ISSN: 0973-7510
E-ISSN: 2581-690X
Bacillus spp. probiotics used as feed additives can form spores and tolerate the harsh conditions of the human digestive system and are beneficial for the treatment of inflammatory bowel syndrome. Since reports on probiotics and anti-inflammatory properties of Bacillus spp. isolated from the fermented food of Northeast India have not been explored much. The present study focused on Bacillus spp. BN5, AY5, and AN8, possessing these desired properties. In the probiotics study, the isolates were screened for their tolerance to acid and bile salt, auto-aggregation, hydrophobicity, cholesterol assimilation, antibiotic resistance, and antagonistic properties. It was found that these isolates possessed the desirable probiotic traits. The Bacillus spp. culture and their supernatant were also screened for their ability to reduce LPS-induced inflammation in murine macrophage (RAW 264.7) cells. All the Bacillus spp. culture and their supernatant treatments were found to reduced the Nitric oxide (NO) production by LPS-induced cell lines. The supernatant of LPS-induced cell lines were also analyzed to measure the level of inflammatory cytokine production. It was found that the levels of TNF-α, IL-6, and IL-1β were reduced after co-treatment with LPS and Bacillus spp. culture or LPS and Bacillus spp. supernatant. Results suggested that the Bacillus spp. are potential probiotic candidates with anti-inflammatory properties.
Bacillus spp., Traditional Fermentation, Probiotic Properties, Anti-inflammatory Properties
The North-Eastern Region of India, inhabited by different indigenous tribes, is characterized by unique socio-cultural and dietary habits. The people of the region practice their traditional technology for the preparation of fermented foods and beverages. Traditional fermentation of soybean is popular in the region, and the product is known by different names such as ‘Tungrymbai’ in Meghalaya, ‘Hawaijar’ in Manipur, ‘Bekang’ in Mizoram, ‘Akhone’ in Nagaland and ‘Peruyaan’ in Arunachal Pradesh. These products are spontaneously fermented without any starter culture and are known to have rich microbial diversity involved in the fermentation process.1,2 Some of the microbial cells present in these fermented foods are probiotic in nature which is beneficial to humans.3
Analysis of parameters like acid and bile tolerance, cell hydrophobicity, auto-aggregation, antimicrobial properties, and resistance to antibiotics provide a better understanding of probiotic properties and health-promoting benefits. Studies show that Bacillus spp. isolated from fermented food were applied in industrial products as probiotics.4-6 These bacteria are known to produce spores which, when exposed to the extreme environment, are heat stable and resistant to low pH such as gastric juice.4-8 Thus, Bacillus spp. have an advantage over other non-spore-forming bacteria because of their ability to survive in extreme environmental conditions such as high temperatures, acidic environments and dry and undernutrition environments.5,8,9,10
However, in vitro screening for potential probiotic isolates required a specific characteristic, such as resistance to gastric acid and bile salt with adhesion activity to the human epithelial cells. Cell hydrophobicity, autoaggregation, and antimicrobial activity are also required to be tested for probiotics.5,11
Probiotics play an important role in chronic intestinal diseases because of their role in immune system modulation and anti-inflammatory response.12 The use of probiotics not only improves microbial population and gut health but also increases the secretion of mucus that prevents tight junction protein from destruction by decreasing the number of lipo-polysaccharides (LPS).13-15 Higher level of Lipopolysaccharides (LPS) of pathogenic bacteria enhances inflammation and other bowel problem that lead to Inflammatory bowel Syndrome (IBD). In response to this, epithelial cells and macrophages produce NO to induce an immune response. The reduction in NO production by the test isolates can be used as an indication of reduced inflammation by the isolates.16
Understanding the traditionally fermented foods, their microbiology, and the associated activity holds importance from the perspective of the dearth of science on these ethnic products. Thus, the present study was aimed at screening Bacillus spp., which are spontaneously occurring bacteria in traditional foods, for their probiotics properties and also in studying the in vitro anti-inflammatory properties such that they can be bio-prospected for large-scale exploration aided by starter culture fermentation technology.
Isolation of Bacillus spp.
The selected Bacillus spp. from ethnically fermented soybean products, namely, ‘Akhone’ and ‘Bekang’ were isolated using serial dilution techniques and plated on Yeast Malt agar and Nutrient agar. The colonies were randomly selected, and pure cultures were maintained for identification.
Molecular characterization
DNA isolation was performed using a standard kit (HiPurA 96 Bacterial Genomic DNA Purification Kit, Himedia, India) for identification of the isolate. DNA was used for amplification of 16S rRNA using universal primers, 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’GGTTACCTTGTTACGACTT-5’) PCR mixtures (25 µL) contained approximately 40 ng template DNA, 2 µM each of forward primer (27F) and reverse primer (1492R) 1.75 mM of MgCl2 (Taq Buffer), deoxynucleoside triphosphates (250 µM each of dATP, dCTP, dGTP and dTTP) and 0.3 U Taq polymerase. DNA amplification was carried out using the GeneAMP PCR system 9700 (Applied Biosystems, CA, USA), and approximately 1,500 bp were amplified. The amplified DNA samples were sequenced, and the DNA sequences obtained were used to perform blastn analysis with the Ez Taxon database.17 Sequences were submitted to the NCBI BankIt, and the accession numbers were obtained.
Tolerance to acidic condition
Tolerance to low acidic conditions was determined by the method of Nithya and
Halami.18 Bacillus spp. Cultures were incubated in LB broth at 37°C for 18 hours. After incubation, the bacterial cells were harvested by centrifugation at 8000 rpm at 4°C for 15 minutes. The pellets were washed once with Phosphate buffer saline (PBS) of pH 7.2±2 followed by resuspension in PBS of pH 3. Cell suspensions were incubated in a shaker incubator for 0, 1, 2, and 3 hours at 37°C. After the incubation period, the culture was plated on LB agar, and the counts were expressed as CFU/ml.
Percentage of survival was calculated using the formula:
Survival % = [(CFU/ml) of cell survived / (CFU/ml) of initial cells inoculated] × 100
Tolerance to Bile salt
Bacillus spp. cultures were incubated at 37°C for 18 hours followed by centrifugation for 15 minutes at 8000 rpm at 4°C. Pellets were washed and resuspended in 0.85% NaCl. The bacterial suspension was inoculated into LB broth with 0.3% bile salt concentration and incubated for 0, 1, 2, and 3 hours at 37°C. Viable plate counts were determined by plating on LB agar plates after each incubation period.19
Cell hydrophobicity
Cell hydrophobicity of the selected Bacillus spp. was determined according to the method followed by Lee et al.5 with minor modifications. The bacterial culture was incubated for 24 hours and then centrifuged at 13000 rpm for 3 minutes. The Bacillus spp. supernatant was discarded, and the pellet was washed twice and then suspended with PBS buffer saline (pH 7.4). One ml of the cell suspension was measured for optical density (OD) in a spectrophotometer at 600nm, and the value was used as Ao. An equal volume of solvent (xylene) was added and vortexed thoroughly for 5 minutes. The mixture was then allowed to separate into two phases by keeping it undisturbed at 37°C for 30 minutes. The aqueous phase was taken, and absorbance was observed at 600 nm, and the value obtained was taken as A1.
Hydrophobicity (%)= (1 – [A1 / A0] ) × 100
Where, A1 = Absorbance after 30 minutes incubation; Ao = Absorbance at 0 hours.
Auto-aggregation
Auto-aggregation of Bacillus spp. was performed according to the method of Lee et al.5 An overnight bacterial culture was centrifuged at 13000 rpm for 3 minutes. Pellets were washed twice with phosphate buffer saline (pH 7.4). The pellet was then re-suspended in the Bacillus spp. supernatant and vortexed for 50 seconds. Absorbances were measured every 0, 1, 2, and 3 hours at 600nm.
Auto-Aggregation (%)= (A0 – [At / A0] ) × 100
Where, At = Absorbance at 0, 1, 2, 3 hours at 600nm; Ao = Absorbance at 0 hours.
Assimilation of cholesterol
Cholesterol assimilation by the strains was determined as described by Rudel and Morris.20 The bacteria were cultured on LB broth for 18 hours at 37°C in a Shaker incubator. 1% of the bacterial culture was inoculated to the fresh media containing cholesterol (100µg/ml) and 0.2% bile salt and incubated at 37°C in a shaker incubator for 24 hours. Media without cholesterol was used as a control. The cultures were then Centrifuged at 9000 X g at 4°C for 15 minutes. To a 1ml of the bacterial supernatant in a test tube, 1ml of KOH (33% wt./Vol.) and 2ml of absolute ethanol were added, and the mixture was then vortexed for 1 minute, followed by heating at 37°C for 15 minutes and then cooled to room temperature. 2 ml of distilled water and 3 ml of hexane were added and vortexed for 1 minute. The mixture was allowed to stand till the hexane layer was properly separated. 1ml of Hexane layer was transferred to a fresh test tube and heated at 65°C in a water bath till it gets evaporated. Then 2ml of ortho-phthalaldehyde reagent was immediately added to the dried residue and vortexed for 10 minutes. The dissolved mixture was then mixed with 0.5ml of Conc. H2SO4 and vortexed for 1 minute. The mixture was kept in the dark at room temperature for 20 minutes. The absorbance was measured at 550nm in triplicates. The percentage of cholesterol assimilated was determined as follows:
Cholesterol % remained in the sample = [(O.D.of sample with cells) / (O.D.of sample without cells )] × 100
Cholesterol % assimilated=100- cholesterol % remained in sample
Susceptibility to antibiotics
Antibiotic susceptibility for the selected Bacillus spp. was evaluated by the method followed by Nithya and Halami.18 Antibiotic disks like Tetracycline, Vancomycin, Erythromycin, Penicillin-G, Gentamycin, and Chloramphenicol were used to screen for the resistance of bacterial isolates. An overnight culture of bacterial isolates was standardized to an OD range of 0.08 to 0.1 at 600nm wavelength. The cultures were then evenly swabbed onto Mueller Hinton agar plates, and the antibiotic disks were placed with an and incubated for 24 hours. The diameter of the inhibition zone was recorded.
Antagonistic activity
The antagonistic activity was performed by minor modification of the agar well diffusion assay method of Valgas et al.21 Bacillus spp. were tested against E. coli MTCC723, Klebsiella pneumoniae MTCC109, and Staphylococcus aureus MTCC2940. The test culture and Bacillus spp. were incubated for 24 hr at 37°C. Test cultures were standardized to McFarland standard no. 0.5 and uniformly swabbed onto MHA plates. Each agar plate was punched with sterile pipette tips to form wells. One ml of Bacillus spp. suspensions were then centrifuged at 10,000g for 20 minutes. The Bacillus spp. supernatant was then filtered through a 0.2 µm sterile filter. 100 µl of the Bacillus spp. supernatant was then added to each well. A Chloramphenicol disc was used as a reference. The plates were then incubated at 37°C for 24 hours.
MTT assay
The stock RAW 265.7 cells were activated in DMEM supplemented with 10% FBS and 1% P/S solution in the presence of 5% CO2 in a humidified chamber at 37°C for three consecutive transfers, and the cells were grown and passaged till 80% confluence. Then the confluent macrophages were seeded in a 96-well cell culture plate followed by the treatment with Bacillus Spp. at different OD (2, 1.5, 1, and 0.5) and Bacillus spp. supernatant (1, 0.5, and 0.25 mg/mL). The cells were incubated at 37°C under 5% CO2 in a humidified incubator. After 24 hours of incubation, MTT (03-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added to a final concentration of 0.5 mg/mL followed by 4 hours incubation under 5% CO2 in the dark at 37°C. Thereafter, the medium was removed, accompanied by 100 µl DMSO formazan crystal dissolution. The absorbance was measured at 570 nm using a microplate reader (M200 PRO, Tecan Life Science).22
Assessment of Bacillus spp. on LPS-induced inflammation in RAW 264.7 macrophages
Confluent macrophage cells were seeded in a 48-well plate (1×105 cells) and allowed to achieve confluence as described above. Then the cells were treated with 1 µg/mL LPS and each Bacillus spp. culture (0.5 OD) or Bacillus spp. supernatant (0.25 mg/mL) for 16 h in a humidified incubator at 37°C under 5% CO2. This was followed by an estimation of nitrite through the Griess reagent method. The optical density was measured at 540 nm.
Measurement of TNF-α, IL-6, & IL-1β cytokines
The supernatant from the various cell line treatments were collected, and the levels of TNF-α, IL-6, & IL-1β were measured using enzyme-linked immunosorbent assay (ELISA) by using commercially available kits as per the manufacturer’s instructions (Elabscience, USA).
Statistical analysis
The statistical significance among different groups was performed by one-way ANOVA followed by Tukey’s post hoc test. Graph Pad prism 9.0.0(121) Software was used to analyze the data. Statistically, the significance was considered to be P ≤ 0.05.
Identification of Isolates
The marker gene 16S rRNA was sequenced and the isolates were identified as Bacillus siamensis, Bacillus tequilensis and Bacillus subtilis.
Phylogeny
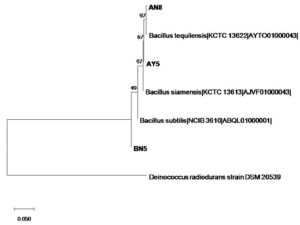
The phylogenetic tree for 16S rRNA gene sequences was constructed using the neighbor-joining method in MEGA-X software with Deinococcus radiodurans taken as an outgroup organism. The dendrogram showed clustering of isolates to the closest match from the database. Isolates, AY5 clustered with Bacillus siamensis, AN8 clustered with Bacillus tequilensis, and BN5 clustered with Bacillus subtilis (Figure 1).
Tolerance to acidic condition and bile salt
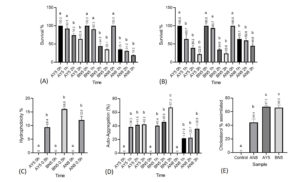
Initial bacterial inoculum, inoculated at 0 hours, served as a control to measure the survival percentage of Bacillus spp. at 1, 2, and 3 hours. It was observed that all the Bacillus spp. tested showed high tolerance to acidic conditions. After 3 hours of incubation at pH 3, isolate AY5 showed the highest survival percentage (63.9%) in acidic conditions, followed by the isolates BN5 (35.1%) and AN8 (19.2%) (Figure 2A).
All the tested Bacillus spp. showed tolerance to 0.3% bile salt concentration after 1, 2, and 3 hours of incubation. After 3 hours of incubation, AN8 showed the highest survival percentage (44.8%), followed by BN5 (23.8%) and AY5 (22.0%) (Figure 2B).
Cell Hydrophobicity
All three isolates showed cell hydrophobicity. The isolate BN5 showed comparatively higher hydrophobicity (16%), followed by AN8 (12%) and AY5 (9.4%) (Figure 2C).
Auto-aggregation
All the tested Bacillus spp. showed a significant auto-aggregation pattern at different intervals of the incubation period. The isolate BN5 showed the highest auto-aggregation of 67.0% after 3 hours incubation period. The isolate AY5 showed significant auto-aggregation of 42.2 % after 3 hours of incubation. Similarly, isolate AN8 showed the highest auto-aggregation of 35.8% after 3 hours incubation period (Figure 2D).
Assimilation of cholesterol
All the tested isolates showed cholesterol assimilation properties. Isolates AY5 showed the highest assimilation of 67.3%, followed by BN5 (66%) and AN8 (44.4%) (Figure 2E).
Figure 2. (A) Acid tolerance, (B) bile salt tolerance, (C) cell hydrophobicity, (D) auto-aggregation and (E) cholesterol assimilation of Bacillus spp. (AY5, BN5 and AN8). Different letters above each bar for graph ‘a’, ‘b’ and ‘c’ represent significant difference between values of the same isolate (p<0.05). For graph (e), letters above each bar represent significant difference between different isolates (p<0.05)
Susceptibility to antibiotics
All the tested isolates are susceptible to Tetracycline, Vancomycin, Erythromycin, Penicillin G, Gentamycin, and Chloramphenicol except for the isolate AN8, which showed resistance against penicillin-G (Table 1).
Table (1):
Susceptibility test of Bacillus spp. against different antibiotics
Isolate name |
Tetracycline (10 mcg; in mm) |
Vancomycin (30 mcg in mm) |
Erythromycin (10 mcg in mm) |
Penicillin G (10 mcg in mm) |
Gentamycin (10 mcg in mm) |
Chloramphenicol (30 mcg in mm) |
|---|---|---|---|---|---|---|
AY5 |
32 |
26 |
29 |
15 |
27 |
37 |
BN5 |
38 |
27 |
32 |
29 |
28 |
33 |
AN8 |
37 |
22 |
31 |
R |
26 |
34 |
*R= resistance
Antagonistic activity
The isolates AY5 showed antagonistic activity against Klebsiella pneumoniae MTCC109. AN8 showed antagonistic activity against Staphylococcus aureus MTCC2940 and E. coli MTCC723. BN5 showed antagonistic activity against E. coli MTCC723, Staphylococcus aureus MTCC2940, and Klebsiella pneumoniae MTCC109.
Effect of Bacillus spp. and Bacillus spp. supernatant on RAW264.7 cell viability
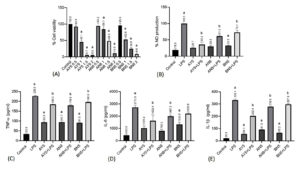
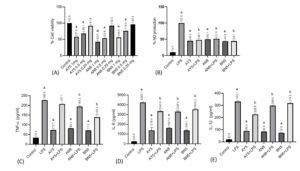
The cell viability after exposing Bacillus Spp. (AY5, BN8 and AN8 and Bacillus spp. supernatant on the RAW 264.7 macrophages at different OD (2, 1.5, 1 and 0.5) and concentrations (1, 0.5, and 0.25 mg/mL), respectively, are illustrated in Figure 3A and Figure 4A. Cells exposed to Bacillus Spp. at 1-2 OD (6X107/ml) showed significant loss of cell viability. In contrast, no loss of viability was observed when the macrophages were treated with Bacillus spp. at 0.5 OD relative to the control cells. In case of Bacillus spp. supernatant, treatment at 0.25 mg/mL gave the highest viability, whereas treatment at 0.5 and 1mg/mL exhibited a loss in cell viability. With this observation, we chose 0.5 OD for Bacillus spp. and 0.25 mg/mL for Bacillus spp. supernatant for subsequent experiments.
Figure 3. Effect of the Bacillus spp. on (A) Cell viability, (B) NO production; (C) TNF-α; (D) IL-6 and (E) IL-1β measured in supernatant of LPS-stimulated RAW 264.7 cells. ‘a’ relative to the control, ‘b’ relative to the LPS (lipopolysaccharide). Data were presented as mean ± SEM; n= 3 and evaluated by one-way ANOVA followed by Tukey’s post hoc test
Effect of Bacillus spp. and its Bacillus spp. supernatant treatment on nitrite synthesis in RAW 264.7 macrophages induced by macrophages
The effect of Bacillus spp. and Bacillus spp. supernatant on LPS-induced inflammation in macrophages was investigated by assessing (nitric oxide) NO response, which is one of the essential inflammatory parameters. It was found that LPS stimulation resulted in a remarkable elevation of NO response ten times higher than control (Figure 3B and Figure 4B). This NO production by LPS-activated cells appeared to be reduced by Bacillus spp. and Bacillus spp. supernatant, suggesting a promising anti-inflammatory activity.
Cytokine analysis in the supernatant of RAW 264.7 cells
The generation of TNF-a, IL-6, & IL-1b by RAW 264.7 macrophages stimulated with LPS for 16 hours was investigated. As shown in the figure, the TNF-a, IL-6, & IL-1b levels were significantly elevated following stimulation of the cells with LPS which was further reduced in all the treatment groups (Figure 3C-3E and Figure 4C-4E).
Figure 4. Effect of the lyophilized Bacillus spp. supernatant on (A) Cell viability; (B) NO production; (C) TNF-α (D) IL-6 and IL-1β measured in supernatant of LPS-stimulated RAW 264.7 cells. ‘a’ relative to the control, ‘b’ relative to the LPS (lipopolysaccharide). Data were presented as mean ± SEM; n= 3 and evaluated by one-way ANOVA followed by Tukey’s post hoc test
The isolates were identified based on their closest match by using the EzTaxon database. The accession numbers for the sequence were obtained from NCBI. 16S rRNA sequences were submitted in the NCBI BankIt database under the Genbank ID: MK641490, MK641489, and MK332360 (Table 2).
Table (2):
Potential isolates and their closest match based on 16S rRNA sequences analysis
Fermented soybean samples |
Isolate Name |
Closest related microorganisms |
Similarity (%) |
Nucleotide bases Submitted |
Genbank Accession number |
|---|---|---|---|---|---|
Akhone |
AY5 |
Bacillus siamensis (AJVF01000043) |
99.85 |
1,378 |
MK641490 |
AN8 |
Bacillus tequilensis (AYTO01000043) |
99.78 |
1,394 |
MK641489 |
|
Bekang |
BN5 |
Bacillus subtilis (ABQL01000001) |
99.85 |
1,364 |
MK332360 |
The ability of the microorganisms to inhabit the gastrointestinal tract is depicted by their ability to withstand the stress condition generated by the physical and chemical mechanism of the host.11,23 Therefore, the ability of Bacillus spp. AY5, BN5, and AN8 to remain viable and tolerate low pH and bile salt concentration is an indication of their ability to thrive under the stressful condition of the gastrointestinal tract.
Cell hydrophobicity is the measurement of the capacity of any bacterial cell to adhere to the hydrocarbon, which holds the ability of the bacterial cells to adhere to the epithelium of the digestive tract, an important property for the first contact between the bacteria and the host cell.24 The ability of the isolates AY5, BN5, and AN8 to adhere to hydrocarbon (xylene) indicates their ability to adhere to the epithelial cells of the human gut.
Auto-aggregation between microorganisms of the same strain is an important factor, especially in the human gut, as it protects the bacteria from external stress, and it also relates to the ability of the bacterial cells to withstand the stress inside the gastrointestinal tract, which is an important property of probiotics.25 The ability of the test isolates, AY5, BN5, and AN8, to auto-aggregate supports their probiotics nature.
Coronary heart disease develops because of elevated cholesterol levels in the blood, and the condition is called Hypercholesterolemia. Therefore, lowering cholesterol levels is important to prevent such diseases.19,26 The capability of these isolates to assimilate cholesterol, especially under stimulated intestinal conditions, indicates their potential as a cholesterol-lowering agent in the future.
The resistance of AN8 to Penicillin-G may collaborate with the findings of Hoa et al.,27 who reported the resistance of Bacillus spp. against Penicillin G, as this may be an intrinsic characteristic useful for Bacillus taxonomy, and such resistance may serve the purpose of restoration of normal microbial flora during antibiotic therapy.28 Thus, the resistance of bacteria to antibiotics does not always constitute a safety concern unless it carries a mobile genetic element like the Tetracycline resistance gene, which may constitute a reservoir of resistance for gut pathogens.
Antagonistic activity against pathogens is one of the most important properties of any isolates when tested for probiotic properties of any probiotic strain.29 These properties indicate the ability of microorganisms to produce antimicrobial compounds that inhibit pathogenic bacteria, which is beneficial for the host.30 Thus, the isolates which showed inhibition properties indicate their ability to inhibit gut pathogens.
In the murine RAW 264.7 macrophages, LPS induces NO production. Furthermore, LPS is well known to cause an inflammatory response by activation of toll-like receptor (TLR4) followed by the release of other pro-inflammatory mediators such as IL-6, IL-1b, and TNF-a. Hence, RAW 264.7 cells are an excellent model for screening and further analysis related to anti-inflammatory properties.31 The cytokines such as TNF-a, IL-6, and IL-1b are pro-inflammatory in-vitro and in-vivo, which are chiefly produced by macrophages and are believed to induce an inflammatory response.32 These findings concord with those of Diao et al.33 and Kim et al.,34 who used an anti-inflammatory study of Bacillus spp. and their polysaccharides using RAW 264.7 cells.
The characterized isolates showed credible probiotic potentials through auto-aggregation, cell hydrophobicity, cholesterol assimilation, antimicrobial properties, and susceptibility to antibiotics. They also showed the ability to withstand stimulated in-vitro stress conditions like bile salt tolerance and acidic condition. The Bacillus spp. and Bacillus spp. supernatant possess potent anti-inflammatory activity by reducing the production of Nitric oxide, pro-inflammatory cytokines, and their mediators. Considering the current findings, it can be concluded that the isolates under study possess the required probiotic properties and anti-inflammatory activity. The study offers scope for exploration of traditionally fermented foods for bioprospection of potential isolates as probiotics and also as starter cultures in traditional fermentation practices.
ACKNOWLEDGMENTS
The authors would like to acknowledge the financial support received from DBT, Govt of India (BT/PR16088/NER/95/69/2015) and Non-NET fellowship (F.24-25/DSW/2020/1753).
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Thokchom S, Joshi SR. Microbial and chemical changes during preparation of traditionally fermented soybean product Tungrymbai of ethnic tribe of Meghalaya. Indian J Tradit Knowl. 2012;11(1):139-142.
- Singh TA, Devi KR., Ahmed G, Jeyaram K. Microbial and endogenous origin of fibrinolytic activity in traditional fermented foods of Northeast India. Food Res Int. 2014;55:356-362.
Crossref - Wang YB, Li JR, Lin J. Probiotics in aquaculture:Challenges and outlook. Aquaculture. 2008;281(1-4):1-4.
Crossref - Cutting SM. Bacillus probiotics. Food Microbiol. 2011;28(2):214-220.
Crossref - Lee S, Lee J, Jin YI, et al. Probiotic characteristics of strains isolated from Korean traditional soy sauce. LWT- Food Sci Technol. 2016;79:518-524.
Crossref - He H, Yu Q, Ding Z, Zhang L, Shi G, Li Y. Biotechnological and food synthetic biology potential of platform strain: Bacillus licheniformis. Synth Syst Biotechnol. 2023;8(2):281-291
Crossref - Wang Y, Zhang H, Zhang L, et al. In vitro assessment of probiotic properties of Bacillus isolated from naturally fermented congee from Inner Mongolia of China. World J Microbiol Biotechnol. 2010;26(8):1369-1377.
Crossref - Setlow P. Germination of spores of Bacillus species: What we know and do not know. J Bacteriol. 2014;196(7):1297-1305.
Crossref - Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extra-terrestrial environments. Microbiol Mol Biol Rev. 2000;64(3):548-572.
Crossref - Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO. Screening for Bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol. 2005;71(2):968-978.
Crossref - Lei J, Ran X, Guo M, Liu J, Yang F, Chen D. Screening, Identification, and Probiotic Properties of Bacillus pumilus from Yak. Probiotics Antimicrob Proteins. 2023.
Crossref - Plaza-Diaz J, Ruiz-Ojeda F, Vilchez-Padial L, Gill A. Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients. 2017;9(9):555.
Crossref - Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation:A Door to the Body. Front Immunol. 2021;12:1-21.
Crossref - Bajagai YS, Yeoh YK, Li X, et al. Enhanced meat chicken productivity in response to the probiotic Bacillus amyloliquefaciens H57 is associated with the enrichment of microbial amino acid and vitamin biosynthesis pathways. J Appl Microbiol. 2023;134(5):lxad5
Crossref - Hashempour-Baltork F, Sheikh M, Eskandarzadeh S, et al. The Effect of Probiotics on Various Diseases and their Therapeutic Role: An Update Review. J Pure Appl Microbiol. 2021;15(3):1042-1058.
Crossref - Singh S, Bhatia R, Khare P, et al. Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate-induced colitis and associated gut microbial dysbiosis in mice. Sci Rep. 2020;10(1).
Crossref - Chun J, Lee J, Jung Y, et al. EzTaxon:a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57(Pt 10):2259-2261.
Crossref - Nithya V, Halami PM. Evaluation of the probiotic characteristics of Bacillus species isolated from different food sources. Ann Microbiol. 2013;63(1):129-137.
Crossref - Shehata MG, El Sohaimy SA, El-Sahn MA, Youssef MM. Screening of isolated potential probiotic lactic acid bacteria for cholesterol-lowering property and bile salt hydrolase activity. Ann Agri Sci. 2016;61(1):65-75.
Crossref - Rudel LL, Morris MD. Determination of cholesterol using o-phthalaldehyde. J Lipid Res. 1973;14:364-366.
Crossref - Valgas C, De Souza SM, Smania EFA, Smania Jr A. Screening methods to determine antibacterial activity of natural products. Braz J Microbiol. 2007;38(2):369-380.
Crossref - Khare P, Maurya R, Bhatia R, et al. Polyphenol rich extracts of finger millet and kodo millet ameliorate high fat diet-induced metabolic alterations. Food Funct. 2020;11(11):9833-9847.
Crossref - Canganella F, Paganini S, Ovidi M, et al. A microbiological investigator on probiotic pharmaceutical products used for human health. Microbiol Res. 1997;152:171-179.
Crossref - Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosi S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003;94(6):981-987.
Crossref - Bao Y, Zhang Y , Zhang Y , et al. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control. 2010;21(5):695-701.
Crossref - Jia B, Zou Y, Han X, Bae JW, Jeon CO. Gut microbiome-mediated mechanisms for reducing cholesterol levels:implications for ameliorating cardiovascular disease. Trends Microbiol. 2023;31(1):76-91.
Crossref - Hoa NT, Baccigalupi L, Huxham A, et al. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl Environ Microbiol. 2000;66:5241-5247.
Crossref - Gueimonde M, Sanchez B, De los Reyes-Gavilan CG, Margolles A. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:202.
Crossref - Fijan S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int J Environ Res Public Health. 2014;11(5):4745-4767.
Crossref - Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:302.
Crossref - Elisia I, Pae HB, Lam V, Cederberg R, Hofs E, Krystal G. Comparison of RAW264.7, human whole blood and PBMC assays to screen for immunomodulators. J Immunol Methods. 2018;452:26-31.
Crossref - Gao G, Zhou J, Zhou J, et al. Divalent cations of magnesium, iron and copper regulate oxidative responses and inflammatory cytokines in RAW 264.7 macrophages. Food Control. 2022;141:109-212.
Crossref - Diao Y, Xin Y, Zhou Y, et al. Extracellular polysaccharide from Bacillus sp. strain LBP32 prevents LPS-induced inflammation in RAW 264.7 macrophages by inhibiting NF-?B and MAPKs activation and ROS production. Int Immunopharmacol. 2014;18(1):12-19.
Crossref - Kim YM, Lee KS, Kim WM et al. Hydrochloric acid-treated Bacillus subtilis ghosts induce IL-1 beta, IL-6, and TNF-alpha in murine macrophage. Mol Cell Toxicol. 2022;18(2):267-276.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.