ISSN: 0973-7510
E-ISSN: 2581-690X
L-glutaminase is a unique enzyme with catalytic activity and the ability to modulate glutamine levels, making it a valuable enzyme with numerous potential applications. L-glutaminase triggers a distinctive reaction by converting L-glutamine into glutamic acid while releasing ammonia concurrently. This enzymatic process holds potential applications across diverse industries, notably in food and pharmaceuticals. The primary objective of the present research was to identify and isolate a fungal strain proficient in L-glutaminase production from soil found in maritime environments. The physical and nutritional conditions were optimized for maximum synthesis of L-glutaminase under solid state fermentation (SSF) and submerged fermentation conditions (SmF). The isolated organism was identified as Fusarium solani-melongenae strain CRI 24 by morphological and 18S rRNA analysis. The optimum carbon source under SmF and SSF was found to be starch (0.2% w/v). Wheat bran as solid substrate among others showed optimum enzyme activity. On the seventh day of incubation, at pH 8.0 and 0.7% L-glutamine concentration under SSF and SmF, the highest enzyme activity was detected. The greatest enzyme activity in SSF was seen at a moisture content of 10%. Fusarium solani-melongenae species produced the enzyme under optimal conditions and 4.20 and 4.73-fold increase (from 0.8 U/mL to 3.61 U/mL and from 0.781 U/mL to 3.69 U/mL) was achieved after optimization in submerged and in solid state fermentation, respectively. The selective isolation and optimization processes described in this work are a promising technique for the industrial production of L-glutaminase and can be applied in the pharmaceutical and food industries.
L-Glutaminase, Fusarium solani-melongenae, Solid State Fermentation, Submerged Fermentation
The most prevalent amino acid in human muscles and plasma is L-glutamine. The L-glutaminase is a hydrolytic enzyme that catalyzes the conversion of L-glutamine to glutamic acid and ammonia, which is then further catabolized to create glutathione, ATP, nucleotides, amino acids, and lipids.1 L-glutaminase has several implications in the pharmaceutical and food sectors. L-glutaminase can be used in the pharmaceutical industry to treat acute lymphocytic leukemia (ALL) as an antioxidant and anticancer drug. L-glutaminase is utilised in food industry for acrylamide breakdown, theanine production, flavour enhancer, soy sauce preparation and numerous additional applications. L-glutaminase is used in biosensors to check the glutamine levels in hybridoma and human cells, which is an important use.2
For over four decades, the most powerful enzymes utilized to cure leukemia have come from bacterial sources, primarily from Erwinia carotovora and Escherichia coli. Glutaminase production has been reported from a wide range of terrestrial bacteria, including E. coli, Pseudomonas sp., Acinetobacter sp., Bacillus sp., and Proteus morganni.3 The majority of them are isolated from soil and the aquatic (marine) environment. L-glutaminases generated by terrestrial microorganisms have a few drawbacks like instability under harsh temperatures, incompatibility with human blood, and potential reason for adverse effects in patients. Therefore, there is a pressing need to look into new enzymatic sources.1
Submerged fermentation (SmF) occurs when the substrate used for fermentation is constantly liquid and contains the nutrients required for growth. Substrates are quickly depleted, hence they must be replaced/supplemented with nutrients on a regular basis. This fermentation procedure is best suited for microorganisms like bacteria which require an excessive amount of moisture. The submerged fermentation (SmF) offers several advantages in terms of high product yield, scalability, control and monitoring, high purity and reduced contamination risk, increased process efficiency, ease of downstream processing, flexibility in strain selection, and reduced production costs. These advantages make SmF a preferred choice for various industrial fermentation processes.4
Solid state fermentation is described as “the fermentation process that occurs in the absence or near absence of free water and uses a solid substrate.” SSF has the following advantages: the process is simple, cost-effective, produces less effluent, minimizes pollution, replicates the natural habitat of some fungi and bacteria, and facilitates downstream processing. Therefore, the present investigation investigated at production of enzymes using SmF and SSF.
The marine environment, characterized by variations in temperature, pH, pressure, salinity, mineral and nutrient concentrations, as well as dynamic interactions among living organisms, serves as the largest and most diverse ecosystem on Earth. Numerous extremophiles have been isolated from this environment and have potential uses in biotechnological operations. The physiological characteristics of the bacteria that flourish in marine environments vary widely. These microorganisms’ enzymes are comparatively more stable than those of analogous terrestrial microorganisms and are resistant to extremes in salinity, pH, temperature, and pressure.5 Since, seawater and human blood plasma are chemically closer, marine microbial enzymes may be suitable for therapeutic applications with little adverse effects. Marine environments, such as seawater, share chemical and physiological similarities with human biological systems, including human blood plasma. Enzymes derived from marine microorganisms have evolved to function in these environments, making them more compatible with human physiology compared to enzymes from other sources.6 Thus, the goal of the current work was to identify marine microorganisms in order to produce L-glutaminase.
Collection of substrate
Saltwater and soil samples were gathered from Mumbai’s Aksa Beach, Maharashtra, India. The seawater was aged for 10-15 days. The collected soil samples were placed in sterile polythene bags, labelled and stored at 4˚C.
Sugarcane bagasse was gathered from the neighbourhood vendors in Bangalore, Karnataka, India. Oil cakes were procured from a coconut oil mill in Chikkaballapura, Karnataka. Other substrates like rice bran and wheat bran, were bought from organic stores. All the substrates were stored in sterile condition to avoid contamination.
L-glutaminase-producing microbe isolation and primary screening of from marine soil The soil samples (1g) were serially diluted from 10-1 to 10-3 and plated on minimum glutamine agar medium (MGA) having minor variations in composition for screening of L-glutaminase producing fungus.7 L-glutamine (0.5%), Agar (15g), magnesium sulphate heptahydrate (0.5g), ferrous sulphate heptahydrate (0.1g), zinc sulphate heptahydrate (1g), potassium dihydrogen phosphate (1g), potassium chloride (0.5g), and phenol red (0.012g) in old sea water made up the fermentation medium.
Inoculum Preparation and Submerged Fermentation
The fungi were inoculated into potato dextrose broth and incubated in a shaker incubator for 7 days at 100 rpm. The submerged fermentation process involves transferring the inoculum into 100 ml of the production medium and incubating it in a shaker incubator at 25 ± 2°C for seven days. The medium had the same components and conditions as that of the screening medium except for agar and phenol. Following the incubation time, L-glutaminase activity was measured by centrifuging the culture for 15 minutes at 10000 rpm at 4°C.
Estimating Protein and Measuring L-glutaminase Activity
L-glutamine was the substrate in the Imada et al. method for testing the activity of the enzyme.1 In order to separate the supernatant, one milliliter (mL) of the culture suspension was centrifuged at 10000 rpm for 15 minutes at 4°C. Prior to being incubated for 30 minutes, 0.1 mL of supernatant was mixed with 0.5 mL of glutamine (0.04 M) and 0.9 mL of 0.5M phosphate water (pH 7). Using distilled water, the amount was increased to 2 mL. The reaction was stopped by adding 0.5 mL of 1.5 M TCA to the mix. It was mixed with 0.2 mL of reaction mixture, 0.4 mL of Nessler’s solution, and 7.5 mL of distilled water. The mix was then left for 20 minutes and absorbance was measured at 470 nm using a Genesys 10S UV-VIS spectrophotometer. A reference curve for ammonium sulfate was used to measure the amount of ammonia that was released. One unit of L-glutaminase activity was equal to the amount of enzyme that could produce 1 µmol/min/mL of ammonia under test conditions. Using Lowry et al. method and bovine serum albumin as a standard, the protein concentration in the cell-free sample was determined.8
Identification of L-glutaminase producing fungi
The fungal strains were identified based on their appearance, lactophenol cotton blue (LPCB) staining was used. Furthermore, the most potent isolate was subjected to 18S rRNA molecular sequencing, conducted in Cellkraft Biotech PVT.LTD Hebbala (Bengaluru, India). On a 1.2% agarose gel, the DNA that was taken out was observed. The ABI 3730xl Genetic Analyzer was used to perform the PCR amplification utilizing the BDT v3.1 Cycle sequencing kit and forward and reverse DNA sequencing reaction of the PCR amplicon with 1F and 4R primers. Aligner software was used to create the consensus sequence of the 753 bp 18S region using forward and reverse sequence data.
Optimization of fermentation conditions for production of fungal L-glutaminase
To obtain the optimum medium parameter for both solid-state fermentation and submerged fermentation, one factor at a time was studied.
Optimizing the culture conditions of submerged fermentation to produce L-glutaminase
The impact of medium’s initial pH on the synthesis of L-glutaminase A digital LI 120 pH meter (ELICO) was used to regulate the pH from3.0 to 9.0 before sterilizing the fermentation media of L-glutaminase synthesis by adding 1 N NaOH or 1 N HCl. Following a 15 minute autoclaving period at 121°C, 1% (v/v) inoculum was added, and a shaker incubator was used to cultivate the mixture for seven days at 25 ± 2°C. Following fermentation, L-glutaminase activity was measured as previously mentioned.
The impact of different incubation period on synthesis of L-glutaminase
The enzyme activity was tested at different intervals, ranging from three to eight days, in order to conduct the time course study. The enzymatic output by the fungus during submerged fermentation was studied throughout a time course at room temperature (25 ± 2°C) and starting pH 7.0. The culture medium that contains an inoculum and glutamine substrate in 0.5 mL (0.04 M). Two mL of the culture broth were taken from the fermentation mixture every day at the same time and filtered through Whatman No. 1 filter paper. The filtrate was then used to find the best time for L-glutaminase productions.
The impact of different carbon source on the synthesis of L-glutaminase
Glucose, sucrose, starch, maltose, and lactose at a concentration of 0.2 percent (w/v) were added, respectively to the medium along with the inoculum and incubated at 25 ± 2°C in a shaker incubator for 7 days in order to examine the effects of various carbon sources during submerged fermentation at pH 7.0. As previously mentioned, the L-glutaminase activity was ascertained.
The impact of substrate concentration on the synthesis of L-glutaminase
The impact of varying L-glutamine concentrations (0.1, 0.3, 0.5, 0.7, and 0.9 (w/v)) with inoculum was investigated by submerged fermentation to examine the influence on enzyme synthesis. For seven days, at a pH of 7, the media was incubated at 25 ± 2°C in a shaking incubator. According to the previous description, the L-glutaminase activity was ascertained.
Optimizing the culture conditions of solid state fermentation to produce L-glutaminase The different physico-chemical factors that Fusarium solani-melongenea uses for solid state fermentation to enhance L-glutaminase output were studied. Twenty grams of every substrate, such as rice bran, wheat bran, oil cakes, and sugar cane bagasse, were weighed, dried at 50°C in a mechanical drier until the moisture content remained constant, and then transferred to 250 mL conical flasks. The substrate was wetted with 10 mL of a salt solution containing 0.6% glucose, 0.1% KH2PO4, 0.05% MgSO4, 0.7 H2O, and 0.05% KCl. After autoclaving for 20 minutes at 121°C, prior to inoculating the flasks with the fungal conidial suspension were allowed to reach room temperature. At room temperature (25 ± 2°C) and initial pH of 7.0, solid state fermentation was carried out. Following incubation enzyme activities under different solid substrate fermentation was determined. The most active enzyme-producing substrate was chosen for additional investigation. L-glutaminase synthesis was assessed in relation to the starting pH (3–9), initial moisture content (10–50% w/v), and incubation duration (3–8 days). Additionally, studies were conducted on the impact of additional carbon sources, such as lactose, 0.2% (w/v), glucose, sucrose, starch, and maltose. L-glutamine was incubated in flasks with varying concentrations (0.1, 0.3, 0.5, 0.7, and 0.9% (w/v)) in production medium in order to determine the ideal substrate concentration. Following incubation, each fermentation batch’s crude enzyme was produced. Triplicates of each test were done, and the average results were published.
Statistical analysis of data Enzyme activity and protein concentration were measured in three separate trials for each treatment. Microsoft Excel 2007 was used to calculate the mean ± standard deviation (SD). One way analysis of variance (ANOVA) was used to compare the results and determine if any significant differences in the mean values was observed, using the IBM SPSS Statistics software.
Isolation and screening of potent L-glutaminase producing microorganisms
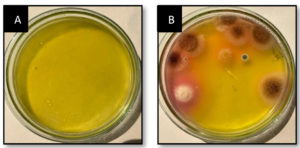
Serially diluted marine soil samples was screened for L-glutaminase production on MGA plates using the spread plate technique. One plate served as a control, with no soil microbes inoculated. In qualitative analysis after 48 hours, the largest and deepest pinkish zones were seen in isolates numbers 1, 2, and 3 on MGA plates supplemented with phenol red (Figure 1). Individual fungal colonies selected and inoculated onto MGA plate for further selection of species that produces maximum activity of L-glutaminase. A total of three fungal isolates were inoculated onto an MGA plate to test their capacity to synthesise extracellular L-glutaminase. The isolated colonies that grew larger than 10 mm after 48 h of screening were selected for secondary screening (Figure 2). Enzymes produced by these active strains were quantified by SmF.
Figure 1. Extracellular fungal L-glutaminase on MGA plate.
(A) Considered as control which consisted of MGA media. (B) Considered as test which consisted of MGA media and marine micro-organism
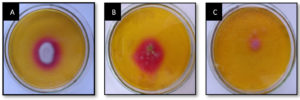
Figure 2. Fungal isolates exhibiting extracellular L-glutaminase on MGA agar plate from marine soil sample.
(A) A, B and C representing different isolates from primary plate exhibiting extracellular fungal L-glutaminase production after 48h of inoculation in to MGA plate
Secondary screening of potent fungus for L-glutaminase from soil
The enzyme activity was evaluated separately from the cell free supernatant of three different isolates for secondary screening. The selected three isolates demonstrated different levels of enzyme production under submerged fermentation. The enzyme production by the selected strains performed in triplicates. Results reported as mean ± standard deviation. The isolates produced an extracellular enzyme and isolate 1 showed highest enzyme activity of 0.86 U/mL, which was selected for the optimization studies. Isolate 2 and isolate 3 showed 0.51 U/mL and 0.43 U/mL enzyme activity, respectively.
Identification of L-glutaminase producing fungi
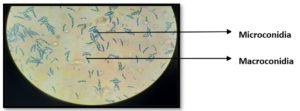
Fungal isolate 1 was morphologically identified by lactophenol cotton blue staining. Due to the observation of septate, hyaline, sickle-shaped macro- and microconidia, the fungus is classified as a Fusarium sp. (Figure 3).
Additionally, the quality of the genomic DNA was examined using a 1% Agarose gel. High-fidelity PCR polymerase was utilized to amplify the approximately 1100 bp 18S fragment. The ABI3130xl platform was utilized for bi-directional sequencing of the PCR product. To locate the fungus and its nearest neighbours, the sequence data was aligned and examined. Phylogenetic Tree Builder uses sequences that have been aligned using a System Software aligner. A distance matrix was produced using the Jukes-Cantor corrected distance model. Thus, the most potent fungal generating L-glutaminase was found to be Fusarium solani-melongenea strain CRI 24 (Figure 4).
Figure 4. The phylogenetic tree and the neighbour joining technique were used to reconstruct the evolutionary history of Fusarium solani-melongenea strain CRI
Optimizing the culture conditions of submerged fermentation to produce L-glutaminase
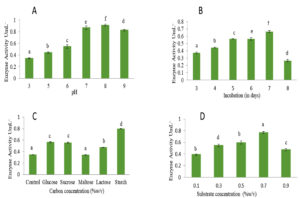
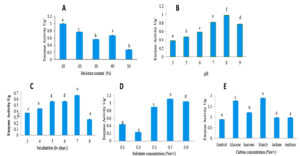
Optimization of cultural conditions for maximum L-glutaminase production were carried out by submerged fermentation and solid-state fermentation. Microorganisms make L-glutaminase best through submerged fermentation. Microorganisms’ production of extracellular L-glutaminase is heavily affected by physical, chemical, and nutritional factors. The effect of pH on L-glutaminase activity was examined over the pH range of 3 to 9. The maximum amount of L-glutaminase, with an activity of 0.913 U/mL, was produced at pH 8.0. (Figure 5A).
The time course study to determine the effect of incubation period under submerged conditions, revealed maximum enzyme production on 7th day with activity of 0.665 U/mL but a significant decrease in its yield on was observed 8th day. Therefore, further optimization steps were carried out by harvesting the fungal culture on 7th day of incubation (Figure 5B). The effect of carbon source on production of L-glutaminase from Fusarium solani-melongenea strain CRI 24 was carried out using glucose, maltose, lactose, starch and sucrose (0.2% (w/v)). Starch was found to be good carbon source compared to others resulting in maximum activity of 0.797 U/mL. (Figure 5C).
The effect of substrate concentration was studied using 0.1% to 0.9% (w/v) L -glutamine. An increase in substrate concentration enhanced the enzyme activity. 0.7% of substrate concentration served as an excellent source resulting in maximum activity of 0.76 U/mL. However, decline in enzyme production observed at 0.9% concentration (Figure 5D). The enzyme was produced by Fusarium solani-melongenea strain CRI 24 under ideal conditions, which included pH 8.0, 0.7% L-glutamine, 0.2% w/v starch as a carbon source, and a 7-days incubation period. Following optimization results in a 4.2-fold increase (from 0.8 to 3.61 U/mL) was obtained.
Figure 5. Optimizing the culture conditions of submerged fermentation to produce L-glutaminase
A: Effect of initial pH on Fusarium solani- melongenea strain CRI 24 L-glutaminase production. B: Effect of incubation time on Fusarium solani- melongenea strain CRI 24 L-glutaminase production. C: Effect of carbon source on Fusarium solani-melongenea strain CRI 24 L-glutaminase production. D: Effect of substrate concentration on Fusarium solani-melongenea strain CRI 24 L- glutaminase production.
Different alphabets on the error bars indicate significant differences at p<0.001
Optimizing the culture conditions of solid-state fermentation to produce L-glutaminase The effect of agro-wastes like rice bran, wheat bran, sugarcane bagasse and oil cakes were studied for L-glutaminase production from Fusarium solani-melongenea strain CRI 24.
Screening of various agro-wastes for L-glutaminase production by SSF revealed that wheat bran as the most suitable substrate with activity of 0.781 U/mL. The second best was rice bran with activity of 0.481 U/mL, followed by sugarcane bagasse with activity of 0.33 U/mL. Whereas oil cakes showed very low enzyme activity of 0.181 U/mL. Therefore, optimization studies were carried using wheat bran as substrate. The effect of moisture level (10% to 50% (w/v)) for L-glutaminase production was assessed and 10% found to be ideal resulting in maximum activity of 0.995 U/mL. (Figure 6A).
L-glutaminase production was tested at pH levels ranging from 3 to 9. The activity was best at pH 8.0, with 0.980 U/mL (Figure 6B). Effect of incubation time under solid-state fermentation, the time course study revealed maximum activity on 7th day with activity of 0.665 U/mL. Therefore, further optimization steps were carried out by harvesting the fungal culture on 7th day of incubation (Figure 6C). The effect of substrate concentration was studied from 0.1% to 0.9% (w/v). It was observed that 0.7% served to be an excellent substrate concentration resulting in maximum activity of 1.10 U/mL (Figure 6D). However, decline in enzyme production was observed at substrate concentration of 0.9%.
Glucose, maltose, lactose, starch, and sucrose 0.2% (w/v) were used to determine the effect of carbon sources on the ability of Fusarium solani-melongenea strain CRI 24 to produce L-glutaminase. The present study revealed starch to be good carbon source of L-glutaminase production with activity of 1.89 U/mL (Figure 6 E). The enzyme was produced by Fusarium solani-melongenea strain CRI 24 under ideal conditions, which included 10% moisture content, pH 8.0, 0.7% L-glutamine, and 0.2% w/v starch as a suitable carbon source supplement during a 7-day incubation period. A 4.73-fold increase (from 0.781 to 3.69 U/mL) was achieved after optimization by SSF.
Figure 6. Optimizing the culture conditions of solid-state fermentation to produce L-glutaminase
A: Effect of moisture concentration on Fusarium solani-melongenea strain CRI 24 L-glutaminase production. B: Effect of initial pH on Fusarium solani-melongenea strain CRI 24 L-glutaminase production. C: Effect of incubation time in days on Fusarium solani-melongenea strain CRI 24 L-glutaminase production. D: Effect of substrate concentration on Fusarium solani-melongenea strain CRI 24
L-glutaminase production. E: Effect of carbon sources on Fusarium solani-melongenea strain CRI 24 L-glutaminase production.
Different alphabets on the error bars indicate significant differences at p< 0.001
This study is the first to report the production of L-glutaminase by the Fusarium solani-melongenea strain CRI 24. Previously, Fusarium solani has been shown to produce the anti-cancer enzyme L-asparaginase.9 Many terrestrial fungi, including Penicillium, Phoma, Aspergillus, Phomopsis, Trichoderma, Cladosporium, Drechslera, Mucor, Chaetomium, and Emericella species, have been shown to produce L-glutaminase.10-16 Apart from terrestrial fungus, marine fungus are also used to produce L-glutaminase. There are relatively few reports available on L-glutaminase production from marine fungi. However, the exploration of marine fungi for L-glutaminase production is an emerging area of research that holds promising potential. Marine soil isolated fungus like Aspergillus sp. ALAA-2000, Beauveria bassiana BTMF S10, Beauveria sp, Aspergillus flavus MTCC 9972, Zygosaccharomyces rouxii NRRL-Y 2547 were reported for L-glutaminase production.3,17-19
Submerged fermentation yields substantial quantities of enzymes. Substrates are rapidly consumed in SmF, which significantly simplifies the subsequent product purification. Congruent with our investigations, submerged fermentation yielded L-glutaminase, as reported from Aspergillus sp. ALAA-2000 and Beauveria bassiana BTMF S10.17,18
According to several studies, fungi are the best organisms for SSF because their hyphal mode of growth makes it easy for them to get into the substrate and their ability to grow even when there isn’t much water present. Therefore, studies with Aspergillus sp. ALAA-2000 and Zygosaccharomyces rouxii NRRL-Y 2547 were used to make L-glutaminase in SSF.17,20
Optimizing the production of L-glutaminase by Fusarium solani-melongenea strain CRI 24 was studied using various substrates, such as rice bran, wheat bran, sugarcane bagasse, and oil cakes in SSF. The maximum L-glutaminase productivity was observed using wheat bran as substrate. Therefore, further optimization studies were carried out using wheat bran as a substrate. This might be because wheat bran has superior mechanical qualities and a high nutritional content, which encourages fungal sporulation. These observations are in agreement with the previous studies like Aspergillus oryzae, Zygosaccharomyces rouxii and Trichoderma Koningii.20-23
Depending on the kind of microbe and substrate utilized in the solid state fermentation process, the moisture content in the process is crucial for the generation of enzymes. When 10% moisture was added to wheat bran medium, Fusarium solani melongenea strain CRI 24 produced the largest amount of L-glutaminase. Further increasing in moisture content resulted in decreased enzyme activity. Maximum L-glutaminase production in Aspergillus flavus KUGF009 using SSF reported at moisture content 50% and further increase in moisture content resulted in decrease in enzyme activity.24
The enzyme activity was highest in both SmF and SSF when the pH was 8.0. The production medium’s pH affected the availability of nutrients and the growth of fungus. Highest L-asparginase enzyme activity were reported at pH 7.0 for few Fusarium genus.9,25 Further marine fungus Beauveria bassianna BTMF S10 exhibited maximum production at pH 7.0 .18
L-glutaminase formation increased significantly on 7th day of production in SmF and under SSF. There was a significant decrease in its yield after eight days of incubation under SmF and SSF. L-glutaminase formation increased significantly on the 6th day and 7th day of fermentation and then gradually decreased production course in Aspergillus versicolor Faesay4 and M. circinelloides due to enzyme degradation.10
Five different carbon sources such as glucose, fructose, lactose, starch and maltose were studied for L-glutaminase production. Fusarium solani-melongenea strain CRI 24 produced highest enzyme activity with starch (0.2%) as substrate; maltose and lactose supplementation decreased enzyme activity. Interestingly, substrate such as sucrose and dextrose were found to be good carbon source for L-glutaminase production by A. versicolor Faesay4, F. oxysporum and T. longibrachiatum.10,26,27
It has been reported that fungi show remarkable selectivity for nitrogen source that is present in the medium. The production of enzymes is significantly impacted by the availability of nitrogen. When compared to the other concentrations, the concentration of 0.7% L-glutamine showed the highest L-glutaminase activity. Previous studies found that the highest yield for enzyme production was achieved when L-glutamine was added at concentrations of 1.5, 1.0%, and 2.5%.10,14,27 Lower concentrations of substrate make fermentation more cost effective.
Fusarium solani-melongenea strain CRI 24, a marine soil fungus, was found to be a highly effective source of the medicinal enzyme L-glutaminase in both SmF and SSF conditions. Fusarium solani-melongenea strain CRI 24 produced 4.20 times as much L-glutaminase (3.61 U/mL) under optimized conditions as compare to non-optimized conditions (0.8 U/mL). The optimized parameters of SmF in MGA medium include a 7-day incubation period, an initial pH of 8.0, starch (0.2%, v/w) as the sole carbon source, and glutamine (0.7%, w/v) as the sole nitrogen source. In this study, wheat bran was used as the substrate in the fermentation medium. This made the process more cost-effective. For the best SSF conditions with wheat bran as the substrate, the medium should have a pH of 8.0 at the start, 10.0% moisture, and 0.2% starch (v/w) as the only carbon source and 0.7% glutamine (w/v) should be used as the only nitrogen source. In comparison to 0.781 U/mL of the enzyme produced under non-optimized circumstances, Fusarium solani-melongenea strain CRI 24 greatly increased the L-glutaminase production to 4.73 fold (3.69 U/mL). So, attempts should be made to optimize the culture characteristics for large scale production of L-glutaminase from marine fungus Fusarium solani-melongenea strain CRI 24. Further, strain improvement, purification and characterization of the enzyme is needed to reach industrial demand and also to understand enzyme stability under extreme conditions. Overall, L-glutaminase has significant importance in biomedical applications, the food industry, biotechnology, enzyme engineering, and diagnostic research. Its unique catalytic activity and ability to modulate glutamine levels make it a valuable enzyme with diverse potential applications. Ongoing research and further exploration of L-glutaminase can uncover additional uses and contribute to advancements in various fields.
ACKNOWLEDGMENTS
The authors express their gratitude to Dayananda Sagar University for furnishing all essential necessities and to Cell Kraft Private Ltd. for giving the facility required to perform 18S rRNA analysis. The authors are thankful to the Deanship of Scientific Research at Najran University for funding this work under the Research Group Funding program grant code NU/RG/MRC/12/14.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
The authors are thankful to the Deanship of Scientific Research at Najran University for funding this work under the Research Group Funding program grant code NU/RG/MRC/12/14.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Imada A, Igarasi S, Nakahama K, Isono M. Asparaginase and glutaminase activities of microorganisms. J Gen Microbiol. 1973;76(1):85-99.
Crossref - Yılmaz D, Karakus E. Construction of a potentiometric glutamate biosensor for determination of glutamate in some real samples. Artif Cells Blood Substit Biotechnol. 2011;39(6):385-339.
Crossref - Sabu A, Keerthi T, Kumar SR, Chandrasekaran M. L-glutaminase production by marine Beauveria sp. under solid state fermentation. Process Biochem. 2000;35(7):705-710.
Crossref - Difference Between. Difference between solid state fermentation and submerged fermentation. Compare the Difference Between Similar Terms. 2019. https://www.differencebetween.com/difference-between-solid-state-fermentation-and-submerged-fermentation. Accessed January 15, 2024.
- Nguyen T, Nguyen V. Characterization and applications of marine microbial enzymes in biotechnology and probiotics for animal health. Adv Food Nutr Resvol. 2017;80:37-74.
Crossref - Iyer PV, Singhal RS. Screening and selection of marine isolate for L-glutaminase production and media optimization using response surface methodology. Appl Biochem Biotechnol. 2009;159(1):233-250.
Crossref - Abd-Alla MH, El-Sayed EA, Rasmey AHM. Biosynthesis of L-Glutaminase by Streptomyces variabilis ASU319 isolated from rhizosphere of Triticum vulgaris. Univers J Microbiol Res. 2013;1(3):27-35.
Crossref - Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chemvol. 1951;193(1):265-275.
Crossref - Fazilath U, Narasimha MK, Srinivas C. Optimization of physiological conditions for L-asparaginase production by endophytic fungi (Fusarium solani) isolated from Tinospora cordifolia (Willd.) Hook. F & Thomson. Euro J Exp Bio. 2016;6(3):37-45.
- Awad MF, El-Shenawy FS, El-Gendy MMAA, El-Bondkly EAM. Purification, characterization, and anticancer and antioxidant activities of L-glutaminase from Aspergillus versicolor Faesay4. Int Microbiol. 2021;24:169-181.
Crossref - Khalil MS, Moubasher MH, El-Zawahry MM, Miche MM. Evaluation of antitumor activity of fungal L-glutaminase produced by Egyptian isolates. Lett Appl Nano BioSci. 2020;9(1):924-930.
Crossref - El-Gendy MMAA, Taher MT, Nageh FA, Fareed HSM. Process optimization of L-glutaminase production; a tumour inhibitor from marine endophytic isolate Aspergillus sp. ALAA-2000. Int J PharmTech Resvol. 2016;9(8):256-267.
Crossref - El-Gendy MMAA, Al-Zahrani SHM, El-Bondkly AMA.Construction of potent recombinant strain through intergeneric protoplast fusion in endophytic fungi for anti-cancerous enzymes production using rice straw. Appl Biochem Biotechnol. 2017;183(1):30-50.
Crossref - El-Gendy MMAA, Yahya SMM, Hamed AR, Soltan MM, El-Bondkly AMA. Phylogenetic analysis and biological evaluation of marine endophytic fungi derived from Red Sea sponge Hyrtios erectus. Appl Biochem Biotechnol. 2018;185(3):755-777.
Crossref - Abu-Tahon MA, Isaac GS. Purification, characterization and anticancer efficiency of L-glutaminase from Aspergillus flavus. J Gen Appl Microbiol. 2019;65(6):284-292.
Crossref - Unissa R, Sudhakar M, Reddy ASK, Sravanthi KN. A review on biochemical and therapeutic aspects of glutaminase. Int J Pharm Sci Res. 2014;5:4617-4634.
- Ahmed A, Taha TM, Abo-Dahab NF, Hassan FS. Process optimization of L-glutaminase production; a tumour inhibitor from marine endophytic isolate Aspergillus sp. ALAA-2000. J Microb Biochem Technol. 2016;8:256-267.
- Keerthi T, Suresh P, Sabu A, Rajeevkumar S, Chandrasekaran M. Extracellular production of L-glutaminase by alkalophilic Beauveria bassiana BTMF S10 isolated from marine sediment. World J Microbiol Biotechnol. 1999;15:751-752.
Crossref - Sathish T, Uppuluri K, Chari PVB, Kezia D. Sequential optimization methods for augmentation of marine enzyme production in solid-state fermentation: L-glutaminase production a case study. Adv Food Nutr Res. 2016;78:95-114.
Crossref - Kashyap P, Sabu A, Pandey A, Szakacs G, Soccol CR. Extracellular L-glutaminase production by Zygosaccharomyces rouxii under solid-state fermentation. Process Biochem. 2002;38(3):307-312.
Crossref - Koibuchi K, Nagasaki H, Yuasa A, Kataoka J. Molecular cloning and characterization of a gene encoding glutaminase from Aspergillus oryzae. App Microbiol Biotechnol. 2000;54:59-68.
Crossref - Yano T, Ito M, Tomita K, Kumagai H, Tochikura T. Purification and properties of glutaminase from Aspergillus oryzae. J Ferment Technol. 1988;66:137-143.
Crossref - El-Sayed ASAF. L-glutaminase production by Trichoderma koningii under solid-state fermentation. Indian J Microbiol. 2009;49(3):243-250.
Crossref - Nathiya K, Nath SS, Angayarkanni J, Palaniswamy MJ. Optimised production of L-glutaminase: A tumour inhibitor from Aspergillus flavus cultured on agroindustrial residues. Afr J Biotechnol. 2011;10(63):13887-13894.
Crossref - Hosamani R, Kaliwal BB. Isolation, molecular identification and optimization of fermentation parameter for the production of L-asparaginase, an anticancer agent by Fusarium equiseti. Int J Microbiol Resvol. 2011;3(2):108-119.
Crossref - Olarewaju MO, Nzelibe HC. Statistical optimization of L- glutaminase production by Trichoderma species under solid state fermentation using African locust beans as substrate. Afr J Biochem Res. 2019;13(6):73-81.
Crossref - Hamed SR, Al-wasify RS. Production and optimization of L-glutaminase from a terrestrial fungal Fusarium oxysporum. Int J PharmTech Res. 2016;94:233-241.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.