified as A. fumigatus which are known to be ideally used as the source of active compounds. This study exhibited that both isolate UNJCC F100 and isolate UNJCC F106 have an optimal amylase production condition at 30°C and pH 6. The two isolates are potentially used for amylase production, referring to the specific environmental condition. However, to generate a higher amount with higher amylase activity, other external variables such as medium used, inoculum concentration, and fermentation method are important to consider further for a larger application. Liquid fermentation might result a higher activity and yield of amylase in aerobic conditions.
ACKNOWLEDGMENTS
The authors would like to express gratitude to the Directorate of Research and Community Service, Ministry of Research, Technology, and Higher Education Indonesia for funding this study under the Grant of Higher Education Applied Research (PTUPT). The research grant is on behalf of Dr. Dalia Sukmawati with the title of “Application of probiotic yeasts for the development of prototypes for export quality of Indonesian cocoa production centers. We also would like to thank to Laboratory of Microbiology and Universitas Negeri Jakarta Culture Collection (UNJCC) for the facilities provided to run this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DS conceptualized the study. SR implemented methodology and collected resources. DS, DD, NF and NIR performed data analysis. NF investigated the study. HAE facilitated the study. DD and NF performed the experiments. DS supervised the study. DS, DD, NF, NIR and DJD wrote the original draft. DJD formatted the article. DS, DJD and HAE reviewed and edited the article. SR, DJD and HAE performed final proofreading. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Li W, Huang D, Wang B, et al. Changes of starch and sucrose content and related gene expression during the growth and development of Lanzhou lily bulb. PLoS ONE; 2022;17(1): e0262506.
Crossref - Wingler A, Henriques R. Sugars and the speed of life—Metabolic signals that determine plant growth, development and death. Physiologia Plantarum. 2022;174(2):e13656.
Crossref - Yalcin HT, Corbaci C. Isolation and characterization of amylase producing yeasts and improvement of amylase production. Turkish Journal of Biochemistry. 2013;38(1):101-108.
Crossref - Partil AG, Khan K, Aishwarya S, et al. Fungal Amylases and Their Industrial Applications. In: Abdel-Azeem AM, Yadav AN, Yadav N., Sharma M. (eds) Industrially Important Fungi for Sustainable Development. Fungal Biology. Springer, Cham. 2021.
Crossref - Saibaba KVN. Applications of Microbes in Food Industry. In: Inamuddin, Ahamed MI, Prasad R. (eds) Application of Microbes in Environmental and Microbial Biotechnology. 2022: 323–338 Environmental and Microbial Biotechnology. Springer, Singapore. 2022.
Crossref - Dailin DJ, Abd Manas NH, Wan Azlee NI, et al. Current and Future Applications of Phytases in Poultry Industry: A Critical Review. J Adv Vet Bio Sci Tech. 2018;3(3):65–74.
Crossref - El Sayed EA, Omar HG, Abdel Galil S, El Enshasy HA. Optimization of fed-batch cultivation for extracellular α-amylase production by Bacillus amyloliquefaciens in submerged culture. J Sci Ind Res. 2016;75:480-486. http://nopr.niscpr.res.in/handle/123456789/35152
- El Enshasy HA, El Sayed EA, Suhaimi N, Abd Malek R, Esawy M. Bioprocess optimization for pectinase production using Aspergillus niger in submerged cultivation system. BMC Biotechnol. 2018;18(71):1-13.
Crossref - Dellanerra D, Risandi A, Sunari A, Sukmawati D, Husna SN Al, El-Enshasy HA. Screening and characterization of amylolitic mold originated from ghost crab (Ocypode sp.) in Cidaon, Ujung Kulon National Park, Indonesia. (Proceding of the work of the International Scientific Conference) AIP, 2020. 2019.
Crossref - Sukmawati D, Saidah N, Handayani KT, Rahayu S. The characteristics of fungi contaminating chicken feed in Tegal, Bogor, West Java. Asian J Agric Biol. 2018;6(4):472–480. http://www.ijstr.org/paper-references.php?ref=IJSTR-1119-24428
- Batista BN, Matias RR, Oliveira RLE, Albuquerque PM. Hydrolytic enzyme production from acai palm (Euterpe precatoria) endophytic fungi and characterization of the amylolytic and cellulolytic extracts. World J Microbiol Biotechnol. 2022;38(2):30.
Crossref - Sukmawati D, Miarsyah M. Pathogenic activity of Fusarium equiseti from plantation of citrus plants (Citrus nobilis) in the village Tegal Wangi, Jember Umbulsari, East Java, Indonesia. Asian J Agric Biol. 2017;5(4):202–213.
- Bhat MP, Kumar RS, Almansour AL. Characterization, antimicrobial activity and anticancer activity of Pyrostegia venusta leaf extract-synthesized silver nanoparticles against COS-7 cell line. Appl Nanosci. 2022.
Crossref - Netlak P, Imsabai W. Role of carbohydrates in petal blackening and lack of flower opening in cut lotus (Nelumbo nucifera) flowers. Agric Nat Resources. 2016;50(1):32-37.
Crossref - Beltagy EA, Abouelwafa A, Barakat KM Bioethanol production from immobilized amylase produced by marine Aspergillus flavus AUMC10636. The Egyptian Journal of Aquatic Research. 2022;1687-4285.
Crossref - Afzal H, Shazad S, Nisa SQU. Morphological identification of Aspergillus species from the soil of larkana district (Sindh, Pakistan). Asian J Agric Biol. 2013;1:105–117.
- Sukmawati D, Andrianto MH, Arman Z, et al. Antagonistic activity of phylloplane yeasts from Moringa oleifera Lam. leaves against Aspergillus flavus UNJCC F-30 from chicken feed. Indian Phytopathology. 2020;73(2):79-88.
Crossref - Tamura K, Glen S, Sudhir K. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–3027.
Crossref - Balkan B, Balkan S, Ertan F. Optimization of parameters for α-amylase production under solid state fermentation by Trichothecium roseum. Rom Biotechnol Lett. 2011;16(5):6591–6600. https://www.rombio.eu/rbl5vol16/16%20Bilal_Balkan.pdf
- Powthong P, Sripean A, Suntornthiticharoen P. Screening of active antimicrobial and biological enzymes of microbial isolated from soil in Thailand. Asian J Pharm Clin Res. 2017;10(4):73-78.
Crossref - Aktar MB, Islam MS, Rahman MM. Determination of alpha-amylase activity of Streptomyces spp. isolated from Bangladeshi soils. Int J Interdiscip Multidiscip Stud. 2014;1(10):167-170. http://www.ijims.com/uploads/dccb616cb83be37eb807oc26.pdf
- Harnowo D, Indriani FC, Susanto Gwa, Prayogo Y, Mejaya IMJ. Biodiversity conservation through sustainable agriculture, its relevanve to climate change: a review on Indonesia situation. IOP Conference Series: Earth and Environmental Science 911 012066, 2nd International Conference on Sustainable Cereals and Crops Production System in the Tropics. Makassar, Indonesia. 2021:23-25
- Wulandari TP, Sukmawati D, Kurniati TH. Isolation and selection of amilolitic ending from nangka fruit (Artocarpus heterophyllus Lam.). Bioma. 2018;13(1):37-42.
Crossref - Goldbeck R, Pereira G, Andrade CCP. Screening and identification of cellulase producing yeast-like microorganisms from Brazilian biomes. African J Biotechnol. 2012;11(53):11595-11603. https://www.ajol.info/index.php/ajb/article/view/128950
- Mostafa NM, El-Dahshan O, Singab ANB. Pyrostegia venusta (Ker Gawl.) Miers: A Botanical, Pharmacological and Phytochemical Review. Medicinal and Aromatic Plants. 2013;2(3):1-6.
Crossref - Pankaj T, Bisht MS, Pathak A, Barh A, Sharma GB. Optimization of amylase production from the fungal isolates of Himalayan region Uttarakhand, Indonesia. Ecol Environ Conserv. 2015;21(3):1517–152. http://www.envirobiotechjournals.com/article_abstract.php?aid=6361&iid=200&jid=3
- Padmini ND, Bhattacharya S, Das A, Rajan SS. Solid-State Fermentation and Characterization of α-Amylase from a Rhizospheric Isolate of Aspergillus flavus associated with Mangifera indica. Sch Res Libr Ann Biol Res. 2012;3(8):4082–4090.
- Asadullah M. Amilolytic bacteria isolation from bekatul and ability tests for production of rough amilase enzymes on various types of production media. (Doctoral dissertation, Universitas Islam Negeri Maulana Malik Ibrahim). 2014.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241–6246.
Crossref - Sukmawati D, Setyaningsih A, Handayani KT, et al. Isolation and characterization of aflatoxigenic Aspergillus spp. from maize of livestock feed from Bogor. (Proceding of the work of the International Scientific Conference) IOP. 2018;434(1):12105.
Crossref - Dharmayanti I. Filogenetika Molekuler: Metode Taksonomi Organisme Berdasarkan Sejarah Evolusi. Wartazoa. 2011;21(1):1-10
- Smith T. Plant Systematics by Michael G. Simpson. Systematic Botany. 2016;31(3):631-632.
- Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78:63-139.
Crossref - Staton JL. Understanding phylogenies: Constructing and interpreting phylogenetic trees. J South Carolina Acad Sci. 2015;13(1):24–29.
- Chilakamarry CR, Sakinah AMM, Zularisam AW, et al. Advances in solid-state fermentation for bioconversion of agricultural wastes to value-added products: Opportunities and challenges. Bioresour Technol. 2022;343:126065.
Crossref - Karri S, Talla SG, Dholpuri S. Screening and production optimisation of alpha amylase from Aspergillus strains by using solid state fermentation. Int J Curr Microbiol Appl Sci. 2014;3:623-631.
- Manivannan S, Madhavi P, Bhuvaneswari S. Production and Optimization of α -Amylase from Aspergillus flavus under Solid State Fermentation. Int J Pharm Sci Drug Res. 2015;7:298-303.
Crossref - Onipe OO, Jideani AIO, Beswa D. Composition and functionality of wheat bran and its application in some cereal food products. Int J Food Sci Technol. 2015;50(12):2509–2518.
Crossref - Kaur H. Optimization of α-amylase and glucoamylase production in solid state fermentation of deoiled rice bran (DRB) by Rhizopus oryzae. Int J Pure Appl Biosci. 2015;3:249–256. http://www.ijpab.com/form/2015%20Volume%203,%20issue%206/IJPAB-2015-3-6-249-256.pdf
- Sakthi SS, Kanchana D, Saranraj P, Usharani G. Evaluation of amylase activity of the amylolytic fungi Aspergillus niger using cassava as substrate. International Journal of Applied Microbiology Science. 2012;1:24-34.
- Garriga M, Almaraz M, Marchiaro A. Determination of reducing sugars in extracts of Undaria pinnatifida ( harvey ) algae by UV-visible spectrophotometry ( DNS method ). Desarrollo e innovacion en ingenieria. 2017;444.
- Nwagu TN, Okolo BN. Growth Profile and Amylolytic Activity of a Thermophilic Fungus Aspergillus fumigtus Isolate from Soil. Asian J Biotechnol. 2011;3(1):46-57.
Crossref - Ratnasri P, Lakshmi BKM, Devi KA, Hemalatha KP. Isolation, Characterization of Aspergillus Fumigatus and Optimization of Cultural Conditions for Amylase Production. Int J Res Eng Technol. 2014;3(2):457–463.
Crossref - Kanti A. Potentials of kapang Aspergillus niger, Rhizopus oryzae and Neurospora sitophila as producing ezim phitase and amilase in fermented soybean substrate. Bul Peternak. 2017;41(1):26.
Crossref - Qiang L, Chai L, Tong N, Yu H, Jiang W. Potential carbohydrate regulation mechanism underlying starvation-induced abscission of tomato flower. Int J Mol Sci. 202;23(4):1952.
- Sukmawati D, Denika D, Risandi A. Screening the capabilities of Indonesia indigenous mold in producing cellulose enzyme. IOP Conf. Ser.: Mater. Sci. Eng. 2018;434(1):012125.
Crossref - El Enshasy HA, Kandiyil SK, Malek R, Othman NZ. Microbial Xylanases: Sources, Types, and Their Applications. Microbial Enzymes in Bioconversion of Biomass. 2016;3:151-213.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.