ISSN: 0973-7510
E-ISSN: 2581-690X
In the present research, bioflocculant was produced from Bacillus subtilis using molecular methods. The structural and functional features of the bioflocculant were determined that it consisted of 64% carbohydrate and 18% protein. Production medium for bioflocculant mainly comprised Sucrose 16 g; Peptone 2 g; MgSO4·7H2O 0.3 g were statistically optimized to increase the bioflocculant production. They were further characterized by Field emission scanning electron microscopy (FE-SEM), and Fourier-transform infrared spectroscopy (FTIR).Therefore, they could be considered as a suitable method for treating municipal wastewater based on high flocculating rate.
Bioflocculant, Wastewater, RSM, Kaolin, Treatment, COD, BOD.
Bioflocculant is a polymeric substance secreted by a large group of microorganism such as bacteria, fungi etc.1-3. They are the macromolecular polymer such as polysaccharide, nucleic acid, and glycoprotein4. The bioflocculant consists of monosaccharide units such as mannose, rhamnose, or glucose5. Generally, flocculants are divided into the three components such as (i) inorganic flocculants (ii) organic flocculants (iii) synthetic flocculants. However, these flocculant causes environmental toxic issues, and health issues to animals and humans6. On comparing the bioflocculant secreted by the microorganism, with synthetic flocculants7,8, they are non-toxic, harmless and biodegradable. Therefore, research has been focused recently on bioflocculant.
Many bioflocculant produced from various microorganisms has been described such as Proteus mirabilis9, Bacillus firmus10, Azotobacter and Bradyrhizobium11, Streptomyces and Cellulomonas3, and Aspergillus12. Bioflocculant have been extensively used to treat brewery wastewater, meat processing wastewater, river water, starch wastewater13,14, molasses wastewater15, dye solutions16, and to purify drinking water at low temperature17.
In this study, a novel bioflocculant-producing strain was isolated from the sewage water and was identified as bacillus subtilis by 16S rDNA sequence analysis. Further, research has focused to optimize the medium composition to enhance bioflocculant production via Response surface methodology (RSM). The major components of the bioflocculant were studied. Various factors (dosage of the bioflocculant, time, pH, electrolytes) affecting the flocculation process were investigated to find out the flocculating activity in Kaolin suspension. Finally, bioflocculant developed from bacillus subtilis was used in the municipal wastewater to reduce BOD, COD and other physico-chemical parameters.
Bioflocculant producing bacterial strain isolation
Sewage sample was collected from the drainage mixed into the Cauvery in an air-tight bottle. Serial dilutions were carried out from processed water sample in the nutrient broth. Distinct colonies in their morphological characteristics were selected and inoculated into 50 ml fermentation medium and kept it for 48 hrs at 37°C with continuous shaking. After the incubation, culture broth were analyzed to observe the flocculating activity. Based on flocculating activity, the strain was selected, then stored at 4°C.
Identification of strain
The selected strain was inoculated into LB medium for 16 hrs at 37°C with continuous stirring at 150 rpm. The isolated genomic DNA from the selected strain was further subjected to PCR analysis. The primers used were 52 -AGAGTTTGATC (C/A) TGGCTCAG-32 (forward) and 52 TACGG(C/T)TACC TTGTTACGACTT-32 (reverse). The PCR program consists of 30 cycles with 94°C (1 min), 55°C (30 s), and 72°C (1.5 min)18. The sequence obtained was compared with the NCBI database and was identified as Bacillus subtilis.
Media component optimization with RSM
Plackett–Burman (PB) strategy is the tool used to recognize significant components that affect the bioflocculant production. In the study, six medium components were studied such as Sucrose, Peptone, KH2PO4, K2HPO4, NaCl, and MgSO4. All the variables were evaluated in 12 trials. Selected three components (Sucrose, Peptone, and MgSO4) were optimized by RSM. The optimized result was subjected to analysis of variance (ANOVA).
Production and purification of the bioflocculant
Bacillus subtilis was inoculated into the medium designed and incubated at room temperature with continuous stirring for 72 hrs19. The bioflocculant derived from Bacillus subtilis strain was partially purified based on the previously described method20. The broth was centrifuged at 12000 rpm for 10 min. Chilled ethanol (1:3) was added with the supernatant followed by centrifugation at 12000 rpm for 10 min. The precipitate obtained was then dissolved in deionized water.
Structural analysis of the bioflocculant
Phenol sulfuric acid method were used to determine the sugar content of the bioflocculant9. Similarly, protein content of bioflocculant was determined by the Lowry method21. They were exposed to Fourier transform infrared spectroscopy analysis (FT-IR; Cary 630, Agilent, and USA). Further, it was subjected to SEM (SUPRA 55 SAPPHIRE; Germany)22.
Determination of the flocculating activity
Kaolin suspension (4.0 g/L) was added with varying amounts of bioflocculant solution (2–100 mg/L) and 9 mM CaCl2 solution (5 mL) at pH 7.0. The flocculating activity was then determined according to the literature23. Effects of pH (3-9), effect of time (0-300 min), effect of dosage (0-100 mg/L) and effects of different cations on the flocculating activity were examined to determine the maximum flocculation. KCl, NaCl, MgCl2, CaCl2 were tested as cationic sources.
Application in Wastewater Treatment
The experimental tanks were filled with 50 L of sewage water. 50 mg/L of bioflocculant and 10 mg/L CaCl2 solution were added with the sewage to evaluate its effect on the wastewater. Untreated raw wastewater was considered as the control sample. Finally, parametric measurements such as chemical oxygen demand (COD), suspended solids (SS), and biological oxygen demand (BOD) were determined before and after the treatment. The experiment was repeated thrice and its flocculating activity was determined.
Flocculating activity (%) = ((A-B)/A)x100;
A and B were the OD550 of the untreated and treated solution.
Bioflocculant producing bacterial strain isolation
Total of 45 different morphological strains was screened from the sewage water. Strain showing best flocculating activity in Kaolin suspension was selected for further research. The strain had been subjected to the nucleotide sequence and confirmed to be as Bacillus subtilis. The nucleotide sequence had been submitted to GenBank (MF285078). A phylogenetic tree was constructed based on the comparison with similar sequences (Fig. 1). The strain isolated was Gram-positive, aerobic, rod-shaped bacteria24,25.
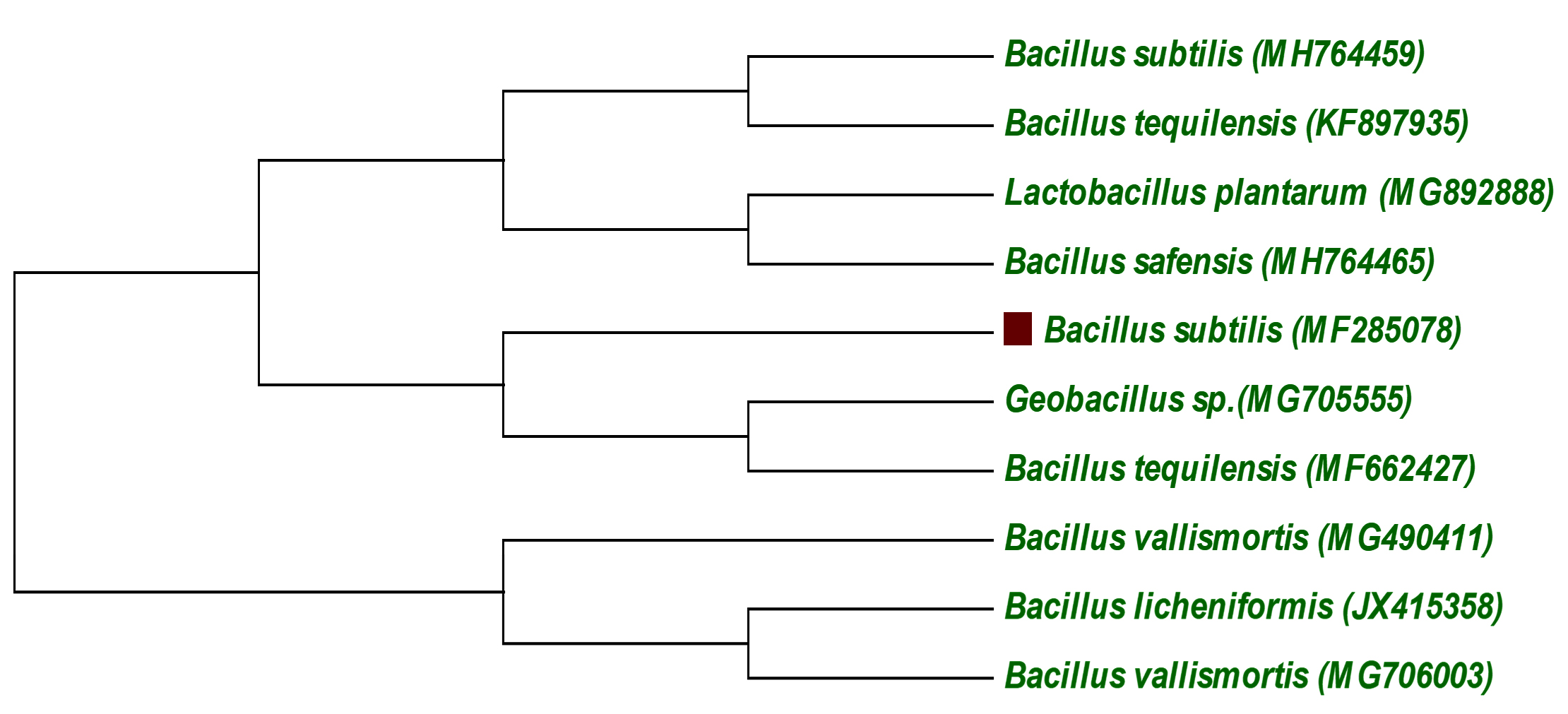 Fig. 1. Neighbor-joining phylogenetic tree based on 16S rDNA gene sequences of the strain bacillus subtilis
Fig. 1. Neighbor-joining phylogenetic tree based on 16S rDNA gene sequences of the strain bacillus subtilis Media component optimization for bioflocculant production
In this study, Plackett–Burman strategy was used to find out the components that were mainly responsible for bioflocculant production. The components were Peptone 3 g; Sucrose 12.5 g; K2HPO4 0.5 g; KH2PO4 0.5 g; MgSO4·7H2O, 0.3 g; NaCl, 0.02g; (Table 1). PB design had selected only three components based on Pareto chart (Fig. 2). The three components (Sucrose 16 g; Peptone 2 g; and MgSO4·7H2O 0.3 g) were further optimized by RSM. Central Composite Design (CCD) for RSM studies framed a total of 20 experiments with the highest yield of 4.86 (Table 3). The results obtained were further subjected to RSM. The second-order response surface model is depicted in Table 2. Form the obtained value, adequacy of the model was represented by high regression coefficient value R2 =0.9354 obtained, with respect to enhance the bioflocculant production. The model proved that coefficient of adjusted value (R2= 0.8772) was also slightly similar3,26.
Table (1):
PB design for the screening of critical media components involved in bioflocculant production
S.No |
Sucrose |
MgSO4 |
KH2PO4 |
K2HPO4 |
Peptone |
Sodium chloride |
Yield (g/L) observed value |
Yield (g/L) predicted value |
|---|---|---|---|---|---|---|---|---|
1 |
10 |
0.3 |
2.5 |
5 |
1.5 |
0.02 |
3.80 |
3.80000 |
2 |
12.5 |
0.3 |
2.5 |
5 |
0.5 |
0.2 |
2.20 |
2.26667 |
3 |
10 |
0.5 |
0.5 |
3 |
0.5 |
0.2 |
3.60 |
3.63333 |
4 |
10 |
0.5 |
2.5 |
5 |
0.5 |
0.2 |
3.00 |
3.20000 |
5 |
10 |
0.5 |
2.5 |
3 |
1.5 |
0.02 |
2.50 |
2.90000 |
6 |
10 |
0.3 |
0.5 |
5 |
1.5 |
0.2 |
3.23 |
3.23333 |
7 |
12.5 |
0.3 |
0.5 |
3 |
1.5 |
0.2 |
1.00 |
1.40000 |
8 |
12.5 |
0.3 |
0.5 |
3 |
0.5 |
0.02 |
4.00 |
4.53333 |
9 |
12.5 |
0.5 |
2.5 |
3 |
1.5 |
0.2 |
0.50 |
0.06667 |
10 |
12.5 |
0.5 |
0.5 |
5 |
0.5 |
0.02 |
3.00 |
2.90000 |
11 |
12.5 |
0.3 |
2.5 |
3 |
0.5 |
0.02 |
3.20 |
3.26667 |
12 |
12.5 |
0.5 |
0.5 |
5 |
1.5 |
0.02 |
0.50 |
1.60000 |
Table (2):
Analysis of variance showing fitted quadratic polynomial model for optimization of bioflocculant production
Source |
DF |
Seq SS |
Adj SS |
Adj MS |
F |
P |
|---|---|---|---|---|---|---|
Regression |
9 |
16.3358 |
16.3358 |
1.81509 |
16.08 |
0.000 |
Linear |
3 |
7.1558 |
7.1558 |
2.44688 |
21.68 |
0.000 |
Sucrose |
1 |
0.5044 |
0.48428 |
0.48428 |
4.29 |
0.065 |
Peptone |
1 |
0.7992 |
0.9927 |
0.99269 |
8.80 |
0.014 |
MgSO4 |
1 |
5.8522 |
5.8522 |
5.85220 |
51.85 |
0.000 |
Square |
3 |
2.2622 |
2.2622 |
0.75408 |
6.68 |
0.009 |
Sucrose * Sucrose |
1 |
1.9433 |
2.1435 |
2.14354 |
18.99 |
0.001 |
Sucrose * Peptone |
1 |
0.3183 |
0.3051 |
0.30507 |
2.70 |
0.131 |
MgSO4*MgSO4 |
1 |
5.8522 |
5.8522 |
5.85220 |
51.85 |
0.000 |
Interaction |
3 |
6.9178 |
2.30593 |
2.30593 |
20.43 |
0.000 |
Sucrose*Peptone |
1 |
4.6208 |
4.6208 |
4.62080 |
40.94 |
0.000 |
Sucrose* MgSO4 |
1 |
0.9522 |
0.9522 |
0.95220 |
8.44 |
0.016 |
Peptone * MgSO4 |
1 |
1.3448 |
1.3448 |
1.34480 |
11.92 |
0.006 |
Residual Error |
10 |
1.1286 |
1.1286 |
0.11286 |
||
Lack-of-Fit |
5 |
0.2387 |
0.2387 |
0.04773 |
2.56 |
0.163 |
Pure Error |
5 |
0.0933 |
0.0933 |
0.01867 |
||
Total |
19 |
17.4645 |
R-Sq = 93.54%; R-Sq (pred) = 74.58%; R-Sq (adj) = 87.72%
 Fig. 2. Pareto chart responsible for selecting main components with respect to increase the yield of bioflocculant production for the strain Bacillus subtilis
Fig. 2. Pareto chart responsible for selecting main components with respect to increase the yield of bioflocculant production for the strain Bacillus subtilis However, sucrose was an important factor since it had a statistical value of about 99% (Table 4). Three dimensional (3-D) response surface curves represented the interaction of substrates on the bioflocculant production (Fig. 3). Component interactions (Sucrose* Sucrose; Peptone* Peptone; MgSO4* MgSO4; Sucrose* Peptone; Peptone* MgSO4; MgSO4* MgSO4) showed significant values for bioflocculant production (Table 2). Further, the contour plot suggested that the interaction between the corresponding variables were significant (Table 2 and Fig. 4). Yield of obtained bioflocculant was 4.86 which was comparable with the predicted production obtained27.
Table (3):
CCD matrix for critical media components showing observed and predicted values for the production of bioflocculant
S. No |
Sucrose |
Peptone |
MgCl2 |
Yield (g/L)observed value |
Yield (g/L) predicted value |
|---|---|---|---|---|---|
1 |
12.0000 |
2.00000 |
0.300000 |
3.80 |
2.54587 |
2 |
14.0000 |
1.25000 |
0.400000 |
2.20 |
1.45692 |
3 |
16.0000 |
2.00000 |
0.500000 |
3.60 |
4.58213 |
4 |
14.0000 |
1.50000 |
0.400000 |
3.00 |
2.74568 |
5 |
12.0000 |
1.25000 |
0.400000 |
2.50 |
3.28796 |
6 |
14.0000 |
1.00000 |
0.400000 |
2.23 |
3.30164 |
7 |
14.0000 |
1.25000 |
0.568179 |
1.00 |
0.99224 |
8 |
16.0000 |
2.00000 |
0.300000 |
4.86 |
3.86979 |
9 |
14.0000 |
1.75000 |
0.400000 |
2.00 |
2.14092 |
10 |
17.3636 |
1.25000 |
0.400000 |
3.50 |
3.12291 |
11 |
14.0000 |
1.25000 |
0.231821 |
3.20 |
3.17359 |
12 |
12.0000 |
2.00000 |
0.500000 |
0.80 |
3.05435 |
13 |
12.0000 |
0.50000 |
0.300000 |
3.80 |
1.86280 |
14 |
16.0000 |
0.50000 |
0.500000 |
2.20 |
2.77654 |
15 |
12.0000 |
0.50000 |
0.500000 |
2.80 |
3.27155 |
16 |
10.6364 |
1.25000 |
0.400000 |
3.00 |
2.54587 |
17 |
14.0000 |
2.25000 |
0.400000 |
2.90 |
1.45692 |
18 |
16.0000 |
0.50000 |
0.300000 |
2.18 |
4.58213 |
19 |
14.0000 |
-0.01134 |
0.400000 |
2.10 |
2.74568 |
20 |
14.0000 |
2.51134 |
0.400000 |
3.00 |
3.28796 |
Table (4):
Regression analysis showing critical media components in bioflocculant production
Media |
Estimated Co-efficient |
t-Value |
p-Value |
|---|---|---|---|
Sucrose |
-0.8525 |
-5.67 |
0.002 |
MgSO4 |
-0.4025 |
-2.68 |
0.044 |
KH2PO4 |
-0.0525 |
-0.35 |
0.741 |
K2HPO4 |
0.0358 |
0.24 |
0.821 |
Peptone |
-0.6642 |
-4.42 |
0.007 |
Sodium chloride |
-0.3308 |
-2.20 |
0. 079 |
 Fig. 3. 3-D response surface graphs derived from CCD showing the interactive effects of different factors on the production of bioflocculant. (a) Interaction between Sucrose and MgCl2; (b) Interaction between Peptone and MgSO4; (c) Interaction between Sucrose and Peptone.
Fig. 3. 3-D response surface graphs derived from CCD showing the interactive effects of different factors on the production of bioflocculant. (a) Interaction between Sucrose and MgCl2; (b) Interaction between Peptone and MgSO4; (c) Interaction between Sucrose and Peptone.  Fig. 4. Contour plots for the bioflocculant obtained from CCD showing interactions between three different factors.
Fig. 4. Contour plots for the bioflocculant obtained from CCD showing interactions between three different factors. Factors influencing the flocculating activity of bioflocculant
Effects of bioflocculant dosage, temperature, metal ions and pH on the flocculating rate were determined (Fig. 5). Bioflocculant activity could be achieved by altering the charges of the solution. Since bioflocculant was negatively charged nature in solution, positively charged ions were required to carry over the flocculating process (Fig. 5(a)). Therefore, different ions such as CaCl2, NaCl, MgCl2, FeSO4, and HgCl2 were examined to achieve higher flocculating activity. Similar kind of work has been reported with other strains14,28.
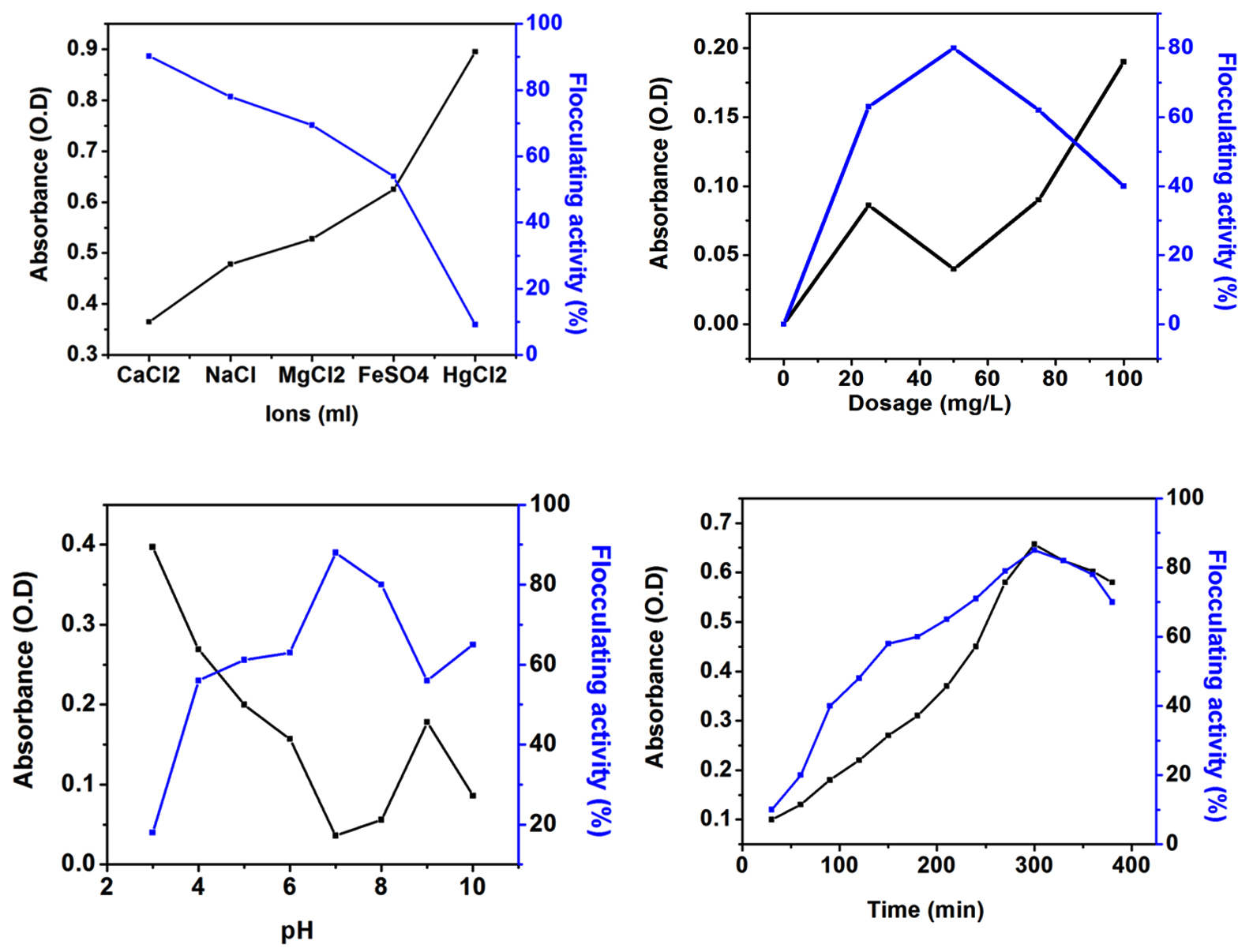 Fig. 5. Effect on the conditions of bioflocculant production; a) Effect of dosage on bioflocculant production; b) Effect of ions on bioflocculant production; c) Effect of pH on bioflocculant production; d) Effect of time on bioflocculant production.
Fig. 5. Effect on the conditions of bioflocculant production; a) Effect of dosage on bioflocculant production; b) Effect of ions on bioflocculant production; c) Effect of pH on bioflocculant production; d) Effect of time on bioflocculant production. The flocculating rate was tested in the range of 0-100 mg/L, and the maximum flocculating rate was observed at an optimum bioflocculant dosage of 50 mg/L (Fig. 5(b)). It can also be observed that a higher or lower amount of bioflocculant caused poorer flocculating rate. Bioflocculant activity could not be reached maximum level when the bioflocculant dosage was inappropriate16.
The effect of pH on the flocculating activity was studied (Fig. 5(c)). The reaction was carried out with varying pH (3-10). The maximum of 90 % flocculating activity was attained at the range of 7. Increasing and decreasing the pH would lead to the lower flocculating activity. Similarly, the effect of time on flocculating activity was also studied (Fig. 5(d)). The reaction was carried out by increasing the time from 0 to 400 min. The maximum 90% flocculating activity was obtained when it reached 300 min9,29.
Structural and functional features of the bioflocculant
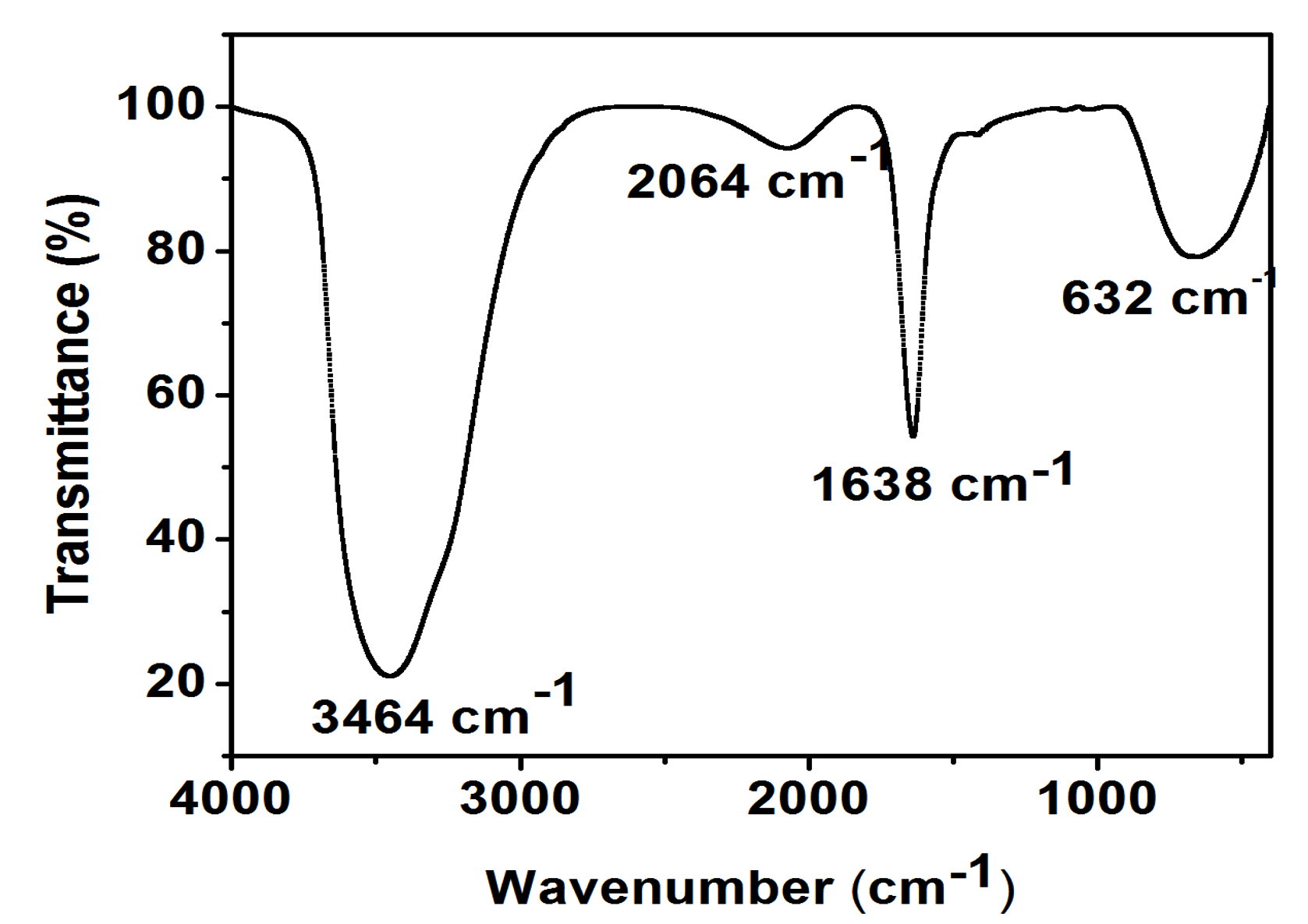
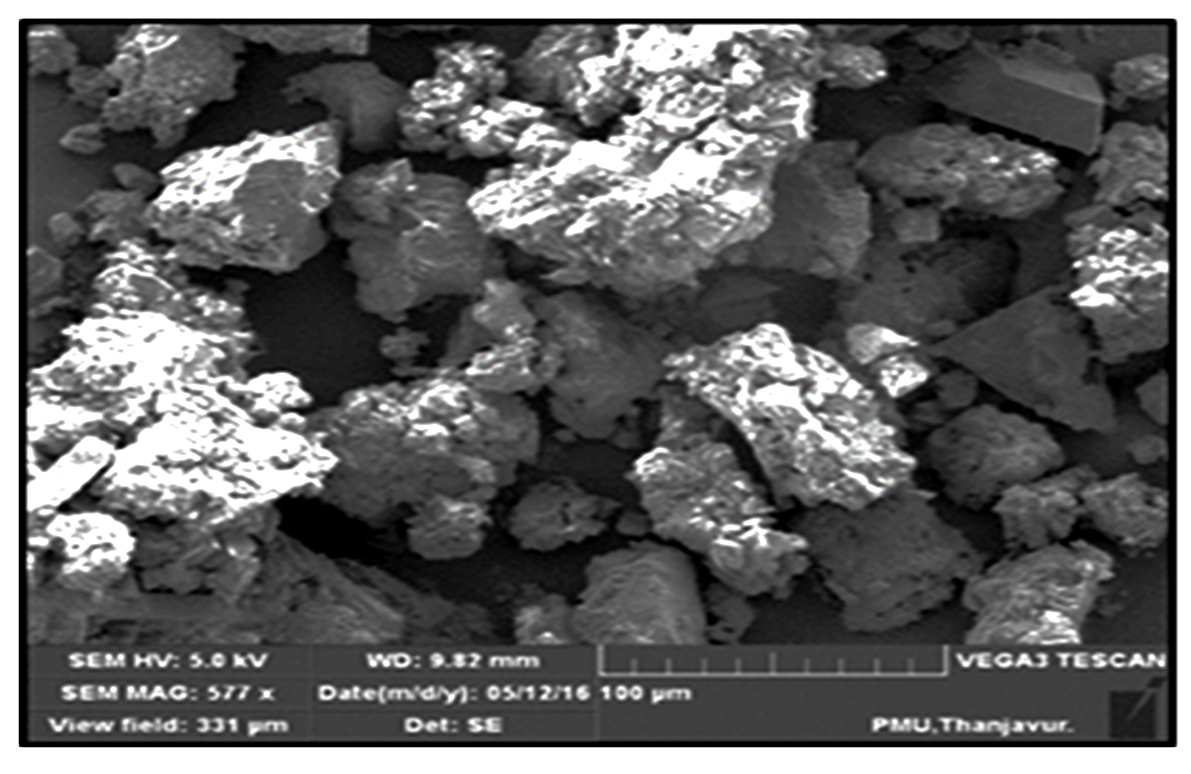
Composition of the bioflocculant was determined as 64% total sugar and 18% protein. The functional group analysis of the bioflocculant derived from bacillus subtilis found out the spectrum at 3464 cm-1 which was the characteristic of -OH, and -NH2 groups. The spectrum displayed at 1638 cm-1 was associated to the group of –COO– ion and thus, showing the presence of uronate in this polysaccharide30. The weak absorption peak identified at 1048 cm-1 was related to all sugar derivatives31. The spectrum indicated the presence of a characteristic peak of 632 cm-1 which showed the functional group of C-H bond. Therefore, -OH, -COO–, and NH2 groups were identified in the bioflocculant molecules (Fig. 6). SEM image of the bioflocculant appeared as an irregular shaped structure (Fig. 7).
Treatment of municipal wastewater
The trials were performed to find out the efficiency of bioflocculant for municipal wastewater6,32. The overall flocculating percentage of the bioflocculant was almost 90% with the dose of the bioflocculant (50 mg/L) (Fig. 8). The higher level of parametric studies indicated organic pollution in the samples. The treatment has shown the reduction of parameters such as BOD, COD, etc., that confirmed the removal efficiency of the bioflocculant.
Isolated bioflocculant-producing strain were identified as Bacillus subtilis. It was composed of mostly polysaccharides and proteins. From the result, it was confirmed that amino, hydroxyl, and carboxylate groups identified in the bioflocculant molecules. They had a high flocculating ability at a minimal dosage requirement (50 mg/L), pH of 7 against Kaolin clay and municipal wastewater. Based on the outstanding features of the bioflocculant, it could be exploited in bioremediation. Further studies are required to carry out the gene responsible for flocculation.
Acknowledgements
Author would like to thank the Department of Science and Technology Promotion of University Research and Scientific Excellence (DST-PURSE; DST Sanction Order No- SR/FT/LS- 113/2009) for providing instrument facility, and Centre of excellence in life science, Bharathidasan University, Trichy.
Funding
None.
Data Availability
All data generated during the study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Agunbiade M., Pohl C., Ashafa O. Bioflocculant production from Streptomyces platensis and its potential for river and waste water treatment. Braz. J. Microbiol., 2018; 49:731-41.
Crossref - Aljuboori A.H.R., Uemura Y., Osman N.B., Yusup S. Production of a bioflocculant from Aspergillus niger using palm oil mill effluent as carbon source. Bioresour. Technol., 2014; 171:66-70.
Crossref - Deng S., Bai R., Hu X., Luo Q. Characteristics of a bioflocculant produced by Bacillus mucilaginosus and its use in starch wastewater treatment. Appl. Microbiol. Biot., 2003; 60:588-93.
Crossref - Dermlim W., Prasertsan P., Doelle H. Screening and characterization of bioflocculant produced by isolated Klebsiella sp. Appl. Microbiol. Biot., 1999; 52:698-703.
Crossref - Gong W.-X., Wang S.-G., Sun X.-F., Liu X.-W., Yue Q.-Y., Gao B.-Y. Bioflocculant production by culture of Serratia ficaria and its application in wastewater treatment. Bioresour. Technol., 2008; 99:4668-74.
Crossref - Guo J., Liu J., Yang Y., Zhou Y., Jiang S., Chen C. Fermentation and kinetics characteristics of a bioflocculant from potato starch wastewater and its application. Sci. Rep., 2018; 8, 3631.
Crossref - He N., Li Y., Chen J. Production of a novel polygalacturonic acid bioflocculant REA-11 by Corynebacterium glutamicum. Bioresour. Technol., 2004; 94:99-105.
Crossref - Humudat Y.R., Al-Naseri S.K., Kadhim S.A. Production of highly efficient bacterial flocculant in water treatment. International Journal., 2014; 2:297-301.
- Kanmani P., Lim S.T. Synthesis and structural characterization of silver nanoparticles using bacterial exopolysaccharide and its antimicrobial activity against food and multidrug resistant pathogens. Process Biochem., 2013; 48:1099-106.
Crossref - Labille J., Thomas F., Milas M., Vanhaverbeke C. Flocculation of colloidal clay by bacterial polysaccharides: effect of macromolecule charge and structure. J. Colloid. Interface Sci., 2005; 284:149-56.
Crossref - Li X.-M., Yang Q., Huang K., Zeng G.-M., Liao D.-X., Liu J.-J., Long W.-F. Screening and characterization of a bioflocculant produced by Aeromonas sp. Biomed. Environ. Sci., 2007; 20:274-8.
- Li Z., Zhong S., Lei H.-y., Chen R.-w., Yu Q., Li H.-L. Production of a novel bioflocculant by Bacillus licheniformis X14 and its application to low temperature drinking water treatment. Bioresour. Technol., 2009; 100:3650-6.
Crossref - Liu J., Ma J., Liu Y., Yang Y., Yue D.,Wang H. Optimized production of a novel bioflocculant M-C11 by Klebsiella sp. and its application in sludge dewatering. Int. J. Environ. Sci.., 2014; 26:2076-83.
Crossref - Liu W., Wang K., Li B., Yuan H., Yang J. Production and characterization of an intracellular bioflocculant by Chryseobacterium daeguense W6 cultured in low nutrition medium. Bioresour. Technol., 2010; 101:1044-8.
Crossref - Liu W., Yuan H., Yang J., Li B. Characterization of bioflocculants from biologically aerated filter backwashed sludge and its application in dying wastewater treatment. Bioresour. Technol., 2009; 100:2629-32.
Crossref - Luo L., Zhao Z., Huang X., Du X., Wang C.a., Li J., Wang L., Xu Q. Isolation, identification, and optimization of culture conditions of a bioflocculant-producing bacterium Bacillus megaterium SP1 and its application in aquaculture wastewater treatment. Biomed Res. Int., 2016.
Crossref - Nie M., Yin X., Jia J., Wang Y., Liu S., Shen Q., Li P.,Wang Z. Production of a novel bioflocculant MNXY1 by Klebsiella pneumoniae strain NY1 and application in precipitation of cyanobacteria and municipal wastewater treatment. J. Appl. Microbiol., 2011; 111:547-58.
Crossref - Nwodo U.U., Green E., Mabinya L.V., Okaiyeto K., Rumbold K., Obi L.C., Okoh A.I. Bioflocculant production by a consortium of Streptomyces and Cellulomonas species and media optimization via surface response model. Colloids Surf. B. Biointerfaces., 2014; 116:257-64.
Crossref - Pathak M., Sarma H.K., Bhattacharyya K.G., Subudhi S., Bisht V., Lal B., Devi A. Characterization of a novel polymeric bioflocculant produced from bacterial utilization of n-hexadecane and its application in removal of heavy metals. Front. Microbiol., 2017; 8:170.
Crossref - Ramamoorthy S., Gnanakan A., Lakshmana S.S., Meivelu M., Jeganathan A. Structural characterization and anticancer activity of extracellular polysaccharides from ascidian symbiotic bacterium Bacillus thuringiensis. Carbohyd. Polym., 2018; 190:113-20.
Crossref - Rasulov B.A., Li L., Liu Y.-H., Mohamad O.A., Xiao M., Ma J.-B., Li W.-J. Production, characterization and structural modification of exopolysaccharide-based bioflocculant by Rhizobium radiobacter SZ4S7S14 and media optimization. 3 Biotech, 2017; 7:179.
Crossref - Rasulov B.A., Pattaeva M.A., Yili A., Aisa H.A. Polysaccharide-based bioflocculant template of a diazotrophic Bradyrhizobium japonicum 36 for controlled assembly of AgCl nanoparticles. Int. J. Biol. Macromol., 2016; 89:682-8.
Crossref - Salehizadeh H., Shojaosadati S. Extracellular biopolymeric flocculants: recent trends and biotechnological importance. Biotechnol. Adv., 2001; 19:371-85.
Crossref - Salehizadeh H., Shojaosadati S. Removal of metal ions from aqueous solution by polysaccharide produced from Bacillus firmus. Water Res., 2000; 37:4231-5.
Crossref - Salehizadeh H., Vossoughi M., Alemzadeh I. Some investigations on bioflocculant producing bacteria. Biochem. Eng. J., 2000; 5:39-44.
Crossref - Sam S., Kucukasik F., Yenigun O., Nicolaus B., Oner E.T., Yukselen M.A. Flocculating performances of exopolysaccharides produced by a halophilic bacterial strain cultivated on agro-industrial waste. Bioresour. Technol., 2011; 102:1788-94.
Crossref - Shahadat M., Teng T.T., Rafatullah M., Shaikh Z., Sreekrishnan T., Ali S.W. Bacterial bioflocculants: A review of recent advances and perspectives. Chem. Eng. J., 2017; 328:1139-52.
Crossref - Wan C., Zhao X.-Q., Guo S.-L., Alam M.A., Bai F.-W. Bioflocculant production from Solibacillus silvestris W01 and its application in cost-effective harvest of marine microalga Nannochloropsis oceanica by flocculation. Bioresour. Technol., 2013; 135:207-12.
Crossref - Xiong Y., Wang Y., Yu Y., Li Q., Wang H., Chen R., He N. Production and characterization of a novel bioflocculant from Bacillus licheniformis. Appl. Environ. Microbiol., 2010; 76:2778-82.
Crossref - Yim J.H., Kim S.J., Ahn S.H., Lee H.K. Characterization of a novel bioflocculant, p-;KG03, from a marine dinoflagellate, Gyrodinium impudicum KG03. Bioresour. Technol., 2007; 98:361-7.
Crossref - Zheng Y., Ye Z.-L., Fang X.-L., Li Y.-H., Cai W.-M. Production and characteristics of a bioflocculant produced by Bacillus sp. F19. Bioresour. Technol., 2008; 99:7686-91.
Crossref - Zhong C., Xu A., Chen L., Yang X., Yang B., Hong W., Mao K., Wang B., Zhou J. Production of a bioflocculant from chromotropic acid waste water and its application in steroid estrogen removal. Colloids Surf. B. Biointerfaces., 2014; 122:729-37.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.