ISSN: 0973-7510
E-ISSN: 2581-690X
Human milk can be an important source for obtaining potential probiotics strains for newborns in order to establish the beneficial gut microbial community and development of immune system. The aim of the study was to explore potential human breast milk probiotics and to carry out their in vitro biosafety assessment. The study obtained three isolates namely, SP1B, B2Enr and SP1 which showed potential probiotic activities compared to standard probiotic Lactobacillus plantarum. In addition, these isolates were found to be safe through various in vitro biosafety aspects. The molecular identification by16srDNA sequencing revealed that SP1B and B2Enr belong to Bacillus cereus (MK210172) and Staphylococcus epidermidis (MK210234), respectively. For the first time, the study suggests that these bacterial strains may come in the category of probiotics and can be considered further after in vivo biosafety assessments.
Probiotics; Biosafety assessment; Human breast milk; 16srDNA sequencing.
The WHO has defined Probiotics as ‘Live microorganisms which when administered in adequate amounts confer a health benefit on the host’1. (FAO/WHO, 2002). In particular, Lactobacilli and Bifidobacteria have been implicated as probiotics in many food supplements2,3. Probiotics promote health physiological functions by surviving and colonizing into the gut4. This bacterial colonization into the gut may regulate the immune system and health status of the infants5,6. The first bacterial colonizers in breast-fed infants are facultative anaerobes that include Enterococci, Staphylococci, Streptococci, Lactobacilli and Enterobacteria as well as strict anaerobe Bi dobacteria7.
The breast milk protects mother and infants from many infectious diseases and is a natural source of potential probiotics strains8. Earlier, the milk from breast was considered as sterile; however, later many studies suggested that the milk contains many beneficial bacteria which enhance neonate’s immune system and protect against many gut disorders. The probiotics isolated from breast milk have shown to possess antimicrobial compounds which inhibit the growth of pathogenic organisms. 9 The common bacterial genera found in breast milk are Bifidobacterium, Lactobacillus, Clostridium, Ralstonia, Staphylococcus, and Streptococcus. 8
A potential probiotic strain must possess good acid tolerance and bile tolerance properties in addition to the antimicrobial properties against pathogenic bacteria. In addition, the good cell surface hydrophobicity % of probiotics ensures attachment to the gut epithelium which enhances the host interaction. 10 Moreover, the bacterial auto-aggregation results in gut bacterial homeostasis11 and the co-aggregation property of probiotics is also crucial for prevention of colonization of host surfaces by pathogens12. Apart from these potential probiotic characteristics they must have GRAS (Generally Regarded as Safe) property as a safety concern for consumption by the host. The assessment of safety aspects of probiotics can be addressed by in vitro and in vivo tests. In particular, the in vitro safety assessment includes antibiotic resistance, mucin degradation, biogenic amines production, deconjugation of bile salts, hemolytic activity and gelatinase production properties of the probiotic test cultures.
Since, the mother’s milk is beneficial to the neonate and may possess such kind of probiotics; the present study was focused to explore potential probiotic bacteria of human breast milk samples and to investigate the probiotic properties along with their in vitro biosafety aspects.
Collection of Sample
Total four human breast milk samples were collected from healthy volunteer mothers. The mothers had full-term normal pregnancy without any maternal perinatal problems. The study plan was carried out in accordance with the 1964 Helsinki Declaration and also approved by the Institutional-Human Research Ethical Committee (IHREC), Maliba Pharmacy College, Uka Tarsadia University, Bardoli, Gujarat, India. All women volunteers were aware about importance of the study and written consent was obtained.
Isolation of probiotic Bacteria
The milk samples were serially diluted with peptone water (10-1, 10-2 & 10-3) and the aliquots were plated on MRS agar. All the plates were incubated at 37°C for 3 days.
Evaluation of probiotic characteristics of the isolates
Acid and Bile Tolerance activity
The isolates obtained were further grown in MRS-broth and cells were harvested. The cells were suspended in PBS (pH 7. 4); which then subjected to serial dilutions using PBS (pH 3. 0) and kept for different time durations (0hr, 2hrs, 4hrs and 24 hrs). The aliquots were plated on MRS agar followed by incubation at 37°C for 24-48 hrs. The CFU/ml was calculated for each of these plates and the growth on MRS agar indicated the acid tolerance of the isolates.
The MRS agar was prepared using different concentrations (0. 3%, 0. 5%, 1. 0%, 1. 5%) of Cholic acid. The serial dilution of cell suspension was prepared and aliquots were plated on Cholic acid-MRS agar followed by incubation at 37°C for 24-48 hrs. CFU/ml was then calculated for each of these plates and the growth on Cholic acid- MRS Agar was used to designate the bile tolerant property.
Antibacterial Activity
The cell-free neutralized supernatants (CFNS) were used for assessing the antibacterial activity. The cultures were grown in MRS-broth for 18 hrs at 37°C to obtain CFNS. The supernatant pH was adjusted to 6. 5-7. 0 using 1N NaOH. The supernatant is then heated at100°C for 5 min. and cooled down followed by storage at -20°C. The neutralized CFNS were then checked for its antibacterial activity against Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, and Proteus vulgaris using the agar well diffusion method.
Cell surface hydrophobicity
The isolates were grown in MRS-broth; cells were harvested, washed with PBS and suspended in five ml Phosphate Urea Magnesium (PUM) buffer. Initial O. D. (ODInitial) of the cell suspension was taken at 610 nm. Three ml bacterial suspension was mixed with one ml of respective hydrocarbons followed by incubation at 37°C for 10 min. It was then vortexed for 120 secs and kept undisturbed at 37°C for one hour to allow phase separation. The aqueous phase was carefully removed after one hour with a Pasteur pipette. The O. D. was measured using spectrophotometer and hydrophobicity percentage (H%) was calculated by the following formula13:
H % = (1 – A1/A0) X 100 [A1 is initial O. D. and A0 is final O. D. ]
Auto aggregation property
The cells were freshly grown in MRS-broth at 37°C, harvested and washed twice with PBS. The cells were then suspended in PBS and initial absorbance (Absinitial) was taken at 600nm. The cell suspension was centrifuged and pellet was resuspended in equal volume of broth removed at first step. The mixture was then allowed to stand for 2 hrs at 37°C. Further, one ml of the upper suspension was taken to measure the absorbance (Absfinal) by using broth as reference. The aggregation % was calculated by the following formula14:
Aggregation %= (Absinitial – Absfinal) / Absfinal X 100
Co-aggregation property
The indicator organisms were grown in nutrient broth and the isolates were grown in MRS-broth at 37°C. The cells were pelleted down, washed twice with PBS and resuspended in PBS. The O. D. was taken at 600nm. The probiotics were mixed with pathogenic organisms followed by incubation at 37°C for 24 hrs. Further, the absorbance was taken at 600 nm and the percentage of co-aggregation was calculated as [(Apathogen + Aprobiotic)/2 – (Amix)/(Apathogen + Aprobiotic)/2] ׳ 100 [Apathogen and Aprobiotic refers to absorbance in the tubes containing either the pathogen or the probiotics respectively; Amix refers to absorbance of the mixture of both at 24hrs]15.
Assessment of in vitro biosafety aspects of isolates
Biogenic amines and Gelatinase production
The biogenic amines production of isolates was assessed as mentioned previously16. The isolates were grown overnight at 37°C in MRS-broth (supplemented with 2g/l final concentration of different amino acids such as histidine, arginine, phenylalanine, tryptophan, and lysine). After 3-5 days of incubation, 0. 2 ml of the suspension was mixed with two ml of modified decarboxylase broth followed by incubation for 3 days under anaerobic condition. The presence of biogenic amines was indicated when purple color changes to yellow and again turned to purple.
Gelatinase production of the isolates was assessed as described by Eatson & Gasson17. The isolates were grown in MRS-broth at 37°C and streaked on Todd-Hewitt agar plates containing 30gm/liter of gelatin. The plates were placed at 4°C for 5 hours after the incubation. The protein hydrolysis was assessed by zones of turbidity around the colonies.
Mucin degradation and Hemolytic activity
The isolates were grown in MRS-broth at 37°C. Ten micro liter of viable cultures were inoculated on the surface of medium B with some modifications. All the plates were incubated at 37°C for 72 hours under anaerobic condition. Mucin degradation was confirmed upon staining with 0. 1% w/v amido black in 3. 5M acetic acid (for 30 min) and washing with 1. 2M acetic acid which resulted in a discolored zone around the colony.
The hemolytic activity was checked as mentioned previously18. The isolates were grown in MRS-broth at 37°C and then streaked onto blood agar plates followed by incubation of 24 – 48 hrs. After incubation period colonies were checked for clear zones to be reported as a-hemolysis, b-hemolysis or g-hemolysis.
Bile salts deconjugation and Antibiotic resistance
Bile salts deconjugation was assessed as mentioned previously19. The isolates were grown in MRS-broth at 37°C and then inoculated on the MRS agar plates (supplemented with 0. 05% w/v L-cysteine and 0. 5% w/v sodium salts). All the plates were incubated at 37°C for 72 hrs under anaerobic condition. The bile salt deconjugation was confirmed by the presence of bile acid precipitation around the colonies.
The disc diffusion method was used for assessing antibiotic resistance of the isolates20. The freshly grown cultures were spreaded onto Muller-Hinton Agar (MHA) plates. The antibiotic multidiscs were then placed and plates were incubated at 37°C for 2 days. The zone of inhibition surrounding the disc was measured in mm, and the isolates were tagged as susceptible, moderately susceptible and resistant to the respective antibiotics.
Molecular identification
The selected probiotic isolates were subjected to genomic DNA isolation and 16srDNA PCR was performed using the forward primer: 5’ AGAGTTTGATCCTGGCTCAG3’ and reverse primer: 5’AAGGAGGTGATCCAGCCGCA3’. The PCR products were then sent for 16srDNA sequencing. The DNA sequences were BLAST from the existence microbial DNA database and Phylogenetic trees were evaluated.
Statistical Analysis
For the cell surface hydrophobicity, auto-aggregation and co-aggregation properties of the isolates, one way ANOVA was carried out using Duncan analysis test in IBM SPSS Statistics for Windows, version XX (IBM Corp. , Armonk, NY, USA). For each sample, the results were expressed as mean±SD.
Evaluation of the isolates for probiotic properties
Total 114 isolates were obtained from the human breast milk samples. The isolates were further subjected to assessment of their probiotic characteristics.
Acid and Bile Tolerance Activity of Isolates
The present study isolates were found to be resistant to pH 3. 0 during 0 hr, 2 hrs, 4 hrs and 24 hrs. However, seven isolates were found to possess good acid tolerance at pH 3. 0 as indicated by CFU/ml (Table 1). Moreover, it was found that the isolate B2ENr showed maximum acid tolerance as compared to that of standard probiotic L. plantarum.
The bile tolerance property was showed by all the isolates at 0. 5% Cholic acid whereas, some of the isolates showed tolerance upto 1% Cholic acid (Table 1). Interestingly, SP1S isolate was found to be more bile tolerant and capable of tolerating 1. 5% Cholic acid as compared to L. plantarum.
Table (1):
Acid and Bile tolerance properties of the different isolates.
| Isolates | CFU/ml (pH 3) | Bile concentration (Cholic acid) | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 hr | 2 hrs | 4 hrs | 24 hrs | 0.3% | 0.5% | 1.0% | 1.5% | |
| L. plantarum | 289 × 105 | 237 × 105 | 175 × 105 | 112 × 105 | 245.5 × 104 | 166 × 104 | 148 × 104 | <30 |
| SP1 | 128 × 105 | 118 × 105 | 93.3 × 105 | 76 × 105 | 88 × 104 | 90 × 104 | 97 × 104 | No growth |
| SP2 | 175 × 105 | 141 × 105 | 108.3 × 105 | 67 × 105 | 228 × 104 | 222 × 104 | 103 × 104 | <30 |
| SP3 | 140 × 105 | 103 × 105 | 87 × 105 | 42 × 105 | 150 × 104 | 88 × 104 | No growth | No growth |
| SP1B | 109.3 × 105 | 98 × 105 | 81.6 × 105 | 57 × 105 | 293 × 104 | 152 × 104 | 78 × 104 | No growth |
| SP1M | 168 × 105 | 102 × 105 | 74 × 105 | 28 × 105 | 148 × 104 | 52 × 104 | 32 × 104 | <30 |
| SP1S | 282 × 105 | 191 × 105 | 89 × 105 | 39 × 105 | 235 × 104 | 202 × 104 | 107 × 104 | 37 × 104 |
| B2Enr | 190 × 105 | 180 × 105 | 177 × 105 | 163 × 105 | 89 × 104 | 123 × 104 | 124 × 104 | <30 |
Antibacterial activity of Isolates
All the isolates showed inhibitory effect on the growth of all test microorganisms used except SP2 and SP1M which did not show antibacterial activity against P. aeruginosa and P. vulgaris respectively; as suggested by the diameter of inhibitory zones (Table 2). However, E. coli was found to be highly susceptible to the antibacterial action of SP1, SP2, SP3, SP1B, SP1M and B2Enr. SP1, The SP1B and SP1S exhibited maximum antibacterial activity against P. vulgaris whereas SP1, SP1S and B2Enr showed good antibacterial activity against P. aeruginosa. The S. aureus growth was highly susceptible to antibacterial action of SP1M, SP1, SP2, SP1S and B2Enr.
Table (2):
Antibacterial activity of isolates against different indicator microorganisms
| Isolates | Diameter of zone of inhibition (in mm) against indicator bacteria | |||
|---|---|---|---|---|
| E. coli | P. aeruginosa | S. aureus | P. vulgaris | |
| SP1 | 25 | 13 | 19 | 29 |
| SP2 | 25 | 00 | 15 | 13 |
| SP3 | 23 | 8 | 10 | 11 |
| SP1B | 24 | 7 | 9 | 25 |
| SP1M | 24 | 7 | 25 | 00 |
| SP1S | 19 | 12 | 16 | 18 |
| B2Enr | 28 | 10 | 16 | 12 |
Cell surface hydrophobicity, Auto-aggregation and Co-aggregation properties of Isolates
The evaluation of hydrophobicity % of all the isolates suggested that most of the probiotic isolates possess good surface hydrophobicity as compared to standard probiotic L. plantarum (Table 3). However, few isolates showed poor adhesion ability as suggested by less hydrophobicity %. The SP3 isolate showed the highest hydrophobicity with xylene and B2Enr showed highest hydrophobicity with chloroform.
Table (3):
Cell surface hydrophobicity, auto-aggregation and co-aggregation properties of the different probiotics isolates.
| Isolates | Cell surface hydrophobicity (%) | Auto- aggregation
(%) |
Co-aggregation (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Xylene | Chloroform | E. coli | Bacillus sp. | Bacillus cereus | Vibrio mimicus | Candida albicans | S. aureus | P. aeruginosa | ||
| SP1 | 25.00±3.00c | 52.66±2.08c | 74.24±0.73d | 95.07±1.05a | 75.97±1.65e | 49.36±1.02d | 98.53±1.00a | 75.46±1.05a | 52.26±1.12b | 99.31±1.09a |
| SP2 | 16.33±0.57e | 42.03±0.95d | 81.23±1.11b | 8.68±0.12e | 9.10±0.11h | 21.33±1.25g | 17.10±1.50f | 16.43±0.80h | 14.30±1.11e | 62.20±1.25e |
| SP3 | 61.33±1.52a | 26.83±0.76f | 53.00±1.00e | 71.18±1.06b | 81.36±1.58d | 39.06±0.90f | 98.46±2.56a | 63.10±2.81c | 12.80±1.83e | 99.46±0.83a |
| SP1B | 19.00±1.00d | 31.20±1.25e | 78.13±0.91c | 60.35±0.55b | 93.32±1.63b | 84.26±1.25a | 61.46±1.19d | 28.16±1.06g | 15.20±1.57e | 86.36±1.09c |
| SP1M | 33.33±1.52b | 56.00±1.00b | 81.05±0.75b | 47.75±12.39c | 62.58±2.03f | 52.26±1.25c | 87.50±0.60b | 43.93±1.62e | 29.10±0.95d | 96.56±1.05b |
| SP1S | 16.83±0.76de | 31.00±1.00e | 85.00±2.00a | 28.66±12.45d | 56.10±1.55g | 42.60±1.55e | 54.43±1.10e | 53.28±0.85d | 46.50±1.17c | 98.46±2.56ab |
| B2Enr | 33.00±1.00b | 83.00±2.64a | 77.00±1.00c | 20.51±1.02d | 85.40±1.15c | 66.26±0.75b | 71.30±1.25c | 41.33±1.04f | 45.10±1.70c | 70.46±0.89d |
| Lactobacillus plantarum | 12.16±0.76f | 31.00±1.00e | 76.93±1.71c | 99.25±1.05a | 98.61±1.20a | 41.33±1.20e | 99.20±0.75a | 71.50±1.24b | 72.03±0.98a | 99.56±0.66a |
*Results were presented as mean ± standard deviation. Data was analyzed using one way ANOVA.
Values with different lower case letters are significantly differed i.e. p< 0.05 according to Duncan analysis Test.
Further, the auto-aggregation property was assessed and analyzed by Duncan analysis test which indicated that all the isolates possess good auto-aggregation property (p < 0. 05; Table 3). Interestingly, the SP1S was found to possess highest auto-aggregation property among all the isolates and as compared to the standard probiotic L. plantarum.
The co-aggregation property was also found to be good for all the isolates with pathogenic test organisms (p < 0. 05; Table 3). The isolates SP1, SP3, SP1M, and SP1S showed highest co-aggregation % with Pseudomonas aeruginosa which was comparable to that of L. plantarum. However, the statistical analysis showed that SP2 isolate exhibit less co-aggregation property with all the tested microbes including Vibrio mimicus.
Assessment of in vitro biosafety aspects of selected probiotics
Biogenic amines and Gelatinase production by isolates
The SP1 isolate did not produce any biogenic amines against arginine, phenylalanine, tryptophane, lysine amino acids, but it produced biogenic amines against histidine (Table 4). The LB isolate was not found to produce biogenic amines against all the amino acids used. The SP1B and L. plantarum showed biogenic amines production against all the amino acids whereas B2Enr did not produce biogenic amines against all amino acids except the arginine.
Table (4):
Biogenic amines (BA) production by probiotic isolates
Amino acids |
SP1 |
SP2 |
SP3 |
SP1B |
SP1M |
SP1S |
B2Enr |
LB |
L. plantarum |
Histidine |
+ve |
-ve |
+ve |
+ve |
+ve |
-ve |
-ve |
-ve |
+ve |
Arginine |
-ve |
+ve |
+ve |
+ve |
+ve |
+ve |
+ve |
-ve |
+ve |
Phenylalanine |
-ve |
+ve |
+ve |
+ve |
-ve |
+ve |
-ve |
-ve |
+ve |
Tryptophane |
-ve |
-ve |
-ve |
+ve |
-ve |
-ve |
-ve |
-ve |
+ve |
Lysine |
-ve |
-ve |
-ve |
+ve |
-ve |
-ve |
-ve |
-ve |
+ve |
*+ve :BA production; -ve : No BA production
Further, all the isolates were checked for their geletinase production property (Table 5b). None of the probiotic isolates showed gelatinase production, as no zone of clearance was found surrounding the colonies on Todd-Hewitt agar plates.
Mucin degradation and Hemolytic activity of Isolates
The mucin degradation property was exhibited by only two probiotic isolates SP1M and SP1S which showed clear zones around colonies on medium B (Fig. 1). The other isolates namely SP1, SP2, SP3, B2Enr, LB and Lactobacillus plantarum did not show mucin degradation.
 Fig. 1. Mucin degradation by probiotic isolates: SP1M and SP1S showed mucin degradation as observed by clear zone around the colonies. Pseudomonas aruginosa was used as positive control for mucin degradation.
Fig. 1. Mucin degradation by probiotic isolates: SP1M and SP1S showed mucin degradation as observed by clear zone around the colonies. Pseudomonas aruginosa was used as positive control for mucin degradation.The b-hemolytic activity was exhibited by two probiotic isolates namely SP2 and SP3 as indicated by yellow zones around the colonies (Fig. 2). The other isolates namely SP1, SP1M, SP1S, B2Enr, LB and Lactobacillus plantarum did not show any hemolysis.
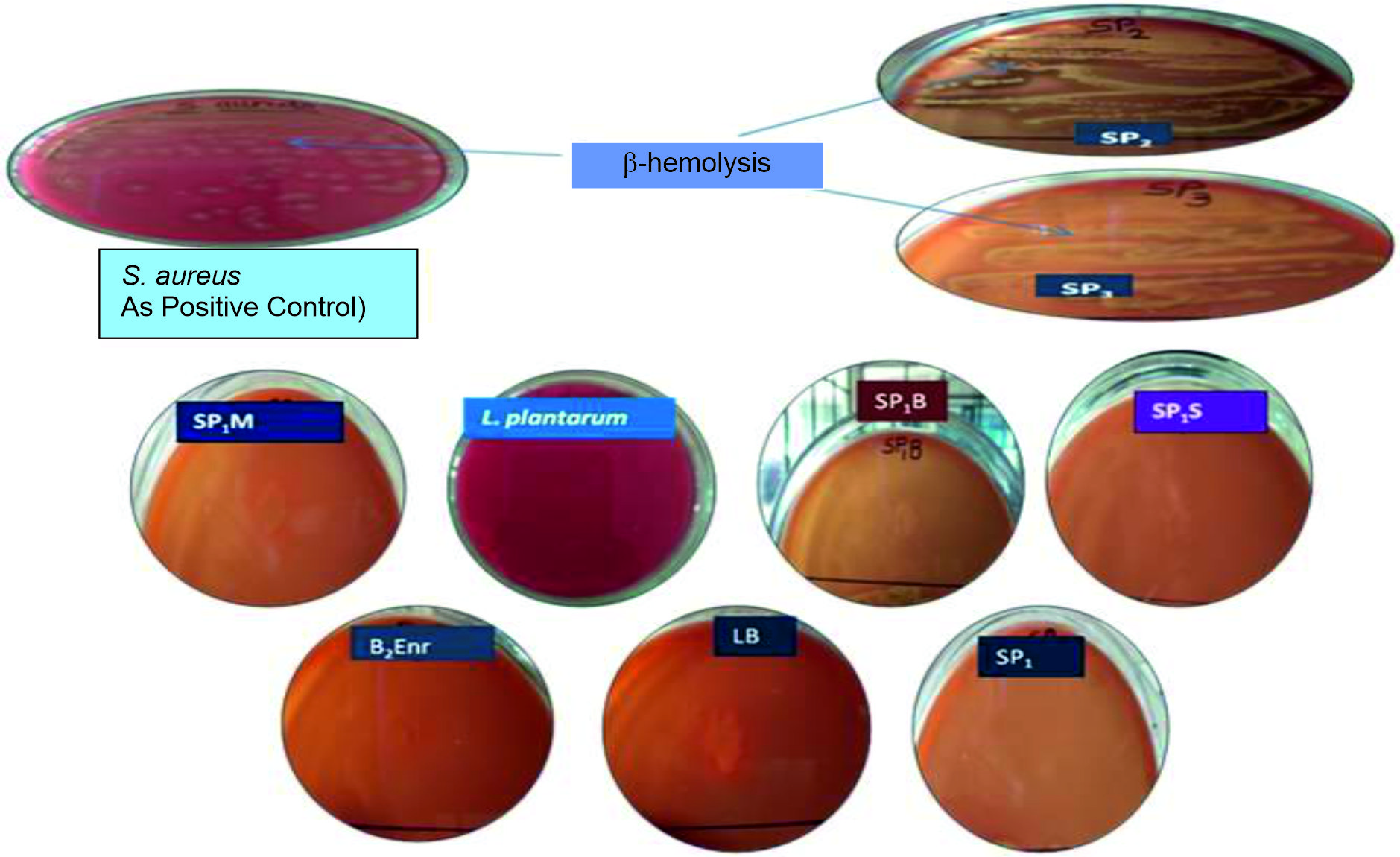 Fig. 2. Hemolytic activity of probiotic isolates: SP2 & SP3 showed b-hemolysis. S. aureus was used as positive control culture for b-hemolysis.
Fig. 2. Hemolytic activity of probiotic isolates: SP2 & SP3 showed b-hemolysis. S. aureus was used as positive control culture for b-hemolysis.Deconjugation of bile salts and Antibiotic resistance of isolates
None of the probiotic isolates were found to exhibit deconjugation property for bile salts, as no precipitation was observed for the colonies (Table 5b).
Table (5):
Comparison of probiotic properties and in vitro biosafety aspects of different isolates.
| (a) Comparison of probiotic properties | ||||||
|---|---|---|---|---|---|---|
| Isolates | Acid tolerance | Bile tolerance | Antibcterial activity | Auto-aggregation | Cell surface hydrophobicity | Co-aggregation |
| SP1 | ++ | +++ | +++ | +++ | ++ | +++ |
| SP2 | +++ | +++ | ++ | +++ | ++ | + |
| SP3 | ++ | ++ | +++ | +++ | ++ | +++ |
| SP1B | +++ | +++ | +++ | +++ | ++ | +++ |
| SP1M | ++ | ++ | ++ | +++ | ++ | ++ |
| SP1S | ++ | ++ | +++ | +++ | ++ | +++ |
| B2Enr | +++ | +++ | +++ | +++ | ++ | +++ |
| L. plantarum | +++ | +++ | +++ | +++ | ++ | +++ |
| *+: Good; ++: very good; +++: Excellent | ||||||
| (b) Comparison of in vitro biosafety aspects | ||||||
| Isolates | Antibiotic resistance | Mucin degradation | Biogenic amine production | Hemolytic activity | Gelatinase production | Deconjugation of bile salts |
| SP1 | + | + | + | + | + | + |
| SP2 | + | + | + | – | + | + |
| SP3 | + | + | + | – | + | + |
| SP1B | + | + | + | + | + | + |
| SP1M | + | – | + | + | + | + |
| SP1S | + | – | + | + | + | + |
| B2Enr | + | + | + | + | + | + |
| L. plantarum | + | + | + | + | + | + |
* +: Considered as biosafe; – : Considered as non biosafe
The antibiotic discs of ampicillin, kanamycin, erythromycin, penicillin-G, vancomycin, rifampicin were used for assessing antibiotic resistance. All the isolates were found to be resistant to penicillin-G. However, they showed susceptibility to ampicillin, kanamycin, erythromycin, vancomycin and rifampicin. The SP1B was moderately susceptible to erythromycin (Fig. 3).
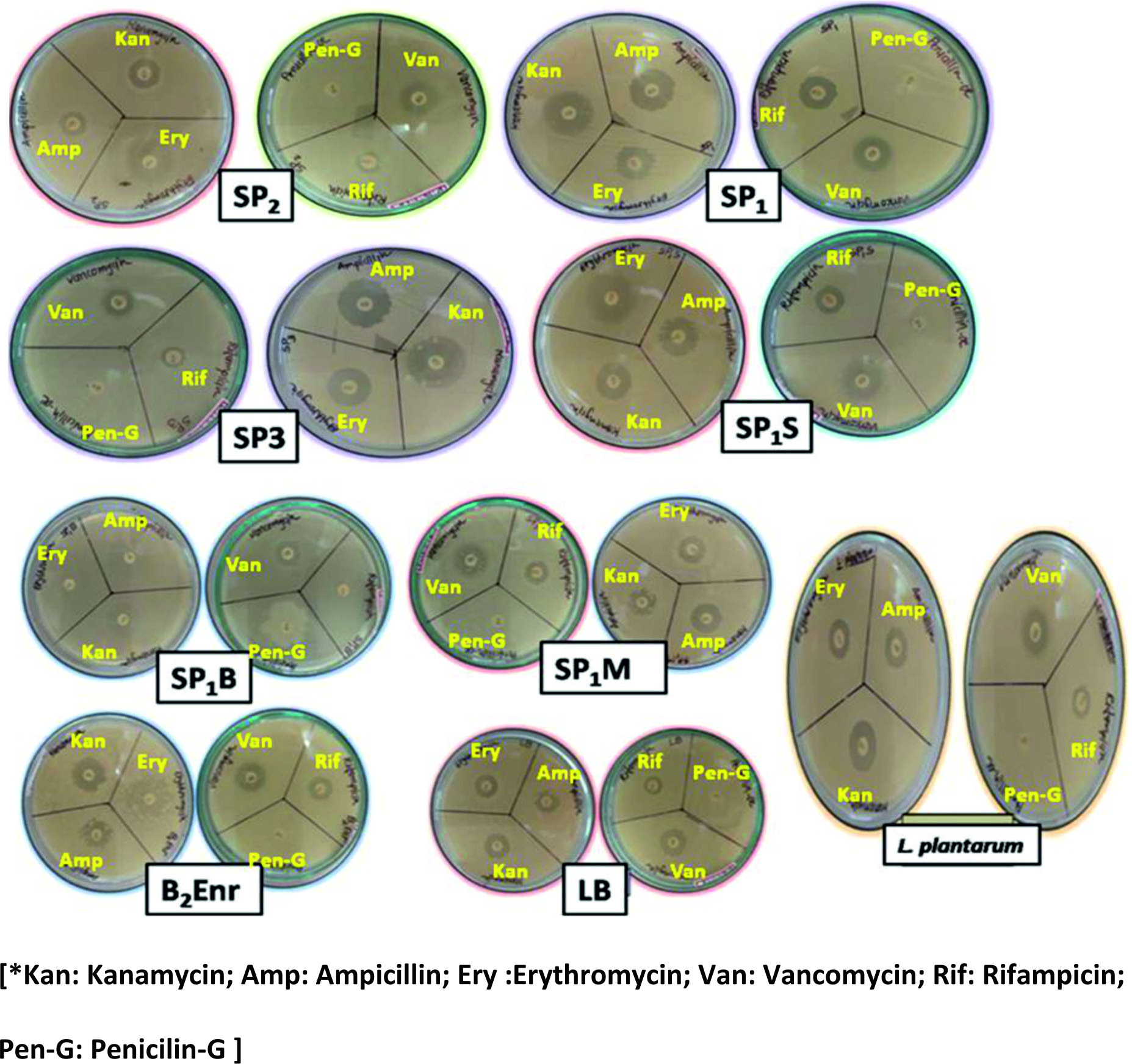 Fig. 3. Antibiotic resistance of probiotic isolates shown on Muller Hinton Agar plates. All the probiotic isolates were resistant to Penicillin-G antibiotic. The probiotic isolates were susceptible to Kanamycin, Erythromycin, Vancomycin, Ampicillin, Rifampicin antibiotics.
Fig. 3. Antibiotic resistance of probiotic isolates shown on Muller Hinton Agar plates. All the probiotic isolates were resistant to Penicillin-G antibiotic. The probiotic isolates were susceptible to Kanamycin, Erythromycin, Vancomycin, Ampicillin, Rifampicin antibiotics.Comparison of probiotic properties and in vitro biosafety aspects of the isolates
Further, the probiotic properties and in vitro biosafety aspects were compared among the isolates (Tables 5a & b, respectively). The comparison of probiotic properties revealed that SP1B and B2Enr exhibited excellent probiotic characterisitics among the isolates which were also comparable to the standard probiotic L. plantarum as well. The comparison of in vitro biosafety aspects of the isolates suggested that SP1B, SP1 and B2Enr can serve as biosafe probiotics, since they passed all the in vitro biosafety assessment criteria used in the present study.
Cultural characteristics and Molecular identification of isolates
The cultural and biochemical aspects were also studied for the selected seven isolates (Table S1 & S2). The SP1M, SP2, B2Enr, SP1, and SP1S were revealed as gram positive cocci, whereas SP3 and SP1B were found to be gram positive bacilli.
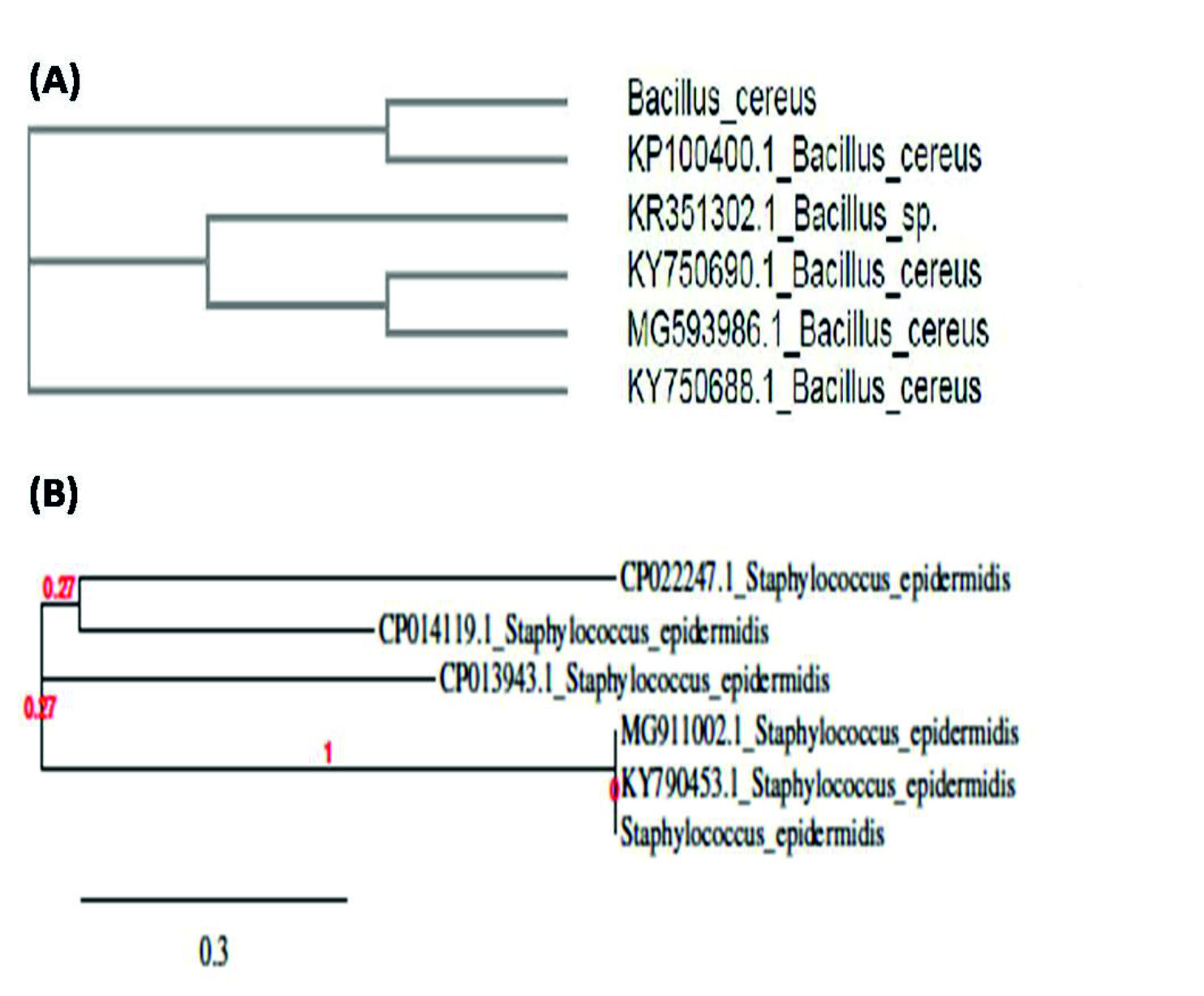
The molecular identification of selected probiotic isolates (SP1B & B2Enr) which passed the in vitro biosafety aspects was carried out by 16srDNA sequencing. The results revealed the SP1B isolate as Bacillus cereus (MK210172) and B2Enr as Staphylococcus epidermidis (MK210234). The 16srDNA sequences were submitted to GenBank-NCBI and the accession numbers MK210172 and MK210234 were obtained for Bacillus cereus and Staphylococcus epidermidis, respectively. The phylogenetic analyses of the probiotic isolates (SP1B and B2Enr) have been shown in Fig. 4a & b.
The breast milk is crucial and fulfills the nutritional requirements for newborns. The human breast milk contains over 700 different types of bacteria, including the genera, Bifidobacteria Micrococci, Lactobacilli, Staphylococci, Streptococci, Enterococci and Lactococci21. Moreover, it also contains prebiotics such as human milk oligosaccharides, which promotes the growth and activity of bacteria22. According to analysis of women who take probiotics during pregnancy reduce their child risk of developing allergies. The bacteria isolated breast milk such as Lactobacillus fermentum, Lactobacillus rhamnosus, Lactobacillus gasseri and Enterococcus feacium have been considered as potential probiotic bacteria23. Thus, the probiotics isolates of breast milk can be of significant use in different human health conditions and particularly for malnourished children.
The present study evaluated probiotic characteristics as well as biosafety aspects of the isolates obtained from the human breast milk samples. Since, probiotics are administrated orally; they must resist the low pH of the gastric juice in the stomach. Hence, acid tolerance is one of the important probiotic properties. Previously, probiotic bacteria isolated from human breast milk [L. crispatus, L. fermentum, L. gasseri, Lactobacillus rhamnosus (KF477283) and Lactobacillus casei (KF477282)] showed good acid tolerant property at pH 324,25. We found total seven isolates showing tolerance to acidic condition (pH 3) with different time durations. The isolate B2Enr showed better acid tolerance property as compared to the standard probiotic L. plantarum. The secretion of bile extract into the duodenum directly hampers probiotic bacteria. The physiological human bile concentrations range from 0. 3% to 0. 5%. Hence, the bile tolerance property of the probiotics must be assessed. Previously, human breast milk isolate L. rhamnosus demonstrated 80% survival rate when subjected to 1. 0% bile concentration. 26 Interestingly, SP1S isolate from the present study was able to tolerate bile salt up to 1. 5% as compared to L. plantarum; whereas, SP2, B2Enr and SP1 showed tolerance upto 1%.
The antimicrobial activity against pathogens is also an important attribute for the selection of potential probiotics to maintain a healthy microbial homeostasis in the GIT. Previously, human breast milk isolates, Pediococcus pentosaceus and Lactobacillus casei showed good antibacterial activity27,28. In the present study, all isolates showed antibacterial activity against the indicator microorganisms except SP2 and SP1M. The E. coli was found to be highly susceptible to the antibacterial action of the isolates. The antibacterial action of SP1, SP1B and SP1S was found to be effective against P. vulgaris, whereas SP1, SP1S and B2Enr showed good antibacterial activity against P. aeruginosa. The S. aureus growth was highly susceptible to antibacterial action of SP1M, SP1, SP2, SP1S and B2Enr. These results suggest that the isolates possess good antibacterial activity which can vary according to the type of probiotic strain and the pathogenic organism.
Furthermore, the probiotics should possess good cell surface hydrophobicity, auto-aggregation as well as co-aggregation properties with different pathogenic strains. For the attachment of bacteria to host tissue, the hydrophobic outermost surface renders a competitive advantage and also important for bacterial colonization in the human GIT12,29. Moreover, to assess the colonization potential of the organism the hydrophobicity to different hydrocarbons has been considered as an in vitro biochemical marker. 30 Our results suggested that SP3 possesses highest affinity that is 61% to xylene as compared to standard probiotic strains Lactobacillus plantarum. With chloroform, B2Enr showed highest affinity (i. e. 83%). The other probiotic isolates also exhibited good affinity with these hydrocarbons indicating that they have good cell surface hydrophobicity. Previous studies have reported that the probiotics showed highest affinity for xylene and relatively more affinity for n-hexadecane in comparison to other strains31,32. In addition, study by Yadav et al. 31, suggested that their isolates have good aggregation property. In the present study, the auto-aggregation property of SP1S, SP1M and SP2 was found to be the highest (i. e. 85%, 81. 05% and 81. 23% respectively). Moreover, the co-aggregation with pathogenic microbes is also important for probiotics since it decreases the activity of the pathogens. Our results of co-aggregation tests are in accordance with the previous studies. 12,29 The isolates were found to co-aggregate with Escherichia coli, Bacillus sp. , Bacillus cereus, Candida albicans, Vibrio mimicus, Staphylococcus aureus and Pseudomonas aeruginosa. The SP1, SP3 and SP1S isolates showed maximum co-aggregation ability with Pseudomonas aeruginosa. The SP3 exhibited 98% co-aggregation property with Vibrio mimicus and the SP1 had 95. 07 % and 98. 53% co-aggregation ability with Escherichia coli and Vibrio mimicus respectively. The SP1B had 93. 32% co-aggregation ability with Bacillus sp. and the SP1M had 96. 56% co-aggregation property with Pseudomonas aeruginosa.
The probiotics must have GRAS property in order to consider it for human consumption and therefore must undergo for in vitro and in vivo biosafety assessment. The present study addressed the different in vitro biosafety aspects. The antibiotic resistance is also a crucial criterion for biosafety. The probiotic must not contain any transferable antibiotic resistance gene. The probiotic bacteria such as Lactobacilli have been found susceptible to penicillin and ampicillin, whereas resistant to vancomycin33. Previously, Lactobacillus sp. was reported to be highly resistant to ciprofloxacin, fusidic acid, metronidazole, streptomycin, sulfadiazine, kanamycin, gentamicin, nalidixic acid, bacitracin, cefoxitin and vancomycin33,34. In the present study antibiotics such as ampicillin, kanamycin, erythromycin, penicillin-G, vancomycin and rifampicin were used. All probiotic isolates were resistant to penicillin-G; however, they were all susceptible to other antibiotics used in the study. The SP1B was found to be moderately susceptible to erythromycin. Earlier, Muסoz-Atienza et al. 35 reported that their probiotic strains including Pediococci strains were resistant to erythromycin, tetracycline, ciprofloxacin, norfloxacin, rifampicin, ampicillin, penicillin, gentamycin, streptomycin etc. In another study, the isolates were sensitive to erythromycin, bacitracin, rifampicin, chloramphenicol, ofloxocin, novobiocin and clindamycin; however, they showed high resistance to polymixin B, cefuroxime, vancomycin, kanamycin gentamycin, cefazolin, ampicillin, amikacin and cephalothin32.
The biogenic amines (BA) are low molecular weight compounds impicated in various biological activities. The food containing higher amount of BA causes human ailments leading to vomiting, hypertension, palpitations, and headache36. The decarboxylase or deiminase activity of some probiotics converts amino acids into BA. Moreover, the amino acids catabolism by probiotics may affect quality and safety of foods. Hence, probiotics should not produce large amount of BA36. Previously, study by Singh et al. 32 suggested that none of their probiotic strains produced BA from the amino acids used, hence they can be considered as safe according to BA production aspect. In this study, most of the probiotic isolates were not found to produce BA when subjected to amino acids such as Histidine, arginine, tryptophane, lysine and phenylalanine. The isolate B2Enr did not produce BA in the presence of all amino acids except arginine. However, the SP1B and L. plantarum produced BA against all the amino acids. In particular, SP1 did not produce BA in the presence of arginine, lysine, tryptophane, and phenylalanine, however, it produced BA in the presence of histidine. The SP2 and SP1S isolates did not produce BA in the presence of histidine, tryptophane and lysine, but they produced BA using arginine and phenylalanine precursors. SP1M did not produced BA using phenylalanine, tryptophane, lysine but it produced BA in the presence of histidine and arginine. The SP3 produced BA in the presence of histidine, arginine, phenylalanine but it did not produce BA by using lysine and tryptophane. Hence, SP1 and B2Enr can be considered as biosafe because they did not produce BA when subjected to different amino acids precursors. However, the isolates which could produce the BA may be further subjected to quantitative evaluation of BA through HPLC to determine the level of BA production.
Furthermore, the mucin degradation is an important criterion for biosafety assessment of probiotics. The probiotic should not degrade mucin. In the present study, except two probiotic isolates SP1M and SP1S, all probiotic isolates did not degrade mucin. Hence, SP1M and SP1S cannot be considered as safe. In one previous study, none of the probiotic isolates degraded mucin32. In addition, the hemolytic activity of bacteria is an indication of pathogenicity. Probiotics must not show hemolytic activity. In this study, all probiotic isolates did not show hemolysis on blood agar, except the two probiotic isolates SP2 and SP3 which showed ג-hemolysis. Hence, SP2 and SP3 cannot be considered as safe. One previous study suggested that Bacillus clausii UBBC07 did not show hemolytic activity and can be considered as safe probiotic37. Similarly, in another study32 none of their isolates showed hemolytic activity. The gelatinase production is also an indication of bacterial virulence38. Probiotics must not produce gelatinase. In the present study, none of the probiotic isolates produced gelatinase and our results are in accordance with the previous study32. The deconjugation of bile salts exerted by microbes may promote many alterations in physiochemical properties. Hence, probiotics should not deconjugate bile salts present in intestine39. In the present study, none of the probiotic isolates showed deconjugation of bile salts and our results are in line with those reported previously32.
Further, we compared the in vitro biosafety aspects of all our isolates which revealed that three probiotic isolates namely, SP1B, B2Enr and SP1 can be considered as safe as they passed all above mentioned criteria of biosafety aspects. Moreover, these isolates possess potent probiotic properties among other isolates. In addition, these probiotic isolates were found to exhibit good cell surface hydrophobicity, good auto-aggregation as well as good co-aggregation property with pathogenic organisms. The molecular characterization of SP1B and B2Enr by 16srDNA sequencing suggested SP1B as Bacillus cereus (MK210172) and B2Enr as Staphylococcus epidermidis (MK210234). Among the currently used probiotic products, mostly probiotic strains are bacterial spore formers such as genus Bacillus, which has been shown to prevent GIT disorders40. The B. cereus has been used as a potential probiotic in human medicine and livestock production as well41. The B. cereus CenBiot was proposed as a suitable candidate for probiotic elaboration42 and was examined in farms where it controlled diarrhoea and feed conversion in pigs43. In the EU two Bacillus products have been licensed for use in animals viz. Toyocerin and BioPlus 2B44,45; wherein the Toyocerin consisting of B. cereus var toyoi was found extremely safe for animal use. Though, till now S. epidermidis has been considered as an opportunistic pathogen, the recent studies reveal that S. epidermidis plays an important role in skin homeostasis via suppressing inflammatory cytokines and producing antimicrobial molecules to inhibit skin pathogens46. In addition, one recent study has reported the strong skincare effect of a probiotic skin product consisting of S. epidermidis47. Moreover, study by Wang et al. 48 reported that S. epidermidis inhibits the growth of Propionibacterium acnes and can be implicated as probiotics in acne vulgaris. Recently, a review article has highlighted the role of S. epidermidis, Lactobacillus and Bifidobacterium sp. in the treatment of atopic dermatitis49. (Mottin and Suyenaga, 2018). However, many of these studies were conducted in vitro, and more detailed research should be performed in order to prove the efficacy and safety of these probiotics.
Overall, the present study found that the three isolates namely SP1B (B. cereus; MK210172), B2Enr (S. epidermidis; MK210234) and SP1 obtained from human breast milk can be considered as potential probiotics. These isolates have shown better probiotic activities as compared to standard probiotic L. plantarum. Though, previously B. cereus and S. epidermidis were considered as opportunistic pathogens; the present study findings along with the other above mentioned studies suggest the use of these bacterial strains to be safe and beneficial. However, these bacterial strains must be assessed further for in vivo biosafety aspects using animal models for its consideration of human and/or animal use.
Biogenic amines (BA); Mucin 2 (MUC2); Cell-free neutralized supernatants (CFNS); Muller-Hinton Agar (MHA); Gastrointestinal tract (GIT).
Acknowledgments
We would like to thank all the volunteers who participated in this study and provided the milk samples. We are thankful to Uka Tarsadia University, Bardoli, Gujarat, India for providing necessary research facilities to conduct the study.
Conflicts Of Interest
The authors declare that there is no conflict of interest.
Authors’ Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
None.
Data Availability
The datasets generated and/or analysed during the current study are available in the GenBank-NCBI database repository, Accession No: MK210172 (Bacillus cereus); MK210234 (Staphylococcus epidermidis).
Ethics Statement
All procedures performed in this study involving human participants were in accordance with the ethical standards of the Institutional-Human Research Ethical Committee (HREC), Maliba Pharmacy College, Uka Tarsadia University, Bardoli, Gujarat, India and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All subjects signed informed consent.
- FAO/WHO. Guidelines for the evaluation of probiotics in food. 2002.
- Gourbeyre P. , Denery S. & Bodinier M. Probiotics, prebiotics, and synbiotics: impact on the gut immune system and allergic reactions. J. Leukoc. Biol. , 2011; 89: 685-695.
- Macpherson A. J. , Harris N. L. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. , 2004; 4: 478-485.
- Guarner F. & Malagelada J. R. Gut flora in health and disease. Lancet, 2003; 361: 512-519.
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999; 69: 1046S-1051S.
- Dwivedi M. , Kumar P. , Laddha N. C. & Kemp E. H. Induction of Regulatory T Cells: A Role for Probiotics and Prebiotics to Suppress Autoimmunity. Autoimmunity Rev. , 2016; 15(4): 379-392.
- Favier C. F. , DeVos W. M. & Akkermans A. D. Development of bacterial and bi dobacterial communities in feces of newborn babies. Anaerobe, 2003; 9: 219-229.
- Fernבndez L. , Langa S. , Martםn V. , Maldonado A. , Jimיnez E. , Martםn R. & Rodrםguez J. M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. , 2013; 69(1): 1-10.
- Olivares M. , Diaz-Ropero M. P. , Martin R. , Rodriguez J. M. & Xaus J. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. , 2006; 101: 72-79.
- Kumar M. , Ghosh M. & Ganguli A. Mitogenic response and probiotic characteristics of lactic acid bacteria isolated from indigenously pickled vegetables and fermented beverages. World J. Microbiol. Biotechnol., 2012; 28: 703-711.
- Rickard A. H. , Gilbert P. , High N. J. , Kolenbrander P. E. & Handley P. S. Bacterial coaggregation: an integral process in the development of multi-species bio lms. Trends Microbiol. , 2003; 11: 94-100.
- Garcםa-Cayuela T. , Korany A. M. , Bustos I. , deCadiסanos LPG, Requena T, Pelבez C et al. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. , 2014; 57: 44-50.
- Geertsema-Doornbusch G. I. , Van der Mei H. C. & Busscher H. J. Microbial cell surface hydrophobicity the involvement of electrostatic interactions in microbial adhesion to hydrocarbons (MATH). J. Microbiol Methods. , 1993; 18(1): 61-68.
- Tomas M. & Nader M. Effect of culture conditions on growth and autoaggregation ability of vaginal Lactobacillus johnsoni CRL 1294. J. Appl. Microbiol. , 2005; 99: 1383-1391.
- Handley P. S. , Harty D. W. , Wyatt J. E. , Brown C. R. , Doran J. P. & Gibbs A. C. A comparison of the adhesion, coaggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J. Gen. Microbiol. , 1987; 133: 3207-3217.
- Bover-Cid S. & Holzapfel W. Improved screening procedure for biogenic amine production. Int. J. Food Microbiol. , 1999; 53: 33-41.
- Eatson T. J. & Gasson M. J. Molecular Screening of Enterococcus Virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Environ. Microbiol. , 2001; 67(4): 1628-1635.
- Romanenko L. A. , Uchino M. , Kalinovskaya N. I. , Mikhailov V. V. Screening of antimicrobial, hemolytic activities. Microbiol. Res. , 2008; 163: 633-644.
- Noriega L. , Cuevas I. , Margolles A. & de los Reyes-Gavilan C. G. Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. , 2006; 16: 850-855.
- Bauer A. W. , Kirby W. M. M. , Sherris J. C. & Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. , 1966; 45(4): 493-496.
- Cabrera-Rubio R. , Collado M. C. , Laitinen K. , Salminen S., Isolauri E. , Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. , 2012; 96(3): 544-51.
- Martin R. , Langa S. , Reviriego C. , Jimenez E. , Marin M. L. , Xaus J. , et al. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. , 2003; 143: 754-758.
- Martin R, Langa S, Reviriego C, Jimenez E, Marin LM, Olivares M, et al. The commensal microflora of human milk: New perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. , 2004; 15: 121-127.
- Kozak K. , Charbonneau D. , Sanozky-Dawes R. & Klaenhammer T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut. Microbes. , 2015; 6(6): 341-351.
- Kavitha J. R. & Devasena T. Isolation, Characterization, Determination of Probiotic Properties of Lactic Acid Bacteria from Human Milk. IOSR J Pharm. Biol. Sci. , 2013; 7(3): 01-07.
- Rajoka M. S. R. , Mehwish H. M. , Siddiq M. , Haobin Z. , Zhu J. , Yan L. , et al. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT – Food Sci. Technol. , 2017; 84: 271-280.
- Osmanagaoglu O. , Kiran F. & Ataoglu H. Evaluation of in vitro Probiotic Potential of Pediococcus pentosaceus OZF Isolated from Human Breast Milk. Probiotics Antimicrob. Proteins, 2010; 2(3):162-74.
- Shokryazdan P. , Sieo C. C. , Kalavathy R. , Liang J. B. , Alitheen N. B. , Jahromi M. F. , et al. Probiotic Potential of Lactobacillus Strains with Anti-microbial Activity against Some Human Pathogenic Strains. BioMed Res. Int. , 2014; 2014: 1-16.
- Di Bonaventura G. , Piccolomini R. , Paludi D. , D’orio V. , Vergara A. , Conter M. & Ianieri A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food contact surfaces: relationship with motility and cell surface hydrophobicity. J. appl. Microbiol. , 2008; 104(6): 1552-1561.
- Rosenberg M. , Gutnick D. & Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell surface hydrophobicity. FEMS Microbiol. Lett. , 1980; 9(1): 29-33.
- Yadav R. , Puniya A. K. & Shukla P. Probiotic Properties of Lactobacillus plantarum RYPR1 from an Indigenous Fermented Beverage Raabadi. Front. Microbiol. , 2016; 7: 1683.
- Singh T. P. , Malik R. K. & Renuka G. K. Safety assessment and evaluation of probiotic potential of Lactobacillus reuteri strains under in vitro conditions. Int. J. Curr. Microbiol. Appl. Sci. , 2014; 3(2): 335-348.
- Blandino G., Milazzo I. & Fazio D. Antibiotic susceptibility of bacterial isolates from probiotic products available in Italy. Microb. Ecol. Health Dis. 2008; 20: 199-203.
- Kastner S. , Perreten V. , Bleuler H. , Hugenschmidt G. , Lacroix C. & Meile L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. , 2006; 29(2): 145-55.
- Muסoz-Atienza E. , Gףmez-Sala B. , Araתjo C. , Campanero C. , Del Campo R. , Hernבndez P. E. , et al. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. , 2013; 13(1): 15.
- Lonvaud-Funel A. Biogenic amines in wines: role of lactic acid bacteria. FEMS Microbiol. Lett. , 2001; 199: 9-13.
- Lakshmi S. G. , Jayanthi N. , Saravanan M. & Sudha Ratna M. Safety assesment of Bacillus clausii UBBC07, a spore forming probiotic. Toxicol Rep. , 2017; 4: 62-71.
- Thurlow L. R. , Thomas V. C. , Narayanan S. , Olson S. , Fleming S. D. & Hancock L. E. Gelatinase Contributes to the Pathogenesis of Endocarditis Caused by Enterococcus faecalis. Infect. Immun. , 2010; 78(11): 4936-4943.
- Dunne C. , O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr., 2001; 73: 386S-92S.
- Hong H. A. , Duc L. H. & Cutting S. M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. , 2005; 29: 813-835.
- Elshaghabee F. M. F. , Rokana N. , Gulhane R. D. , Sharma C. & Panwar H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. , 2017; 8: 1490.
- Gil-Turnes C. , Freitas dos Santos A. , Weykamp da Cruz F. , Monteiro A. V. Properties Of The Bacillus Cereus Strain Used In Probiotic CenBiot. Revista de Microbiologia, 1999; 30: 11-14.
- Zani J. L. , da Cruz F. W. , dos Santos A. F. & Gil-Turnes C. Effect of probiotic CenBiot on the control of diarrhoea and feed efficiency in pigs. J. Appl. Microbiol. , 1998; 84: 68-71.
- SCAN. Assessment by the Scientific Committee on Animal Nutrition (SCAN) of a microorganism product: Esporafeed Plus. European Commission, Health and Consumer Protection Directorate-General. (SCAN) Scientific Committee on Animal Nutrition. 1999, Available from http://europa. eu. int/comm/food/fs/sc/ scan/out39_ en. pdf.
- SCAN. Report of the Scientific Committee on Animal Nutrition on product BioPlus 2B for use as feed additive. European Commission, Health and Consumer Protection Directorate-General. (SCAN) Scientific Committee on Animal Nutrition. 2000, Available from: http://europa. eu. int/comm/food/fs/sc/scan/out49_en. pdf.
- Lai Y. , Di Nardo A. , Nakatsuji T. , Leichtle A. , Yang Y. , Cogen A. L. et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. , 2009; 15: 1377-1382.
- Dekio I. Clinical effect of novel probiotic product for the skin containing Staphylococcus epidermidis isolated from customers. Conference Proceedings of IPC 2016. Paper presented at the International Scientific Conference on Probiotics and Prebiotics, 2016; Budapest (p. 21. ).
- Wang Y. , Kuo S. , Shu M. , Yu J. , Huang S. , Dai A. , et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol., 2014; 98(1): 411-424.
- Mottin V. H. M. & Suyenaga E. S. An approach on the potential use of probiotics in the treatment of skin conditions: acne and atopic dermatitis. Int. J Dermatol., 2018; 57(12): 1425-1432.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.