ISSN: 0973-7510
E-ISSN: 2581-690X
Urinary tract infections (UTIs) are regarded as one of the most serious infections worldwide. Uro Pathogenic E. coli (UPEC) accounts for nearly 80% of UTI infections in females. This study investigated the antibacterial and antibiofilm effects of Lactobacillus acidophilus (l. acidophilus) and Lactobacillus plantarum (lb. plantarum) on multidrug-resistant E. coli obtained from urine samples. Complete bacteriological identification was conducted on 45 E. coli isolated from 80 urine samples of females with UTIs. Antibiotic susceptibility test was performed on all isolates by nine antibiotics. Ten out of the 45 isolates exhibited multidrug resistance (MDR). L. acidophilus and Lb. plantarum showed marked inhibition of MDR E. coli isolates on agar by a diffusion method (16 ± 0.04: 23 ± 0.05 mm). Moreover, L. acidophilus and Lb. plantarum strains inhibited the ability of UPEC to form a biofilm by 56.3% and 39.63%, respectively. The expression of biofilm genes of E. coli are as follows: csgA, crl, csgD showed remarkable downregulation after treatment with probiotics suspension: 0.00364: 0.19078 fold, 0.0005: 0.1894 fold, and 0.0490: 0.0883 for L. acidophilus, respectively. On the other hand, downregulation of biofilm gene expression for csgA, crl, csgD after treatment with Lb. plantarum suspension were expressed by fold changes as follows: 0.0769: 0.3535 fold, 0.05440: 0.12940 fold, and 0.06745: 0.4146, respectively. These findings show that L. acidophilus and Lb. plantarum exhibit potent antibacterial and antibiofilm action against MDR UPEC at both genotypic and phenotypic levels, and appear to be a promising solution in therapeutic applications for recurrent and persistent UTIs.
Urinary tract infections (UTIs), uropathogenic E. Coli, Antibacterial, antibiofilm, Lactobacillus acidophilus, Lactobacillus Plantarum
Enterobacteriaceae are a common intestinal flora of the digestive system in humans, but this type of bacteria can cause extraintestinal urinary tract infections (UTIs).1 The misuse of antibiotics has led to the increased occurrence of resistant isolates worldwide. Nowadays, the emergence of MDR E. coli has contributed to a significant problem, as there are limited therapeutic options for these pathogens, therefore leading to increased levels of both morbidity and mortality. There is an imperious need for new alternative therapeutics to treat the emergence of MDR E. coli.2
UPEC biofilm is responsible for persistent infection and resistance to antibiotics. Biofilms are groups of microorganisms that may be mono-or multispecies, embedded in a matrix of extracellular polymeric substances (EPS). This matrix is composed of exopolysaccharides, proteins, DNA, RNA, lipids,3 and signaling molecules (e.g., autoinducers), which enable communication to occur between bacterial cells through a phenomenon known as quorum-sensing (QS).4 The biofilm matrix acts as a block for the entry of chemical agents, immune molecules, and pH changes in the surrounding environment.5 In addition, the matrix promotes antibiotic resistance and facilitates the spread of resistant genes. Many genes encode for the process of E. coli biofilm formation.
Curli, a kind of amyloid fimbriae, aids the adherence to the urinary bladder and biofilm development. Curli fimbriae assist in cell adhesion, biofilm development, and aggregation in E. coli to a surface.6 The main curli subunit csgA, which is important for adhesion to host components, is encoded by the csgBAC operon.7 The csgA gene was demonstrated to cause extremely drug-resistant UPEC in clinical isolates.8 Curli acts as a glue to adhere the bacterial cells to numerous serum proteins and the extracellular matrix.9
Several non-antibiotic approaches have recently emerged, such as the use of immunomodulators, herbal extracts, hormonal, and biological therapeutics like probiotics. Probiotics offer many advantages over other therapeutics, as they are generally recognized as safe (GRAS).10 Lactobacilli are important members of the probiotic family. Lactobacilli bacteria demonstrate many antimicrobial mechanisms, including competition with pathogenic bacteria for their binding sites and nutrition, stimulation of the protective immune response, secretion of inhibitory molecules such as hydrogen peroxide, fatty acids, bacteriocins, and ethanol.11 In addition, Lactobacilli can produce many types of acids that reduce intestinal pH, for instance, acetic acid, lactic acid, and formic acid. Lactobacilli have demonstrated their capability to act against Pseudomonas aeruginosa, Shigella spp., E. coli, Clostridium difficile,12 Staphylococcus aureus, and Streptococcus mutants.13 However, few studies have studied the lactobacilli activity towards MDR Uropathogenic E. coli. Thus, we conducted the present study to discover the different antibacterial and antibiofilm abilities of lactobacilli against UPEC isolates at both phenotypic and genotypic levels.
Microbiological Examination and E. coli Isolation
This study was carried out on 80 female patients exhibiting UTIs, aged 23 to 58 years, recruited from the inpatient department and outpatient clinic of the internal medicine department, Faculty of Medicine for Girls, Al-Zahraa Hospital – Al Azhar University, Cairo, Egypt, during the period of November 2020 to January 2021.
80 urine samples were aseptically obtained and immediately relocated to the microbiology laboratory. MacConkey agar (Oxoid, USA) was used to culture the samples, which were then incubated aerobically for 24 hours at 37°C. 45 E. coli isolates were obtained. The isolated colonies were then fully identified by complete biochemical identification.14 All E. coli strains were kept at −20°C in a growth medium containing glycerol. Probiotics, L. Acidophilus and Lb. plantarum reference strains (ATCC 4356, ATCC 14917) were obtained from the Microbiological Resources Center, Faculty of Agriculture, Ain Shams University, Egypt. Both lactobacilli strains were cultured on Man-Rogosa-Sharpe (MRS) (HiMedia, India) agar and broth medium under anaerobic conditions at 37°C for 48 h with 5% CO2.
Antibiotic Sensitivity Test for Isolated E. coli
Susceptibility testing for all E. coli isolates was carried out according to the disk diffusion (modified Kirby Bauer) assay15 on Mueller–Hinton agar (Merk, Germany) in accordance with the Clinical and Laboratory Standard Institute (CLSI) guidelines in the case of the following antibiotics (Oxoid, UK): Amoxicillin + Clavulanic Acid (AMC 20 + 10 µg), Ceftazidime (CAZ, 30 µg), Ceftriaxone (CRO, 30 µg), Ciprofloxacin (CIP 5 µg), Amikacin (AK 30 µg), Cefotaxime (CTX 30 µg), Ampicillin (AMP 10 µg), Gentamycin (CN 30 µg), and Nitrofurantoin (F). The results were inferred utilizing CLSI guidelines (2019).
Agar Well Diffusion Method for MDR Isolates
All MDR isolates were further subjected to the agar diffusion method to assess the antibacterial actions of L. acidophilus and Lb. plantarum.16 Briefly, the suspension of E. coli bacteria was adjusted to half McFarland and cultured on nutrient agar plates. 100 μL of each probiotic (0.5 McFarland turbidity) was spilled into 6 mm wells, which were then cut with sterile tips in the plates. After incubation for 24h at 37°C, the size of inhibitory zones diameters was determined in millimeters using a ruler.
Antibiofilm Assay
Probiotic isolates were further tested for their antibiofilm activity. A single colony of each E. coli isolate was added to 5 mL of nutritional broth (Oxoid, UK) and cultured for 20 hours at 37°C. The antibiofilm formation activity was tested as described by Jadhav et al.17 The two tested probiotics, alongside their control (broth medium), were put in a 96-well plate (Sigma Aldrich, USA).
10% (v/v) of all probiotics were used as recommended by Medellin-Pena et al.18 40 µL of the tested probiotics were added to the wells in triplicate, except for the negative controls (40µL of broth medium). For each group, 160 µL of E. coli broth cultures were added (broth medium was added by the same volume to control wells instead), reaching an ultimate volume of 200 µL per well. Then, the microtiter plates were closed and incubated at 37°C for 24 h. After incubation, the culture medium was removed, and then all wells were rinsed three times with sterile distilled water to eliminate any attached cells. After allowing the microtiter plate to air dry, it was dyed with 150 µL of 0.1 percent crystal violet. To remove any unabsorbed stain, the stain was allowed at room temperature for 15 minutes before being washed twice with sterile distilled water. To solubilize the crystal violet, ethanol was applied to all wells. The test organisms’ mean optical density absorbance was determined at 595 nm, and the percentage of inhibition was computed using the formula (Eq.):
Percentage inhibition = 100 − (OD595 nm test for positive control well / OD595 nm test for negative control well) × 100)
Polymerase Chain Reaction (PCR) Amplification and DNA Extraction
DNA was collected from the three E. coli isolates most affected using the QIAamp DNA Mini kit (Germany, Qiagen, GmbH). The isolates were examined for the prominence of biofilm genes crl, csgA, and csgD as mentioned in Table 1.
Table (1):
Different primers and its sequences used for detecting biofilm genes.
| Primer | Sequence | Amplified product | Reference |
|---|---|---|---|
| csgA
|
ACTCTGACTTGACTATTACC | 200 bp | 19 |
| AGATGCAGTCTGGTCAAC | |||
| crl
|
TTTCGATTGTCTGGCTGTATG | 250 bp | 19 |
| CTTCAGATTCAGCGTCGTC | |||
| csgD
|
TTATCGCCTGAGGTTATCGTTTGC | 501 bp | 20 |
| TCTTCAGGCTCTATTATTCTTCTGGATAT |
Table (2):
Target genes and SYBR green rt-PCR cycling conditions.
| Target gene | Sequences of Primers | Reverse transcription | Primary Denaturation | Amplification (40 cycles) | ||
|---|---|---|---|---|---|---|
| Secondary denaturation | Annealing | Extension | ||||
| 16S RNA | GACCTCGGTTTAGTTCACAGA | 50˚C 30 min. | 94˚C 15 min. | 94˚C 15 sec. | 55˚C 30 sec. | 72˚C 30 sec. |
| CACACGCTGACGCTGACCA | ||||||
| csgA | ACTCTGACTTGACTATTACC | |||||
| AGATGCAGTCTGGTCAAC | ||||||
| Crl
|
TTTCGATTGTCTGGCTGTATG | |||||
| CTTCAGATTCAGCGTCGTC | ||||||
| csgD
|
TTATCGCCTGAGGTTATCGTTTGC | |||||
| TCTTCAGGCTCTATTATTCTTCTGGATAT | ||||||
Agarose Gel Electrophoresis of PCR Products
By electrophoresis on a 1% agarose gel (Applichem, Germany, GmbH) with 20 μL of PCR products in each well, the PCR products were split up. A gel documentation system was then used to visualize the gel (Alpha Innotech, Biometra).
qRT-PCR (Quantitative, Reverse Transcriptase PCR) for Biofilm Genes
The expression of biofilm genes was analyzed using qRT-PCR. The 16S rRNA housekeeping gene was used as an internal control to ensure that the expression levels of the samples were comparable. The total reaction volume of 25 μL contained 0.25 μL of RevertAid Reverse Transcriptase (200 U/μL) (Thermo Fisher), 0.5 μL of each primer of 20 pmol concentration, 12.5 μL of the 2x QuantiTect SYBR Green PCR Master Mix (Qiagen, Germany, GmbH), 8.25 μL of water, and 3 μL of cDNA template. The reaction was carried out in a Step-one real-time PCR according to the settings listed in Table 2. Amplification curves, as well as Ct values, were evaluated. The variance of gene expression on the RNA of the various samples was assessed, and the Ct of each sample was compared to that of the positive control group using the “ΔΔCt” protocol investigated by Yuan et al.21
Biochemical Identification of UPEC Isolates and Antibiotic Susceptibility Test
The collected isolates produced rose pink colonies when grown on MacConkey agar due to lactose fermentation. All were confirmed as E. coli bacteria by the exhibited biochemical reactions, including methyl red and indole positive, urea and citrate negative, and Motility Indole Ornithine positive. From the 80 urine samples, 45 E. coli isolates were obtained. The characteristics of the female patients who took part in this study are summarized in Table 3. The disc diffusion method was used to test all isolates with nine antibiotics (CRO, AMP, CN, F, CIP, CXM, AK, AMC, CAZ). The antibiotic susceptibility pattern of all isolates is summarized in Table 4. Ten isolates showed multidrug-resistant UPEC to at least one antibiotic in three or more antimicrobial categories. The antibiotic susceptibility pattern of MDR isolates is summarized in Table 5.
Table (3):
Demographic data of female patients included in this study.
Characteristic |
Total [45] |
|---|---|
Age (Years) |
|
1-19 |
6 |
20-29 |
10 |
30-39 |
6 |
40-49 |
4 |
Above 50 |
19 |
Marital Status |
|
Married |
37 |
Single |
8 |
Pregnant Status |
|
Positive |
6 |
Negative |
39 |
Symptoms of UTI |
|
Yes |
33 |
No |
12 |
Table (4):
Antibiotic susceptibility results for all E. coli isolates.
Antibiotic |
Conc. (µg/disc) |
Resistant (R) No.* (%#) |
Intermediate (I) No* (%#) |
Susceptible (S) No* (%#) |
|---|---|---|---|---|
Gentamycin (CN) |
10 |
16 (35.5) |
3 (6.5) |
26 (58) |
Ampicillin (AMP) |
10 |
26 (57.7) |
1(2.3) |
18 (40) |
Amikacin (AK) |
30 |
7 (15.5) |
13 (29) |
25 (55.5) |
Cefuroxime (CXM) |
30 |
17 (37.8) |
8(17.8) |
20 (44.4) |
Ciprofloxacin (CIP) |
5 |
8 (17.8) |
5(11.1) |
32(71.1) |
Nitrofurantoin (F) |
300 |
11(24.4) |
14(31.2) |
20(44.4) |
Ceftazidime (CAZ) |
30 |
18(40) |
14(31) |
13(29) |
Ceftriaxone (CRO) |
30 |
13(29) |
5(11) |
27 (60) |
Amoxicillin |
20 |
33(73.3) |
4(9) |
8(17.7) |
# expressed as percent regarding all E.coli isolates for each antibiotic tested.
* Designates number of E.coli isolates.
*Denotes: (R) Resistant, (I) Intermediate, (S) Susceptible.
Table (5):
The most resistant E.coli isolates to different antibiotics.
Code |
CN |
AMP |
AK |
CXM |
CIP |
F |
CAZ |
CRO |
AMC |
|---|---|---|---|---|---|---|---|---|---|
E. coli 1 |
S |
R |
S |
R |
R |
R |
R |
R |
R |
E. coli 6 |
R |
R |
S |
R |
R |
R |
R |
R |
R |
E. coli 8 |
R |
R |
R |
R |
S |
S |
R |
R |
R |
E. coli 9 |
R |
R |
S |
R |
R |
R |
R |
R |
R |
E. coli 16 |
R |
R |
I |
R |
R |
R |
R |
R |
R |
E. coli 31 |
R |
R |
R |
R |
S |
R |
R |
R |
R |
E. coli 32 |
R |
R |
S |
R |
R |
R |
R |
R |
R |
E. coli 42 |
I |
S |
R |
I |
R |
R |
R |
R |
R |
E. coli 34 |
S |
R |
S |
R |
R |
R |
R |
R |
R |
E. coli 44 |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S: sensitive, R: resistant, I: intermediate
Antibacterial Activity of L. acidophilus and Lb. plantarum against MDR Isolates
Eight MDR E. coli isolates gave sensitivity to L. acidophilus, and seven out of ten to Lb. plantarum. The inhibition zones range from (16±0.04–23±0.05 mm). The inhibition zones for the different isolates are captured in Table 6.
Table (6):
Antibacterial activity of L. acidophilus and L. Plantarum against MDR isolates.
E.coli Code |
L.Plantarum M SD |
L. Acidophilus M SD |
|---|---|---|
E. coli 1 |
0.0±0 |
18±0.04 |
E. coli 6 |
20±0.08 |
18±0.13 |
E. coli 8 |
18±0.14 |
20±0.08 |
E. coli 9 |
19±0.06 |
19±0.05 |
E. coli 16 |
16±0.04 |
18±0.14 |
E. coli 31 |
0.0±0.12 |
0.0±0 |
E. coli 32 |
22±0.0.06 |
18±0.05 |
E. coli 42 |
0.0±0 |
0.0±0 |
E. coli 34 |
23±0.05 |
22±0.06 |
E. coli 44 |
18±0.06 |
16±0.09 |
Legend—M: mean expressed in mm; SD: standard deviation.
Antibiofilm Assay Results for the MDR E. coli Isolates
The effect of Lactobacillus acidophilus and Lactobacillus plantarum on E. coli‘s preliminary adhesion to biofilm development was observed and reported in Table 7. Overall, the majority of the E. coli isolates tested demonstrated a sufficient ability to create biofilms. L. acidophilus induced the highest inhibition (56.30%) in case of E. coli 44, followed by 45.63%- E. coli 8 and 43.20% – E. coli 42. While L. plantarum caused 39.63 % reduction of E. coli 8 biofilm. L. plantarum had a minor inhibitory effect on E. coli 42 and E. coli 8 isolates (inhibition percentages of 22.68 and 28.84, respectively). In contrast, E. coli 32 biofilm was negatively influenced by both probiotics (Table 7).
Table (7):
Antibiofilm activity of probiotics against MDR E. coli isolates.
| E.coli code | OD595nm (% inhibition) | ||
|---|---|---|---|
| Lactobacillus acidophilus | Lactobacillus plantarum | Control | |
| E. coli 1 | 0.1224±0.08 (24.02) | 0.1764±0.13 (9.49) | 0.1611±0.06 |
| E. coli 6 | 0.1322±0.06 (8.19) | 0.1289±0.06 (10.48) | 0.1440±0.09 |
| E. coli 8 | 0.1296±0.008 (45.63) | 0.1439±0.04 (39.63) | 0.2384±0.08 |
| E. coli 9 | 0.1541±0.07 (27.92) | 0.1934±0.03 (9.54) | 0.2138±0.04 |
| E. coli 16 | 0.1986±0.05 (41.12) | 0.4124±0.006 (0) * | 0.3373±0.08 |
| E. coli 31 | 0.1655±0.06 (0) * | 0.1302±0.007 (21.94) | 0.1668±0.18 |
| E. coli 32 | 0.3044±0.12 (0) * | 0.2614±0.08 (0) * | 0.2520±0.007 |
| E. coli 42 | 0.1426±0.09 (43.20) | 0.1967±0.07 (22.68) | 0.2544±0.006 |
| E. coli 34 | 0.1684±0.08 (29.36) | 0.1739±0.06 (27.05) | 0.2384±0.03 |
| E. coli 44 | 0.1421±0.05 (56.30) | 0.2314±0.04 (28.84) | 0.3252±0.07 |
* Means that when the biofilm biomass of treated E. coli is equal to or greater than the control, there is no inhibition.
Based on the previous results, E. coli 8, E. coli 42, E. coli 44 were used to detect the influence of tested probiotics on the genetic level.
Detection of Biofilm Genes Using Conventional PCR
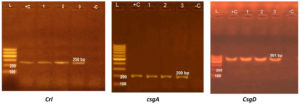
Tested E. coli strains carried crl, csgA, and csgD genes and produced bands at 250, 200, and 501 bp, respectively, as shown in Figure 1.
Figure 1. Agaraose gel electrophoresis for crl, csgA, and csgD genes of three E. coli isolates giving bands at 250, 200, and 501 bp, respectively.
Quantitative Assessment Effect of Probiotic Bacteria on Biofilm Genes of Tested E. coli Isolates Using qRT-PCR
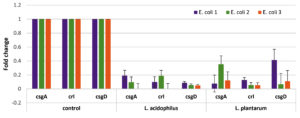
Data in Figure 2 shows the expression of the investigated biofilm gene products (cDNA) before and after treatment with probiotic solutions.
Figure 2. Results of RT-PCR showing the expression of csgA, crL, and csgD in E. coli sp. before and after treatment with Lactobacillus acidophilus and Lactobacillus plantarum.
The fold changes in csgA, crl, csgD gene expression after treatment with L. acidophilus suspension were remarkably downregulated: 0.00364: 0.19078 fold, 0.0005: 0.1894 fold, and 0.0490: 0.0883, respectively. On the other hand, downregulation of biofilm gene expression for csgA, crl, csgD after treatment with L. plantarum suspension were expressed as fold changes: 0.0769: 0.3535 fold, 0.05440: 0.12940 fold, and 0.06745: 0.4146, respectively.
UPEC is a common cause of UTIs. In the present study, ten E. coli isolates were considered to be resistant to several classes of antibiotics. MDR pathogens are considered a serious threat to human health due to their severity and spreading capabilities. The rising antibiotic resistance of UPEC due to the misuse of antibiotics and the biofilm ability of E. coli entail a need for alternative treatment options. As previously published, E. coli resistance to many antibiotics like amoxicillin, clarithromycin, ampicillin, ceftriaxone, ceftazidime, and clavulanic acid) were reported by Abdelhamid et al.2 Also, high percentage of E. coli strains (60.6%) showed resistance to trimethoprim/sulfamethoxazole, ampicillin, tetracycline, and cefazolin.22 Therefore, the emergence of MDR and antibiotic resistance in recent times has led to the development of new alternatives to combat the MDR bacteria. Probiotics are a necessary emerging alternative that contains antimicrobials and antibiofilm properties.23
Probiotics have an inhibitory ability against many pathogenic bacteria both in vitro and in vivo.24 In this study, the antimicrobial activity of two lactobacilli strains, L. acidophilus and Lb. plantarum, was examined for their potential to inhibit the growth of MDR E. coli by utilizing the agar well diffusion method. Eight out of ten MDR E. coli isolates showed sensitivity to L. acidophilus, and seven out of ten to Lb. plantarum. Dawwam et al.25 found that this inhibition potential due to the presence of different bioactive compounds having antimicrobial activities as 9-Octadecenoic acid, Oleic acid, 2,2- Dideutero octadecanal, 1-Hexadecanol, 2-methyl. These compounds were detected in both extracts of L. plantarum and L. Acidophilus using GC-Ms spectroscopy.
Similar results were obtained by Hashem and Abd El-Baky 10 who found that all tested E. coli isolates showed high sensitivity to Lactobacilli supernatants. Moreover, in a study conducted by Ghane et al.,26 seven lactobacilli strains isolated from kefir showed a high inhibitory effect against all UPEC isolates.
In the study of Abdelhamid et al.,2 six types of probiotics were shown to inhibit six MDR E. coli clinical isolates from various diseases, whereas the highest inhibition zone was detected in the case of B. bifidum, L. acidophilus, B. longum, and against three E. coli clinical isolates (inhibition areas were 17.10- 23.10 mm).
Moreover, another study conducted by Tejero-Sarinena et al.27 demonstrated the ability of 15 strains of probiotics, including Bifidobacterium and Lactobacillus, to have the property of antibacterial against gram-negative and gram-positive bacteria.
A common virulence factor of UPEC is the formation of biofilms, which causes persistent and recurring UTIs. Moreover, biofilm formation increases the organism’s resistance to antibiotics.
In this study, 22% of the isolates were capable of biofilm formation. In agreement with the results of the study by Hashem and Abd El-Baky,10 34% of E. coli isolates had formed a biofilm. Also, Karigoudar et al.28 reported that 89.7% of UPEC isolates among the catheterized patients were biofilm-producing.
According to previous studies, probiotics demonstrated their ability to reduce biofilm formation. In the study conducted by Hashem and Abd El-Baky,10 the researchers found that 16 out of 22 cell-free spent media (CFSM) of Lactobacillus isolates from healthy infants (aged 3–6 months), showed a 50% reduction in biofilm formation consisting of the UPEC isolates. In addition, an 80% reduction was observed in four Lactobacillus isolates.
The study of Abdelhamid et al.2 showed that the biofilms formed by MDR E. coli isolates were reduced by L. helveticus and Lb. plantarum CFS (69.49% and 64.57%), respectively. Furthermore, Aboulwafa et al.29 proved the antibiofilm potential of some Lactobacillus strains against E. coli, S. aureus, and Pseudomonas aeruginosa.
According to our study, E. coli isolates harbored csgA, crl, csgD genes. In this regard, Luna-Pineda et al.8; Ochoaet et al.30 showed that UPEC clinical strain obtained from pediatric patients with UTIs was found to contain a high proportion (95%) of the csgA gene. In addition, Sharma et al.31; Ikwap et al.32 mentioned that crl and csgA genes regulate the surface factors of curli fimbriae, which have an important role in mediating the production of exopolysaccharide (EPS) and adhesion of E. coli to biotic and abiotic surfaces.
Lactobacillus strains exhibit various mechanisms that result in a positive effect against pathogens. According to our data, the fold changes in csgA, crl, csgD gene expression after treatment with L. acidophilus suspension were remarkably downregulated than Lb. plantarum. Song et al.33 established that L. rhamnosus L. rhamnosus cells on microcapsules reduced the transcriptional activity of some virulence genes responsible for the regulation of E. coli Qs such as luxS, lsrK, and lsrR, and hence reduce the biofilm formation of E. coli.
Also, L. rhamnosus and L. salivarius significantly downregulated the gene expression of some Streptococcus mutans virulence genes as glucosyltransferases (gtfD, gtfB, and gtfC), which are responsible for glucan biosynthesis and biofilm formation.34,35
Moreover, Matsubara et al.36 and Rossoni et al.37 revealed that L. acidophilus, L. casei, L. fermentum, L. paracasei, and L. rhamnosus downregulated the genes involved in biofilm development, and gluconeogenesis and glycolysis of C. albicans. Another study by Qian et al.38 discovered that the culture supernatant of Lactobacillus sp. reduced the biofilm formation of Gardnerella vaginalis, which is responsible for bacterial vaginosis. Lb. plantarum ZX27 supernatant decreased the expression of genes responsible for biofilm formation, virulence factors, adhesion, metabolism, and antimicrobial resistance. The potential role of probiotics in reducing microbial biofilm by enabling growth inhibition, bacteriocin production, co-aggregation, and adhesion are readily observed.39
Our results support that L. acidophilus and Lb. plantarum have prominent probiotic activity, exhibiting both antibacterial and antibiofilm effects against MDR UPEC at both phenotypic and genotypic levels by downregulating the biofilm encoding genes. This reinforces their use as an alternative non-antibiotic therapy, especially against MDR isolates in recurrent UTIs. Further in vitro and in vivo studies on a larger scale analyzing other probiotic strains remain necessary.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
GED, HFI and MHY designed and directed the project. GED, HFI and IIS performed the Experiments. GED, HFI analyzed the data. GED, HFI wrote the Initial draft. GED reviewed and edited the final manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Toval F, Kohler CD, Vogel U, et al. Characterization of Escherichia coli Isolates from Hospital Inpatients or Outpatients with Urinary Tract Infection. J Clin Microbiol. 2014;52(2):407-418.
Crossref - Abdelhamid A, Esaam A, Hazaa, M. Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm J. 2018;26(5):603-607.
Crossref - Sun F, Qu F, Ling Y, et al. Biofilm-associated infections: antibiotic resistance and novel therapeutic strategies. Future Microbiol. 2.013;8(7):877-886.
Crossref - Corte L, Pierantoni DC, Tascini C, Roscini L, Cardinali G. Biofilm Specific Activity: A Measure to Quantify Microbial Biofilm. Microorganisms. 2019;7(3):73.
Crossref - Ramstedt M, Ribeiro IAC, Bujdakova H, et al. Evaluating Efficacy of Antimicrobial and Antifouling Materials for Urinary Tract Medical Devices: Challenges and Recommendations. Macromol Biosci. 2019;19(5):e1800384.
Crossref - Evans ML, Chapman MR. Curli biogenesis: order out of disorder. Biochim Biophys Acta. 2014;1843(8):1551-1558.
Crossref - Hufnagel DA, Depas WH, Chapman MR. The Biology of the Escherichia coli Extracellular Matrix. Microbiol Spectr. 2015;3(3).
Crossref - Luna-Pineda VM, Ochoa SA, Cruz-Cordova A, et al. Features of urinary Escherichia coli isolated from children with complicated and uncomplicated urinary tract infections in Mexico. PLoS One. 2018;13(10):e0204934.
Crossref - Luna-Pineda VM, Reyes-Grajeda JP, Cruz-Cordova A, et al. Dimeric and Trimeric Fusion Proteins Generated with Fimbrial Adhesins of Uropathogenic Escherichia coli. Front Cell Infect Microbiol. 2016;6:135.
Crossref - Hashem ZS. El-Baky RMA. In vitro inhibition of uropathogenic Escherichia coli biofilm formation by probiotic Lactobacilli isolated from healthy breast fed infants. Novel Research in Microbiology Journal. 2021;5(1):1091-1105.
Crossref - Wadoum RG, Fonteh Florence A, Marie KP, et al. In Vitro Antimicrobial Characterization of Lactobacillus Isolates Towards Their Use as Probiotic Alternatives to Antibiotic Growth Promoters. 2019;4(3):72-86.
Crossref - McFarland LV. Probiotics for the Primary and Secondary Prevention of C. difficile Infections: A Meta-analysis and Systematic Review. Antibiotics (Basel). 2015;4(2):160-178.
Crossref - Wasfi R, El-Rahman OAA, Mansour LE, Hanora AS, Hashem AM, Ashour MS. Antimicrobial activities against biofilm formed by Proteus mirabilis isolates from wound and urinary tract infections. Indian J Med Microbiol. 2012;30(1):76-80.
Crossref - Versalovic J, Carroll K, Funke G, Jorgensen J, Landry M, Warnock D. Manual of clinical microbiology. systems for detection and identification of bacteria and yeasts. ASM Press. 2011.
Crossref - Biemer JJ. Antimicrobial susceptibility testing by the Kirby-Bauer disc diffusion method. Ann Clin Lab Sci. 1973;3:135-140.
- Gonzalez L, Sandoval H , Sacristan N, Castro JM, Fresno JM, Tornadijo ME. Identification of lactic acid bacteria isolated from Genestoso cheese throughout ripening and study of their antimicrobial activity. Food Control. 207;18(6):716-722.
Crossref - Jadhav S, Shah R, Bhave M, Palombo E. Inhibitory activity of yarrow essential oil on Listeria Planktonic cells and biofilms. Food Control. 2013;29(1):125-130.
Crossref - Medellin-Pena MJ, Griffiths MW. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl Environ Microbiol. 2009;75(4):1165-1172.
Crossref - Knobl T, Moreno AM, Paixao R, et al. Prevalence of Avian Pathogenic Escherichia coli (APEC) Clone Harboring sfa Gene in Brazil. ScientificWorldJournal. 2012;2012:437342.
Crossref - Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD Promoter: Interplay between Five Transcription Factors. Microbiology (Reading). 2010;156(8):2470-2483.
Crossref - Yuan J, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. Bioinformatics. 2006;7:85.
Crossref - Li X F, ZJ Liu, X Chen, et al. Study on antibiotic resistance of Escherichia coli and Enterococcus colonized in intestine of neonates from neonatal intensive care unit. Zhonghua Liu Xing Bing Xue Za Zhi. 2017;38(9):1259-1262.
- Fernandes MSM, Lourenco MLMC, Vasconcelos BM, Carneiro VA. Probiotics Lactobacillus strains : A promising alternative therapy against to biofilm-forming enteropathogenic bacteria? 2019;13(28):544-551.
- Yang J, Huang K, Qin S, Wu X, Zhao Z, Chen F. Antibacterial action of selenium-enriched probiotics against pathogenic Escherichia coli. Dig Dis Sci. 2009;54(2):246-254.
Crossref - Dawwam G, Saber I, Yassin M, Ibrahim H. Analysis of Different Bioactive Compounds Conferring Antimicrobial Activity from Lactobacillus plantarum and Lactobacillus acidophilus with Gas Chromatography-Mass Spectrometry (GC-MS). Egyptian Academic Journal of Biological Sciences, G. Microbiology. 2022;14(1):1-10.
Crossref - Ghane M, Babaeekhou L, Ketabi SS. Antibiofilm Activity of Kefir Probiotic Lactobacilli Against Uropathogenic Escherichia coli (UPEC). Avicenna J Med Biotechnol. 2020;12:221-229.
- Tejero-Sarinena S, Barlow J, Costabile A, Gibson GR, Rowland I. Antipathogenic activity of probiotics against Salmonella Typhimurium and Clostridium difficile in anaerobic batch culture systems: is it due to synergies in probiotic mixtures or the specificity of single strains? Anaerobe. 2013;24:60-65.
Crossref - Karigoudar RM, Karigoudar MH, Wavare SM, Mangalgi SS. Detection of biofilm among uropathogenic Escherichia coli and its correlation with antibiotic resistance pattern. J Lab Physicians. 2019;11(1):17-22.
Crossref - Aboulwafa M, Osama D, Elkhatib W, Tawfeik A, Hassouna N. Antimicrobial, Antibiofilm and Immunomodulatory Activities of Lactobacillus rhamnosus and Lactobacillus gasseri against some Bacterial Pathogens. Int J Biotechnol Wellness Ind. 2017;6:12-21.
Crossref - Ochoa SA, Cruz-Cordova A, Luna-Pineda VM, et al. Multidrug- and Extensively Drug-Resistant Uropathogenic Escherichia coli Clinical Strains: Phylogenetic Groups Widely Associated with Integrons Maintain High Genetic Diversity. Front Microbiol. 2016;7:2042.
Crossref - Sharma G, Sharma S, Sharma P, et al. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol. 2016;121(2):309-319.
Crossref - Ikwap K, Larsson J, Jacobson M, et al. Prevalence of adhesin and toxin genes in E. coli strains isolated from diarrheic and non-diarrheic pigs from smallholder herds in northern and eastern Uganda. BMC Microbiol. 2016;16(1):178.
Crossref - Song H, Zhang J, Qu J, et al. Lactobacillus rhamnosus GG microcapsules inhibit Escherichia coli biofilm formation in coculture. Biotechnol Lett. 2019;41(8-9):1007-1014.
Crossref - Lee S-H, Kim Y-J. A comparative study of the effect of probiotics on cariogenic biofilm model for preventing dental caries. Arch Microbiol. 2014;196(8):601-609.
Crossref - Wu C-C, Lin C-T, Wu C-Y, Peng W-S, Lee M-J, Tsai Y-C. Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol Oral Microbiol. 2015;30(1):16-26.
Crossref - Matsubara VH, Wang Y, Bandara HMHN, Mayer MPA, Samaranayake LP. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl Microbiol Biotechnol. 2016;100(14):6415-6426.
Crossref - Rossoni RD, de Barros PP, de Alvarenga JA, et al. Antifungal activity of clinical Lactobacillus strains against Candida albicans biofilms: identification of potential probiotic candidates to prevent oral candidiasis. Biofouling. 2018;34(2):212-225.
Crossref - Qian Z, Zhu H, Zhao D, et al. Probiotic Lactobacillus sp. Strains Inhibit Growth, Adhesion, Biofilm Formation, and Gene Expression of Bacterial Vaginosis-Inducing Gardnerella vaginalis. Microorganisms. 2021;9(4):728.
Crossref - Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic Resistance Related to Biofilm Formation in Klebsiella pneumoniae. Pathogens. 2014;3(3):743-758.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.