ISSN: 0973-7510
E-ISSN: 2581-690X
Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) is increasing in Saudi Arabia. MRSA average rate in the country is considered as high as 38%. This review aims to elucidate the present status of MRSA in Saudi Arabia that may serve as a basis for policy development in controlling the pathogen from spreading among healthcare providers as well as the Saudi community. The literature was systematically reviewed in this study using the PRISMA methodology. Eighteen studies with direct relevance were identified and synthesized for the findings. The incidence of MRSA in Saudi Arabia was fond to be varying considerably from one region to another. The rate of MRSA prevalence of all S. aureus strains in the Western, Central, and Eastern regions was 42%, 32%, 27%, respectively. High incidence in the Western region may be attributed to the presence of the holy Islamic sites which are visited by millions of people annually from all over the world. The nasal carriage of MRSA in Saudi Arabia is projected to be around 25% among all isolated S. aureus strains. MRSA colonization ranged from 18%-76% among healthcare workers in Saudi Arabia. The clonal diversity of isolated MRSA strains was indeed varied. However, it seems that specific genotypes of MRSA are more frequently encountered. Type ST239-III is common with hospital-acquired MRSA whereas types CC80-IV (ST80) and to a lesser extent CC22-IV are prevalent among community-acquired MRSA isolates in Saudi Arabia.
Methicillin-resistant Staphylococcus aureus, MRSA, Systematic Review, Genotypes, Saudi Arabia.
Methicillin-resistant Staphylococcus aureus(MRSA) was first described in the early 1960s1. At that time, it was limited mainly to hospitals (hospital-acquired MRSA, HA-MRSA) or in the context of healthcare. MRSA can result in disastrous clinical outcomes with high mortality and morbidity rates. In the United States alone, the annual death toll caused by MRSA is estimated to be 20,000 cases2.
Limited studies in Saudi Arabia have endeavored to explore risk factors connected with HA-MRSA3,4. Moreover, a diversity of cloned MRSA is starting to proliferate among hospitals in the country. Sustained gathering and molecular descriptions of these strains are vital for full treatment and prevention.5 The appearance of MRSA among previously healthy people in the community, even those not having been exposed to healthcare services, was identified in the 1990s6. Community-acquired MRSA (CA-MRSA) appeared first to be limited to skin, soft tissues and bone infections7. The prevalence of MRSA in Saudi Arabia was noted 10 years ago and unembellished community-acquired infection; was confirmed that the general occurrence is indefinite and diverges among geographical areas in the Kingdom4,8-10.
Resistance among microbes is an unavoidable complication of the use of antibiotics, adding serious challenges to the prevention and treatment of infectious diseases. It emphasizes the need to develop more effective approaches to treating MRSA rather than relying solely on current traditional techniques. The term “methicillin-resistant Staphylococcus aureus” (MRSA) specifies the variants of S. aureus that are resistant to many antibiotics, either in the hospital or a community setting; for example, medications like imipenem, oxacillin, methicillin, cephalosporins, nafcillin, and/or drugs classified as beta-lactamase inhibitors. S.aureus is now resistant to methicillin via the acquisition of an exogenous mobile gene that integrates into its chromosomal DNA. [11] This results in enormous decreased affinity to beta-lactam drugs by changing the binding site of the penicillin-binding protein PBP2a12. Since prevalence of MRSA and their clonal variants have been recorded in scattered studies, this study aims to review the literature that will elucidate the present status of MRSA in Saudi Arabia. It is hoped that the findings from the literature will serve as a helpful tool for policy development to control the spread of the pathogen in any setting in different Saudi regions.
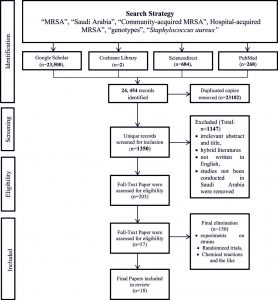
A systematic review of the literature was conducted to synthesize the prevalence of MRSA in Saudi Arabia. The study used the search terms “MRSA”, “Saudi Arabia”, “Community-acquired MRSA”, Hospital-acquired MRSA”, “genotypes”, and “Staphylococcus aureus”among several databases. All searched databases such as Google Scholar (n=23,500), Cochrane Library (n=2), ScienceDirect (n=684), and PubMed (n=268) were housed via PRISMA diagram [Figure 1]. All duplicated copies were then removed (n=23102). All relevant records were retained for another screening (n=1350) and irrelevant abstract and title, hybrid literature, not written in English, and studies not conducted in Saudi Arabia were removed (n=1147). Likewise, relevant studies were retained (n=203). Another screening was done to include those studies written in full text (n=53). All paper articles were carefully re-read for relevance. Finally, 18 full-text articles that fit the topic directly were chosen to synthesize the results.
Prevalence of MRSA in Saudi Arabia
The incidence of MRSA in Saudi Arabia varies considerably from region to region. The rate of MRSA prevalence in the Western, Central, and Eastern regions is 42%, 32%, 27%, respectively.4,13,14 Previously, it was estimated the overall MRSA prevalence rate in the entire country was nearly 30%, representing the second highest incidence among neighboring Gulf countries.15 However, Adam and Abomughaid (2018) recently reported the MRSA rate in the country is considerably higher at 38%.16 In general, it is not definitive what the rates are and it does diverge among different areas. Results from a single study of Riyadh hospitals showed MRSA incidence of entirely S. aureus strains fluctuated between 12% and 49%.17
Massive movements by people to Saudi Arabia (for pilgrimages or migration) are accountable for the widespread distribution of newly developing infections and the appearance of previously undescribed clonal complexes of MRSA. Annually, millions of Muslim people from all over the world visit the country’s holy mosques in Mecca and Medina. This activity certainly contributes to the distribution of MRSA strains throughout the region and even internationally. The main portal of entry to these holy mosques is Jeddah City which probably harbors a wider variety of MRSA clones and species than any other area of the country. A recent meta-analysis-based study showed the highest prevalence of MRSA in the Western region of Saudi Arabia (where the Islamic holy sites are located), with lesser rates in other parts of the country.4,14 However, there is no nationwide analysis of the impact visitors have on the clonality and epidemiology of clinically significant S. aureus strains.
The epidemiological cycle of MRSA is vigorously changing as the incidence of CA-MRSA infections are more reported. S. aureus normally resides on the skin and inside the anterior nares of healthy humans as a commensal microbe, from where it can disseminate into other parts of the body or into the environment causing diverse infections. It is worthwhile to mention that prevalence of MRSA nasal carriage varies greatly from one geographical place to another. For example, reports from India showed nasal MRSA colonization was 3.9% of all nasal staphylococcal strains18, where as it was only 1.5% in the United States.19 The nasal carriage of MRSA in Saudi Arabia is projected to be around 25% among all isolated S. aureus strains.20,21 This increased percentage among community people might be connected to the misuse and overconsumption of antibiotics in the country, in particular beta-lactam drugs.[20]Saudi studies on CA-MRSA are indeed scarce and scattered. Historically, CA-MRSA in Saudi Arabia was first reported in 2001 in the Eastern region of the country22. It was found that CA-MRSA in children was 23.2%-29.8%.21,23 The vast majority of these CA-MRSA pediatric infections were of skin and soft tissues while only 9% represented invasive infections.23 An outbreak of CA-MRSA was reported in 2009 in a neonate nursery without any previous predisposing hospital-associated risk factors in a tertiary clinical facility in Riyadh24.Infected neonates had some features in common like pre-term delivery and underweight; and all24 CA-MRSA infected babies showed mild to moderate skin and soft tissues manifestations24.
CA-MRSA can result in severe invasive infections such as septicemia, necrotizing pneumonia and necrotizing fasciitis25. Clinical conditions like osteomyelitis, deep-seated abscess, pyomyositis, and invasive CNS involvement in a Saudi tertiary hospital were reported25. As stated, investigations on CA-MRSA in Saudi Arabia are limited. More studies need to be conducted as many international reports have confirmed that CA-MRSA is now more prevalent than HA-MRSA infections.Predisposing factors for CA-MRSA infection include direct contact with CA-infected or colonized individuals, crowded living conditions, poor hygiene, sharing of intimate items and contact physical activities26.
Like the incidence of MRSA colonization among people of the community, the incidence in healthcare workers (HCWs) varies from one area to another, whether nationally or internationally. Previous studies showed MRSA colonization among HCWs in Saudi Arabia ranged from 18%-76%27-29.Needless to say, colonized HCWs and hospitalized patients might be implicated in outbreaks of MRSA infections in different wards of any clinical setting. MRSA infections within hospitals spread due to many factors, including undermined infection control policies, prolonged and repeated hospitalization, antibiotics abuse, chronic diseases, poor-sterility surgical procedures, presence of indwelling devices and extreme age. Surgical site infections are among the most common forms of HA-MRSA. Nosocomial MRSA infections are mainly associated with coronary artery bypass graft (CABG) procedures and joint replacement surgery primarily in elderly patients30. The highest global incidence of HA-MRSA were observed in nursery (35.8%), intensive care (19.8%), gynaecological (16.2%) and surgical (11.7%) patients27. A previous study conducted in a Saudi tertiary hospital in Riyadh showed that surgical-associated infections were nearly 7%, and these infections were more pronounced in patients with diabetes and emergency operations.31 HA-MRSA surgical infections represented 50% of all isolated S. aureus strains31. A study released by investigators in the Saudi Security Forces Hospital reported that around 70% of all HA-MRSA infections were of wound, skin and soft tissue origin; septicemia and tracheal infections represented nearly 13% and 18%, respectively32. In a multi-centered retrospective medical chart investigation carried out in Lebanon and Saudi Arabia, the incidence of MRSA skin and soft tissue infections in hospitalized patients was substantially higher than in Lebanon33.HA-MRSA pneumonia was found to be 55% of all MRSA cases, with a prevalence among Lebanese patients < 18years old in contrast to Saudi subjects, where the incidence of HA-MRSA pneumonia was higher in the > 55 years age group33. The mortality rate owing to MRSA pneumonias in Saudi hospitals is between 30-35%34,33. In the south-western area of Saudi Arabia, specifically in Asir Province, HA-MRSA represents 54% of all nosocomial infections caused by S. aureus clinical isolates of which 85% are strikingly multi-drug resistant MRSA strains.35 A study conducted in Taif (west in Mecca Province) found that nosocomial infections accounted for 48% of all infections of hospitalized patients whereas community associated infections were about 52%36. Amongst these infections, investigators noticed the incidence of nosocomial infections in Taif region was 2 per 1000 with HA-MRSA representing 10% of them. The highest incidence of HA-MRSA was observed in wound infections (33.3%) followed by skin and soft tissues at 18.6 % 36. In contrast to Sabra and Abdel-Fattah , Eed et al. noticed that HA-MRSA cases in Taif were 69%, which far outnumbered CA-MRSA (31%)36,37. Most HA-MRSA clinical cases were manifested as sepsis (38%), followed by wound (33%) and lower tract (19%) infections37. HA-MRSA is still a major clinical concern worldwide and, for example, in Europe it causes around 44% of antibiotic-resistant nosocomial infections with a death toll of 22%26.
Some chronic clinical conditions such as diabetes mellitus and haemodialysis (HD) may escalate the prevalence of MRSA colonization and infections38. In diabetic patients, MRSA represents 23% of isolated clinical microbes in wounds pertaining to diabetic foot infections. In the Northern region of Saudi Arabia, 31% of all positive MRSA isolates were taken from diabetic patients;39 keeping in mind that the incidence of foot ulcers in patients with diabetes mellitus is estimated at 6.3% globally.40,41 Hyperglycaemia, impaired nerve function and poor peripheral blood flow in patients with diabetes mellitus complicate the healing of wounds and ulcers and may lead to catastrophic clinical consequences. Both MRSA and MSSA in end-stage renal disease Saudi patients older than 75 years were frequently identified as the nasal variety42. This report rings an alarm for infection control implementers in Saudi Arabia to pay more attention to the elderly undergoing HD as they might be more vulnerable for MRSA-linked vascular access-related septicaemia. MRSA infections in HD patients lead to significantly longer hospitalizations, increased healthcare costs, and higher death rates43.
In the United States, prevalence of MRSA infections in patients with cystic fibrosis (CF) has risen from 12% in 2003 to nearly 26% in 2013, with serious lung function impairment and subsequent mortality 44. In a recent retrospective chart review, MRSA incidence among CF patients in Saudi Arabia was reported to be 11%45. Due to serious MRSA complications in CF patients, its eradication must be considered and it was found that a combination of oral rifampicin and nebulized vancomycin remained one of the successful eradication regimens46.
Circulating MRSA genotypes in Saudi Arabia
MRSA resistance to beta-lactam antibiotics is due to the acquisition of mec genes within a mobile genetic cassette (SCCmec) in the staphylococcal chromosomal DNA, leading to altered penicillin-binding proteins 47,48. Genetically, SCCmec is classified into eight different genotypes (I-VIII) with some divided further into subtypes49,50.Genotyping of the SCCmec provides a strong marker to discriminate HA-MRSA fromCA-MRSA isolates51. It is well established that CA-MRSA strains have SCCmec type IV or V, whereas HA-MRSA strains have SCCmec type I, II or III).52,53 Moreover, the presence of Panton-Valentine leukocidin (PVL), an exotoxin that is lethal to leukocytes, is thought to be highly associated with CA-MRSA while it is absent in HA-MRSA strains.[22]Multilocus sequence typing (MLST) is increasingly becoming a valuable epidemiological tool in identifying microbial infections, and it is based on the sequencing of seven normal housekeeping genes of each microbe. Accordingly, the generated sequences allow each strain a specific designation named sequence type (ST) which help in domestic and global epidemiological studies54. Regarding MRSA strains, the New York/Japan (ST5/SCCmec II) and Brazilian/Hungarian (ST239/SCCmec III) clones are widespread globally and connected to HA-MRSA isolates while the Taiwanese (ST59/SCCmec IV or V), USA300 (ST8/SCCmec IV), European (ST80/SCCmec IV), and USA400 (ST1) clones are always connected to CA-MRSA.55Additionally, MRSA can be further categorized into different clonal complexes (CC). Each distinct CC is comprised of a group of STs in which every ST shares at least five of seven identical alleles with at least one other ST in the group56.
In the Central region of Saudi Arabia (specifically in Riyadh), many CCs of MRSA strains have been reported. One of the earliest studies conducted found that four CCs were abundant among MRSA isolates in Riyadh57. The most predominant strains were CC8/ST239-III (Brazilian/Hungarian Strain), CC22-IV (UK-EMRSA-15/Barnim Epidemic Strain), ST30-IV (Southwest Pacific Clone) and the European CA-MRSA clone CC80-IV57 . ST239-III is a pandemic HA-MRSA strain widely reported in many parts of the world including many countries in the Middle East such as Saudi Arabia.58 HA-MRSA strains isolated from hospitalized cancer patients in Riyadh were all found to be SCCmec type III with a predominance of ST239 (58%) and ST80 (22%).59 It was found that HA-MRSA collected from cancer patients were more multiple-resistant to antibiotics than those isolated from non-cancer hospitalized patients.59 However, Senok et al. found 14 different clones of all tested HA-MRSA strains with the most common being CC80, CC6, CC5, and CC22, respectively.60 Only 9 strains of 117 isolates were ST239 MRSA-III whereas all others were of CA- MRSA lineages as they harboured SCCmec IV and V.60 This finding strongly suggests a shift has occurred in the epidemiological cycle of MRSA, offering a serious evidence that CA-MRSA clones are now predominately implicated within healthcare settings and in hospital-acquired infections. Types of MRSA nasal carriage in Qassim (Central Saudi Arabia) were found to be SCCmec type V (42%) and SCCmec type IVa (23%), while 35% of isolates were non-typeable.20 As for the clonal complexes, CC5, CC8, CC15, CC22, CC25, CC80, and CC188 were characterized. The prevalence of PVL among these CA-MRSA isolates was only 11.5%.20 The latter finding may indicate that PVL detection is currently of little diagnostic value as far as CA-MRSA is concerned. The first documented glycopeptide intermediately susceptible S. aureus (GISA) case was announced in 2009 in Riyadh with a 69-year-old Saudi male patient presenting with severe MRSA sepsis.61 Subsequent investigation of this case demonstrated that the MRSA involved was a genotype of SCCmec type III and was of sequence type ST241, a variant of the notoriously known nosocomial multi-resistant clone ST239.61
In the Western region of Saudi Arabia, five tertiary hospitals in Makkah were screened for MRSA genotypes prevalence.53 Out of 114 MRSA isolates collected, 3% were SCCmec type I; 9% were type II; 47% were type III, and 29% were type IV with only 19% of the isolates harboring the PVL toxin.53 This finding shows that SCCmec type III is still by far the commonest MRSA genotype associated with healthcare settings in Saudi Arabia, similar to many places worldwide. Abulreesh and co-investigators reported that clinical isolates of MRSA collected from different clinical laboratories in Makkah were 100% positive for mecA gene while the strains showed 0% positivity for PVL encoding genes.62 In Taif, HA-MRSA strains were genotyped as SCCmec type I (45%), SCCmec type III (37.5%), and 17.5% as type II, whereas CA-MRSA were 61% type IV and 33% type V.37 Amplification of the coagulase gene and application of restriction fragment length polymorphism (RFLP) showed seven distinct patterns among HA-MRSA; five patterns in CA-MRSA isolates.37 Moussa and co-workers showed MRSA isolates from Jeddah had 42.5%, 38.6%, 15.8% and 2.9% incidence of isolates belonging to SCCmec types V, III, Iva, and Ivc, respectively.62 The prevalence of PVL genes was nearly 38% among all strains tested, mainly associated with soft tissues and wound infections (39.1%) followed by pneumonia and respiratory infections (25%).63
In the Eastern Province of Saudi Arabia, molecular investigations were performed on MRSA isolates from carriers and active infections.64 Strains of MRSA studied were all positive for the genes of mecA, femA, and sa442; SCCmec-IV genotype was by far the most abundant type at 77% among all isolates. Prevalence of SCCmec-V and III was 13% and 9%, respectively.64 Alkharsah and co-workers found that CC80 (ST80, ST1440) lineage was the most commonly encountered strain (35%) followed by the clonal lineage of CC22 (ST22), which was 10% among all MRSA isolates.64 Incidence of PVL showed an overall rate of 59% of all MRSA isolates tested and was more frequently associated with active infections (71%) compared to carrier colonization isolates (47%). In another study conducted in the same province, high clonal diversity was reported among MRSA isolates collected from community members, clinical students and healthcare workers.65 In an agreement with previous studies, El-Mahdy and co-workers stated that SCCmec-IV genotype of collected MRSA was the most common at 45%.65 The most predominant MRSA clonal lineages detected were CC1 t128, CC88 t1339, and CC80.65 Very recently, Alkharsah and colleagues genotyped MRSA strains collected from the Eastern Province using staphylococcal protein A (spa) and pulsed field gel electrophoresis (PFGE) typing methods. 66 Again, the clonal diversity of strains examined was pronounced with 35 spa types and 34 PFGE groups. The most prevalent spa types were t044 (30%), t127 and t304 (6% each), t363 (5%), and t1200 and t002 (4% each).66 It is noteworthy that spa genotype t044 is the CC80 (ST80),a common strain circulating in Saudi Arabia according to the various studies mentioned above.
Based on the data inferred from investigations conducted in different geographical parts of Saudi Arabia, the clonal diversity of isolated MRSA strains is indeed found to be varied. However, it seems that specific genotypes of MRSA such as ST239-III, CC22-IV, and CC80-IV (ST80) are more frequently encountered.ST239-III, carrying the staphylococcal cassette chromosome type III (SCCmecIII), is considered the most successful pandemic HA-MRSA clone associated with healthcare-based infections in all populated global regions.67 This clone with extensive resistance to many antibiotics and hetero-resistant to vancomycin (hVISA) has been reported in isolates from Australia.68 ST239-III has previously been identified in many Middle Eastern countries including United Arab Emirates, Iraq, Saudi Arabia and Turkey.57 CC80-IV MRSA clone represents the European CA-MRSA clone and is observed throughout the Middle East including the Gulf region.64 The widespread prevalence of CC80-IV in the region and in Saudi Arabia probably makes it the most isolated MRSA associated with carrier colonization and infections of community origin. MRSA type CC22-IV (also known as UK-EMRSA-15/Barnim EMRSA) is becoming a pandemic strain in many parts in the world.60 Although this genotype of MRSA is considered a community associated pathogen, its molecular dissection showed presence of antibiotic resistance markers (such as aaca-aphd gene encodes for aminoglycosides resistance), that were previously limited to HA-MRSA isolates. 57 This finding implies that caution should be applied as the antibiogram picture of MRSA isolates has classically been used to differentiate between HA-MRSA and CA-MRSA.
MRSA and its associated infections remain a clinically distressing issue in Saudi Arabia. This microbe is region-sensitive in the country as there are geographical variations of its prevalence in different Saudi provinces. Makkah Province (with the major cities of Makkah, Jeddah and Taif) shows the highest rate (above 40%) of MRSA isolates whereas prevalence in the Eastern Province is estimated at 27%. The most common HA-MRSA strain circulating in Saudi Arabia is SCCmec type III with the sequence type of ST239, while that of frequently encountered CA-MRSA isolates encompass clonal complexes of CC80, CC22, and CC5, which are descendants of genetic lineages having either SCCmec-IV or SCCmec-V chromosomal cassettes. An annual nationwide surveillance report on MRSA prevalence, infections and antibiograms is a necessity to contain its clinical consequences and subsequent economic burden.
Limitation of this study
The main scope of this review is to infer the current epidemiological status of MRSA in Saudi Arabia and the fact susceptibility of the pathogen to antibiotics has not been addressed. Therefore, it would be highly sensible to investigate the antibiotic resistance of MRSA to drugs of choice, whether via original researches on a countrywide scale or by generating a review utilizing already, although, scanty available data. This will certainly add a value particularly in empiric treatment of life-threating MRSA infections.
ACKNOWLEDGMENTS
None.
FUNDING
None.
ETHICS STATEMENT
None.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Shamweel A, Mehr-un-Nisa MM, Shamweel H. (2017). Methicillin resistant Staphylococcus aureus (MRSA) a challenge for health care professionals and patients. ActaScientif Pharm Scis 2017; 1(4):13-22.
- Kavanagh KT. Control of MSSA and MRSA in the United States: protocols, policies, risk adjustment and excuses. Antimicrob Resist Infect Control 2019; 8: 103
Crossref - Al-Anazi AR. Prevalence of methicillin-resistant Staphylococcus aureusin a teaching hospital in Riyadh, Saudi Arabia. Biomed Res, 2009; 20(1):7-14. Available at https://www.alliedacademies.org/articles/prevalence-of-methicillinresistant-staphylococcus-aureus-in-a-teaching-hospital-in-riyadh-saudi-arabia.html
- Adam KM, Abomughaid MM. Prevalence of methicillin-resistant Staphylococcus aureus in Saudi Arabia revisited: A meta-analysis. Open Public Health J, 2018; 11(1): 584-91.
Crossref - Al Yousef S, Taha E. Methicillin-resistant staphylococcus aureus in Saudi Arabia: Genotypes distribution review. Saudi J Med MedSci, 2016; 4(1):2-8.
Crossref - Choo EJ. Community-associated methicillin-resistant Staphylococcus aureus in nosocomial infections. Infect Chemother, 2017; 49(2):158-59.
Crossref - Alaklobi F, Aljobair ., Alrashod A, Alhababi R, Alshamrani M, Alamin W, et al. The prevalence of community-associated methicillin-resistant Staphylococcus aureus among outpatient children in a tertiary hospital: A prospective observational study in Riyadh, Saudi Arabia. Int J PediatrAdolesc Med, 2015; 2(3-4): 136-140.
Crossref - Islam SI, Moore C. Prevalence of methicillin-resistant Staphyloccocusaureus and associated risk factors on admission to a specialist care eye hospital. Ann Saudi Med, 2002; 22(3=4): 153-157.
Crossref - El Amin N, Faidah HS. Methicillin-resistant Staphylococcus aureus in the western region of Saudi Arabia: prevalence and antibiotic susceptibility pattern. Ann Saudi Med, 2012; 32(5):513-16.
Crossref - El-Mahdy TS, Al-Agamy MH, Emara M. Barakat A. Goering RV. Complex clonal diversity of Staphylococcus aureus nasal colonization among community personnel, healthcare workers, and clinical students in the Eastern Province, Saudi Arabia. Biomed Res Int, 2018; 4208762.
Crossref - AlNhazi 2009 cited on p. 2 but missing from final reference list.
- Sharon J.P, Gavin KP. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem, 2015; 84(1):577-601.
Crossref - Al-Hamad AM, Alfaraj AA, Altowaileb J, Al-Shamlan SM, Leskafi H, Alsubeikhy FA. et al. Incidence and antibiotic susceptibility of MRSA infections in a Saudi Arabian Hospital: a 10-year surveillance study. J Infect Dev Ctries, 2018; 12(06):454-61.
Crossref - Al-Zahrani IA, Azhar EI, Jiman-Fatani AA, Siddig LA, Yasir M, Al-Ghamdi AK, Harwood, CR. Impact of mass migrations on the clonal variation of clinical Staphylococcus aureus strains isolated from the Western region of Saudi Arabia. J Infect Public Health, 2019; 12(3):317-22.
Crossref - Aly M, Balkhy HH. The prevalence of antimicrobial resistance in clinical isolates from Gulf Corporation Council countries. Antimicrob Resist Infect Control, 2012; 1(1):26.
Crossref - Goering 2012 is cited on p. 5 but missing from this final list
- Baddour MM, Abuelkheir MM, Fatani AJ, Bohol MF, Al-Ahdal MN. Molecular epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) isolates from major hospitals in Riyadh, Saudi Arabia. Can J Microbiol, 2007; 53(8):931-36.
Crossref - Chatterjee SS, Ray P, AggarwalA, DasA, Sharma M. A community-based study on nasal carriage of Staphylococcus aureus. Indian J Med Res , 2009; 130(6): 742-48.
- Sun J, Yang M, Sreevatsan S, Singer RS, Knutson TP, Marthaler DG, et al. Longitudinal study of Staphylococcus aureus colonization and infection in a cohort of swine veterinarians in the United States. BMC Infect Dis, 2017; 17:690.
Crossref - Abou Shady H., Bakr AA, Hashad ME, Alzohairy MA. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis, 2015; 19(1):68-76.
Crossref - Alaklobi 2015 cited on p. 5, missing from final list.
- Bukharie H. A review of community-acquired methicillin-resistant Staphylococcus aureus for primary care physicians. J Family Community Med, 2010; 17(3):117.
Crossref - Al-Otaibi FE, Bukhari EE. . Community acquired MRSA in outpatient children assisted at university hospital in Saudi Arabia 2005-2008. J Pediatr Infect Dis, 2010; 5(4):369-76. DOI: 10.3233/JPI-2010-0278
- Alsubaie S, Bahkali K, Somily AM, Alzamil F, Alrabiaah A, Alaska A, et al. Nosocomial transmission of community-acquired methicillin-resistant Staphylococcus aureus in a well-infant nursery of a teaching hospital: CA-MRSA in a well-infant nursery. PediatrInt, 2012; 54(6):786-92.
Crossref - Bukhari EE, Al-Otaibi FE. Severe community-acquired infection caused by methicillin-resistant Staphylococcus aureus in Saudi Arabian children. Saudi Med J, 2009; 30(12):1595-600.
- Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill, 2010; 15(41):19688.
Crossref - Iyer AP, Baghallab I, Albaik M, Kumosani T. Nosocomial infections in Saudi Arabia caused by methicillin resistance Staphylococcus aureus (MRSA). Clin Microbial, 2014; 3:3.
Crossref - Al-Humaidan 2015. Cited p. 6, missing from this list.
- Alsulami SO, Al Doghaither H, Iyer AP. Detection of methicillin resistant Staphylococcus areus isolated from nasal carriage of health care workers by polymerase chain reaction. J. Experiment Biol Agricult Sci 2017; 5(3):288-93.
Crossref - Jernigan J. Is the burden of Staphylococcus aureus among patients with surgical-site infections growing? Infect Control HospEpidemiol, 2004; 25(6):457-60.
Crossref - Khairy GA, Kambal AM, Al-Dohayan AA, Al-Shehri M, Zubaidi AM, Al-Naami MY, et al. Surgical site infection in a teaching hospital: A prospective study. Journal of Taibah University Medical Sciences, 2011; 6(2):114-20.
Crossref - Al-Obeid S, Jabri1 L, Shawaf R, Obuli A, Hussni T. Nosocomial infection caused by healthcare-associated (HA) and community-associated (CA) methicillin-resistant staphylococcus aureus (MRSA). EC Microbiology, 2019; 15(6):436-42.
- Moghnieh R, Alothman AF, Althaqafi AO, Matar MJ, Alenazi TH, Farahat F, et al. Epidemiology and outcome of invasive fungal infections and methicillin-resistant Staphylococcus aureus (MRSA) pneumonia and complicated skin and soft tissue infections (cSSTI) in Lebanon and Saudi Arabia. J Infect Public Health, 2017; 10(6):849-54.
Crossref - Madani T, Al-Abdullah N, Al-Sanousi A, Ghabrah T, Afandi SZ, Bajunid HA.Methicillin-resistant Staphylococcus aureus in two tertiary-care centers in Jeddah, Saudi Arabia. Infect Control HospEpidemiol , 2001; 22(4):211-16.
Crossref - Hamid ME. Resistance pattern of coagulase positive Staphylococcus aureus clinical isolates from Asir region, Kingdom of Saudi Arabia. J Microbiol Antimicrob 2011; 3(4):102-8.
- Sabra SM, Abdel-Fattah MM. Epidemiological and microbiological profile of nosocomial infection in Taif hospitals, KSA (2010-2011). World J Med Sci , 2012; 7(1):01-09.
- Eed EM, Ghonaim MM, Hussein YM, Saber TM, Khalifa AS. Phenotypic and molecular characterization of HA-MRSA in Taif hospitals, Saudi Arabia. J Infect Dev Ctries 2015; 9(03):298-303.
Crossref - Yates C, May K, Hale T, Allard B, Rowlings N, Freeman A, et al. Wound chronicity, inpatient care, and chronic kidney disease predispose to MRSA infection in diabetic foot ulcers. Diabetes Care, 2009; 32(10):1907-9.
Crossref - Alanzi F, Al Shammari H, Soomro S. Prevalence of methicillin resistance Staphylococcus aureus (MRSA) infection and incidence of diabetic MRSA patients in Northern region of KSA. Microbiol Infect Dis , 2019; 3(1):1-4.
Crossref - Tentolouris N, Petrikko, G, Vallianou N. Prevalence of methicillin-resistant Staphylococcus aureus in infected and uninfected diabetic foot ulcers. ClinMicrobiol Infect 2006; 12(2):186-9.
Crossref - Armstrong DG, Boulton AJM, Bus, SA. Diabetic foot ulcers and their recurrence. N Engl J Med 2017; 376: 2367-75.
Crossref - Saxena AK, Panhotra BR, Chopra R. Advancing age and the risk of nasal carriage of Staphylococcus aureus among patients on long-term hospital-based hemodialysis. Ann Saudi Med, 2004; 24(5):337-42.
Crossref - Cuervo G, Camoez M, Shaw E. Dominguez MA, Gasch O, Padilla B, et al. Methicillin-resistant Staphylococcus aureus (MRSA) catheter-related bacteraemia in haemodialysis patients. BMC Infect Dis, 2015; 15:484.
Crossref - Muhlebach MS, Beckett V, Popowitch E. Microbiological efficacy of early MRSA treatment in cystic fibrosis in a randomised controlled trial. Thorax 2017; 72: 318-26.
Crossref - Banjar H, AlQahtani H, Al-Mogarri I, et al. (2019). The first report of methicillin-resistant Staphylococcus aureus (MRSA) in cystic fibrosis (CF) patients in Saudi Arabia. Int J Pediatrs Adolesc Med 2019.
Crossref - Chmiel JF, Timothy R, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, et al. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann Am ThoracSoc, 2014; 11(7):1120-29.
Crossref - Becker K, van Alen S, Idelevic EA, , et al. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerging Infect. Dis 2018; 24:242-8.
Crossref - Monecke S, Slickers P, Gawlik D,Muller E, Reissig A, Ruppelt-Lorz A, et al. Molecular typing of ST239-MRSA-III from diverse geographic locations and the evolution of the SCCmec III element during its intercontinental spread. Front Microbiol, 2018; 9:1436.
Crossref - Wong H, Louie L, Lo RY, Simor AE. Characterization of Staphylococcus aureus isolates with a partial or complete absence of staphylococcal cassette chromosome elements. J ClinMicrobiol, 2010; 48:3525-31.
Crossref - Sohail, M., Latif, Z. Molecular typing of methicillin resistance Staphylococcus aureus (MRSA) isolated from device related infections by SCCmec and PCR-RFLP of coagulase gene. Adv Life Sci, 2018; 6(1):34-40.
- Makgotlho P, Kock, MM, Hoosen A, Lekalakala R, Omar S, Dove M, et al. Molecular identification and genotyping of MRSA isolates, FEMS Immunol Med Microbiol, 2009; 57(2):104-15.
Crossref - Al-Hazzani A. Molecular subtyping of multiresistant community-associated methicillin-resistant Staphylococcus aureus isolates in Riyadh, Saudi Arabia. Afr J Microbio Res, 2011; 5(11):1252-60.
Crossref - Asghar AH. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals. Pak J Med Sci, 2014; 30(4): 698-702.
Crossref - Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R,et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U.S.A. 1998; 95: 3140-45.
Crossref - Xiaojuan Y, Shubo Y, Qingping W, Zhang J, Wu S, Rong D, et al. Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Front Microbiol 2018; 9: 197.
Crossref - Seidl K, Leimer N, Palheiros Marques M, Furrer A, Holzmann-Burgel A, Senn G, et al. Clonality and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus at the University Hospital Zurich, Switzerland between 2012 and 2014. Ann Clin Microbiol Antimicrob 2015; 14: 14.
Crossref - Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, Hakawi A, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiology, 2012; 12(1):146.
Crossref - Cirlan M, Saad M, Coman G, Bilal NE, Elbashier AM, Kreft D,et al. International spread of major clones of methicillin resistant Staphylococcus aureus: nosocomial endemicity of multi locus sequence type 239 in Saudi Arabia and Romania. Infect Genet Evol 2005; 5:335-9.
Crossref - Alreshidi MA, Alsalamah AA, Hamat RA, Neela V, Alshrari AS, Atshan SS, et al. Genetic variation among methicillin-resistant Staphylococcus aureus isolates from cancer patients in Saudi Arabia. Eur J ClinMicrobiol Infect Dis, 2013; 32:755-61.
Crossref - Senok A, Ehricht R, Monecke S, Al-Saedan R, Somily A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect 2016; 14:13-8.
Crossref - Al-Obeid S, Haddad Q, Cherkaoui A, Schrenzel J, Francois P. First detection of an invasive Staphylococcus aureus strain (D958) with reduced susceptibility to glycopeptides in Saudi Arabia. J ClinMicrobiol 2010; 48(6):2199-04.
Crossref - Abulreesh H, Organji SR, Osman G, Elbanna K, et al. Prevalence of antibiotic resistance and virulence factors encoding genes in clinical Staphylococcus aureus isolates in Saudi Arabia. Clinical Epidemiology and Global Health 2017; 5(4).
Crossref - Moussa I, Kabli SA, Hemeg HA, Al-Garni SM, Shibl AM. A novel multiplex PCR for molecular characterization of methicillin resistant Staphylococcus aureus recovered from Jeddah, Kingdom of Saudi Arabia. Indian J Med Microbiol, 2012; 30:296-301.
Crossref - Alkharsah KR, Rehman S, Alkhamis F, Alnimr A, Diab A, Al-Ali AK. Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann Clin Microbiol Antimicrob 2018; 17(7).
Crossref - El-Mahdy TS, Al-Agamy, MH, Emara, M, et al. Complex Clonal Diversity of Staphylococcus aureus Nasal Colonization among Community Personnel, Healthcare Workers, and Clinical Students in the Eastern Province, Saudi Arabia. BioMed Research International 2018;
Crossref - Alkharsah KR, Rehman S, Alnimr A, et al. Molecular typing of MRSA isolates by spa and PFGE. Journal of King Saud University – Science 2019, 31(4), 999-100
Crossref - Holden MTG, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S,et al. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J. Bacteriol 2010; 192:888-92.
Crossref - van Hal SJ, Jones M, Gosbell IB, Paterson DL. Vancomycinhetero resistance is associated with reduced mortality in ST239 methicillin-resistant Staphylococcus aureus blood stream infections. PLoS One 2011; 6:e21217.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.