ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus is one of the major pathogen causing infections in human ranging from mild to severe life-threatening conditions. Methicillin-Resistant Staphylococcus aureus (MRSA) is an important nosocomial pathogen with high morbidity and mortality in both hospital and community settings. Total 600 nasal swabs were collected from patient visitors and Healthcare workers. Of these, 184 S.aureus (30.66%) were isolated. All S.aureus isolates screened for MRSA and 73 (39.67%) isolates showed MRSA by Cefoxitin disc diffusion method and PCR. 21 (28.76%) isolates detected pvl gene of the 73 isolated MRSA i.e., CA-MRSA. All MRSA isolates were typed into SCCmec element (I to V). Of these SCCmec type III was found more prevalent than other SCCmec types and 3 isolates were not typeable. MRSA still remains a significant problem in public Healthcare settings. Screening of MRSA among Healthcare Workers and patient visitors is mandatory to prevent the spread of CA-MRSA in hospitals.
MRSA, Patient visitors, Healthcare workers, CA-MRSA
Staphylococcus aureus is one of the major pathogens causing infections in humans ranging from mild, minor infections to severe life-threatening conditions. Staphylococcus aureus is a normal commensal bacterium that typically colonizes the skin and mucosal membrane; especially anterior nares of 20-30% of human population. Endogenous source is a major risk factor of Staphylococcal infections in carriers.1 Nasal carriage of S.aureus is associated with a high risk of infection and with pathogen transmission in health-care settings.2 Methicillin Resistant Staphylococcus aureus (MRSA) is an important nosocomial pathogen with high morbidity and mortality in both hospital and community settings.3
MRSA is induced by mecA gene which is located on Staphylococcal Cassette Chromosome mec (SCCmec). SCCmec is a large mobile genetic element, encoding a low affinity penicillin binding protein 2a (PBP2a).4 It confers resistance to entire class of Beta-lactum antibiotics except ceftaroline and ceftobiprole.5 Twelve different types of SCCmec (I to XII) have been identified till date; of these SCCmec type (I to V) are distributed worldwide.6
MRSA are divided into 2 types which include: 1. Health care associated MRSA (HA-MRSA) and 2. Community acquired MRSA (CA-MRSA). HA-MRSA is found more in hospitalized patients with invasive medical procedures etc and is normally resistant to other beta-lactum antibiotic; whereas CA-MRSA is susceptible to other beta-lactum antibiotics.7 SCCmec type I, II and III are usually distributed in HA-MRSA and SCCmec type IV and V are seen in CA-MRSA.9 The aim of the present study is to identify nasal carriage of MRSA and its associated SCCmec types among the patient visitors and Healthcare workers. This is the first study of its kind in Kolhapur city.
Exclusion Criteria
- Healthcare workers and patient visitors with any respiratory infections, skin infections up to 4 weeks before nasal sample collection.
- Subjects by treated with anti-MRSA ointments and other antibiotics in the last 14 days.
Sample collection
Anterior nasal swabs were collected from Healthcare workers (Nurses, House Keeping Workers, Resident doctors) and patient visitors (Patient’s Relatives, Friends and Care-givers of the patients) by using Hi-chrome sterile cotton swabs. Swabs were immediately inoculated into 5% salt BHIB broth, labeled properly and transported to the laboratory for further processing.
Isolation of Staphylococci aureus was done by using standard microbiological procedure.9 MRSA was screened by using MeRSA chrome agar (Hi media) and Cefoxitin (30 µg) disc diffusion method as per the CLSI guidelines 2020.10
DNA Extraction
DNA was extracted by using boiling lysis method.11
A pure culture of the isolates was obtained by inoculating 4-5 discrete colonies in BHIB and incubating at 37°C for 24 hours.
From this, pure discrete colonies were transferred into a micro-centrifuge tube containing 400μl of PCR water.
The suspension was heated at 100°C for 10 minutes for cell disruption and centrifuged at 6000 rpm for 5 minutes.
After centrifugation, tube was kept in deep freezer (-20°C) overnight.
The supernatant was used as template DNA.
PCR products were separated on a 1.5% agarose gel stained with ethidium bromide (0.5 µg/mL) along with a 100 bp DNA ladder (Hi-Media, Mumbai, India) and electrophorized gel was photographed using a gel imager (Applied Biosystem)
Detection of mecA gene
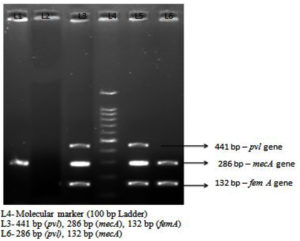
Detection of mecA gene was detected by multiplex PCR method.12 The following cycling conditions and primers (Primers purchased from Syngene Pvt. Ltd.) were used in this study [Table 1 and Table 2]
Table (1):
Primers for mecA, femA and pvl gene.
| Gene | Sequence | Size (bp) | References |
|---|---|---|---|
| mecA | F: 5’-TGCTATCCACC CTCAAACAGG-3’ R: 5’-AACGTTGTAAC CACCCCAAGA-3’ |
286 | 12 |
| femA | F: 5’ – AAAAAAGCAC ATAACAAGCG – 3’ R: 5’ – GATAAAGAAGA AACCAGCAG – 3’ |
132 | |
| pvl | F:5’–ATCATTAGGTAAAAT GTCTGGACATGATCCA–3’ R: 5’– GCATCAASTGTATT GGATAGCAAAAGC– 3’ |
441 |
Table (2):
Cycling Condition of mecA, femA and pvl gene.
Gene |
MecA /fem A/pvl |
|---|---|
Initial denaturation |
94°C 2 mins |
Denaturation |
94°C 45 secs |
Annealing |
55°C 30secs |
Extension |
72°C 45 secs |
No. of Cycles |
35 |
Final Extension |
72°C 2 mins |
Detection of SCCmec typing
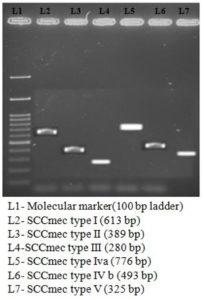
SCCmec types (I to V) were detected by multiplex PCR.13 [Table 3 and Table 4]
Table (3):
Primers for SCCmec types (I to V).
Target |
Sequence |
Size |
|---|---|---|
SCCmec type I |
5’GCTTTAAAGAGTGTCGTTACAGG 3’ 3’ GTTCTCTCATAGTATGACGTCC 5’ |
613 bp |
SCCmec type II |
5’ CGTTGAAGATGATGAAGCG 3’ 3’ CGAAATCAATGGTTAATGGACC 5’ |
389 bp |
SCCmec type III |
5’ CCATATTGTGTACGATGCG 3’ 3’ CCTTAGTTGTCGTAACAGATCG 5’ |
280 bp |
SCCmec type IVa |
5’GCCTTATTCGAAGAAACCG 3’ 3’CTACTCTTCTGAAAAGCGTCG 5’ |
776 bp |
SCCmec type IVb |
5’ TCTGGAATTACTTCAGCTGC 3’ 3’ AAACAATATTGCTCTCCCTC 5’ |
493 bp |
SCCmec type IVc |
5’ ACAATATTTGTATTATCGGAGAGC 3’ 3’ TTGGTATGAGGTATTGCTGG 5’ |
200 bp |
SCCmec type IVd |
5’CTCAAAATACGGACCCCAATACA 3’ 3’ TGCTCCAGTAATTGCTAAAG 5’ |
881 bp |
SCCmec type V |
5’ GAACATTGTTACTTAAATGAGCG 3’ 3’ TGAAAGTTGTACCCTTGACACC 5’ |
325 bp |
Table (4):
Cycling Condition for SCCmec (types Ito V).
| Steps | Temperature and Time | Cycle |
|---|---|---|
| Initial Denaturation | 94°C for 45 secs | 10 Cycles |
| Denaturation | 94°C for 45 secs | |
| Annealing | 65°C for 45 secs | |
| Extension | 72°C for 90 secs | |
| Denaturation | 94°C for 45 secs | 25 cycles |
| Annealing | 55°C for 45 secs | |
| Extension | 72°C for 2 mins | |
| Final Extension | 72°C for 10 mins | |
| Hold | 4°C |
Total of 600 nasal swabs were collected from Healthcare workers and Patient visitors. Of these, 184 S.aureus and 134 Coagulase negative Staphylococci were isolated. 400 nasal swabs were collected from patient visitors. Of these, 200 swabs were collected from in-patient visitors especially those who visited IPDs and 200 swabs were collected from outpatient visitors coming with patients. Distribution of Staphylococci is shown in Table 5
Table (5):
Distribution of S.aureus.
S.aureus |
CONS |
|
|---|---|---|
Inpatient visitors (n=200) |
51 (27.71%) |
39 (29.10%) |
Outpatient visitors (n=200) |
41 (22.28%) |
41 (30.59%) |
Health care workers (n=200) |
92 (50%) |
56 (41.79%) |
Total |
184 (30.66%) |
134 (22.33%) |
Methicillin-resistant S.aureus (MRSA)
73 (39.67%) isolates showed Methicillin-Resistant S.aureus (MRSA). Of these, Healthcare workers showed more MRSA prevalence i.e., 39 isolates (21.66%) followed by In-Patient Visitors 22 (12.22%) and Out-Patient Visitors 12 (6.66%) [Table 6]. femA gene was detected in all MRSA isolates.
Table (6):
Distribution of MRSA.
MRSA |
MSSA |
|
|---|---|---|
Out-Patient Visitors |
12 (6.66%) |
39 (21.66%) |
In-Patient Visitors |
22 (12.22%) |
19 (10.55%) |
Health Care Workers |
39 (21.66%) |
53 (30.55%) |
Total |
73 (39.67%) |
111 (60.32%) |
Panton-Valentine Leukocidin (PVL) gene was tested against the MRSA isolates. Out of 73 (39.67%) MRSA isolates, pvl gene detected 21 (28.76%) isolates. Of these, In-Patient Visitors and Healthcare workers had same prevalence rate. Eight (10.95%) and 5 (6.84%) isolates were detected from Out-patient visitors [Table 7] [Figure 1].
Table (7):
Distribution of pvl gene against MRSA isolates.
Pvl gene detected |
Pvl gene Not detected |
|
|---|---|---|
Out-Patient Visitors |
5 (6.84%) |
12 (16.43%) |
In-Patient Visitors |
8 (10.95%) |
22 (30.13%) |
Health Care Workers |
8 (10.95%) |
39 (53.42%) |
Total |
21 (28.76%) |
52 (71.23%) |
Diversity of Staphylococcal Cassette Chromosome mec (SCCmec) elements among MRSA
73 MRSA isolates were typed for SCCmec (SCCmec Type I to V) [Figure 2]. Of these, SCCmec type III was more prevalent 20 (28.76%), followed by other SCCmec types IV, II, I, V. Three isolates were not typeable. [Table 8]
Table (8):
Diversity of Staphylococcal Cassette Chromosome mec (SCCmec) elements among MRSA.
| Prevalence of SCCmec Types (n=73) | Out-patient Visitors | In-Patient Visitors | Heath care workers | Total | |||
|---|---|---|---|---|---|---|---|
| Pvl negative isolates | Pvl positive isolates | Pvl negative isolates | Pvl positive isolates | Pvl negative isolates | Pvl positive isolates | ||
| SCCmec type III | 4 | 0 | 6 | 0 | 11 | 0 | 21 (28.76%) |
| SCCmec type IVa | 0 | 2 | 0 | 3 | 0 | 3 | 8 (10.95%) |
| SCCmec type IVb | 1 | 2 | 0 | 3 | 0 | 5 | 11 (15.06%) |
| SCCmec type I | 1 | 0 | 3 | 0 | 4 | 0 | 8 (10.95%) |
| SCCmec type II | 5 | 0 | 3 | 0 | 3 | 0 | 11 (15.06%) |
| SCCmec type V | 3 | 0 | 0 | 2 | 0 | 0 | 5 (6.84%) |
| SCCmec type III+ I | 1 | 0 | 1 | 0 | 0 | 0 | 2 (2.73%) |
| SCCmec type III+ II | 0 | 0 | 0 | 0 | 2 | 0 | 2 (2.73%) |
| SCCmec type III+ IV | 0 | 1 | 0 | 1 | 0 | 0 | 2 (2.73%) |
| Non typeable | 1 | 0 | 0 | 0 | 1 | 0 | 3 (4.10%) |
| Total | 17 | 5 | 13 | 9 | 21 | 8 | 73 |
Nasal colonization of S.aureus depends on it’s ability to survive and adapt the host immune system, that may promote or inhibit its growth. In addition, other factors such as age, professional occupation, geographical location also contribute to nasal colonization of S.aureus.13 Nasal carriage of S.aureus is a global phenomenon among healthy human population but the detection of nasal carriage rate of S.aureus is different in different countries. In the present study, 73 (39.67%) MRSA were isolated in nasal colonization and it contributed to 184 (30.66%) of all S.aureus isolated from anterior nares of Healthcare workers and patient visitors. Detection of nasal carriage rate of S.aureus varies depending on the sampling methods, sampling sites and methods of analysis. Some studies reported lower incidence of nasal carriage of S.aureus among healthy individuals eg, Spain (19.1%), Norway (27%), India (27.92%), and Germany (21.9%).1 Other studies reported higher incidence, such as Ukraine (40.2%).14 In India, Karnataka (62.14%),15 Haryana (52.35%)16 and Aligarh (47.62%)17 have also reported higher prevalence.
World Health Organization (WHO) Classified S.aureus as a higher priority pathogen which is resistant to most of the antibiotics used to treat Staphylococcal infections in clinical settings.18 Overall Nasal colonization of MRSA in our study showed 73 (39.67%). WHO reported that international range of nasal carriage of MRSA is approx. 6-18% among Healthcare workers. Our study showed low prevalence of MRSA carriage rate 12 (6.66%) among Healthcare workers as compared to other studies conducted by Sharon Rainy Rongpharpi et al (11.43%), Vinodh Kumaradithyaa A et al. (15.4%).18,19 The variation of MRSA between different studies may be due to the variations in duration of exposure of the patients, personal hygiene of the Healthcare workers and infection control practices in those hospitals.
Prevalence of MRSA among the patient visitors in our study is quite higher [Table 7]. No Indian studies are available in this regard to the best of our knowledge and literature search. MRSA carriage rate among the healthy adult population shows a wide variation as seen in the other studies conducted by Nicola best et al(0.2%),20 Goud et al(16.6%)21. Patient visitors may carry CA- MRSA and transmit it to other Healthcare workers and patients. In the present study, the rate of community acquired MRSA based on the Pvl gene detection is 21 (28.76%). All the 73 MRSA isolates was screened for SCCmec types. Of these, SCCmec type III is more common in our study i.e., 21 (28.76%) followed by other SCCmec types [Table 7] and SCCmec type III is the most prevalent hospital strain in India.24 21 pvl positive MRSA isolates carried SCCmec type IV and V among these, 19 isolates (26.02%) carried SCCmec type IV and 2 (2.73%) isolates carried SCCmec type V which is common in Community acquired MRSA. Hence the rate of CA-MRSA in present study based on pvl and SCCmec type IV and V is 28.76% and HA-MRSA is 54.79% [Table 8]. pvl gene is not a reliable marker for the confirmation of CA-MRSA hence, in the present study confirmation of CA-MRSA was based on the SCCmec type IV and V. pvl gene used in this study as initial screening marker of CA-MRSA.25 Three isolates (4.10%) did not detect pvl gene but it carried SCCmec type V in Out-patient visitors. Hanane Aouati et al23 reported that HA-MRSA carried SCCmec type V of ST34 clones with TSST-1 virulence gene positive strain. This might be due to horizontal gene transfer.23
Two isolates of MRSA carried SCCmec type I + III, SCCmec type III+ II, SCCmec type III+ IV. Co-existence of SCCmec types are not common in S.aureus isolates, this requires further research to identify the identify the reason for co-existence of SCCmec types reason. Three isolates (4.10%) of MRSA are not-typeable. They may belong to other SCCmec types which are reported in other Indian studies.22
The detection of CA-MRSA in hospitals plays an important role which enables close monitoring of the Healthcare environment. This monitoring can prevent the favorable conditions required for proliferation of CA-MRSA, thus reducing the transmission of CA-MRSA in hospital environment.21
MRSA still remains a significant public health problem. Screening of MRSA among Healthcare workers and patient visitors is mandatory to prevent the spread of antibiotic resistance in hospitals. Furthermore, it can reduce the selective pressure for emergence and persistence of MRSA associated with overuse of antibiotics by improving antibiotic prescribing. This will help to improve the importance of hospital infection control practices and its strict implementation in hospitals.
ACKNOWLEDGMENTS
We acknowledge the significant contribution and support of Dean, Medical Superintendent, Administrative officer, PRO, Nursing staff from D.Y. Patil Hospital and Research Institute, Kolhapur and also laboratory technicians from Krisna Laboratory Pvt. Ltd. and D.Y. Patil Hospital and Research Institute, Kolhapur, India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RAC conceived and designed the experiments. AK performed the experiments. RAC, AK and DB analyzed the data and wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, D.Y. Patil Medical College, Kolhapur, India with reference number DYPMCK/209/2019/IEC.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Ahmadi E, Khojasteh M, Mortazavi SM, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus nasal carriage in the West of Iran: a population-based cross-sectional study. BMC Infectious Diseases. 2019;19(1):899.
Crossref - Albrich WC, Harbarth S. Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis. 2008;8(5):289-301.
Crossref - Albarrag A, Shami A, Almutairi A, Alsudairi S, Aldakeel S, Al-Amodi A. Prevalence and Molecular Genetics of Methicillin-Resistant Staphylococcus aureus Colonization in Nursing Homes in Saudi Arabia. Can J Infect Dis Med Microbiol. 2020;2020:2434350.
Crossref - Ghanbari F, Saberianpour S, Esfahani FSZ, Ghanbari N, Taraghian A, Khademi F. Staphylococcal Cassette Chromosome mec (SCCmec) Typing of Methicillin-Resistant Staphylococcus aureus Strains isolated from Community- and Hospital-Acquired Infections. Avicenna J Clin Microbiol Infect. 2017;4(2):42244.
Crossref - Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular haracterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18.
Crossref - Khairalla AS, Wasfi R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep. 2017;7(1):7390.
Crossref - DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375(9725):1557-1568.
Crossref - Amirkhiz MF, Rezaee MA, Hasani A, Aghazadeh M, Naghili B. SCCmec Typing of Methicillin-Resistant Staphylococcus aureus: An Eight Year Experience. Arch Pediatr Infect Dis. 2015;3(4):e30632.
Crossref - Tille, Patricia M., author. Bailey & Scott’s Diagnostic Microbiology. St. Louis, Missouri: Elsevier, 2014
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA : Clinical and Laboratory Standards Institute; 2020
- Dashti AA, Jadaon MM, Abdulsamad AM, Dasht HM. Heat Treatment of Bacteria: A Simple Method of DNA Extraction for Molecular Techniques. Kuwait Medical Journal. 2009;41(2):117-122.
- Abimanyu N, Krishnan A, Murugesan S, Subramanian GK, Gurumurthy S, Krsihnan P. Use of Triplex PCR for rapid detection of PVL and differentiation of MRSA from Methicillin Resistant Coagulase Negative Staphylococci. J Clin Diagn Res. 2013;7(2):215-218.
Crossref - Zhang K, McClure J-A, Elsayed S, Louis T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of Staphylococcal cassette chromosome mec types I to v IN Methicillin resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026-5033.
Crossref - Netsvyetayeva I, Fraczek M, Piskorska K, et al. Staphylococcus aureus nasal carriage in Ukraine: antibacterial resistance and virulence factor encoding genes. BMC Infect Dis. 2014;14:128.
Crossref - Mahesh CB, Ramakant BK, Jagadeesh VS. The prevalence of inducible and constitutive clindamycin resistance among the nasal isolates of staphylococci. J Clin Diagn Res. 2013;7(8):1620-1622.
Crossref - Chatterjee SS, Ray P, Aggarwal A, Das A, Sharma M. A community-based study on nasal carriage of Staphylococcus aureus. Indian J Med Res. 2009;130:742-748.
- Kumar P, Shukla I, Varshney S. Nasal screening of health care workers for nasal carriage if coagulase positive MRSA and prevalence of nasal colonization with Staphylococcus aureus. Biol Med. 2011;3:182-186.
- World Health Organization. Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline. Geneva. 2019. Licence: CC BY-NC-SA 3.0 IGO. ISBN 978-92-4- 000019-3.
- Vinodhkumaradithyaa A, Uma A, Shirivasan M, Ananthalakshmi I, Nallasivam P, Thirumalaikolundusubramanian P. Nasal carriage of methicillin-resistant Staphylococcus aureus among surgical unit staff. Jpn J Infect Dis. 2009;62(3):228-9.
- Goud R, Gupta S, Neogi U, Agarwal D, Naidu K, Chalannavar R, Subhaschandra G. Community prevalence of methicillin and vancomycin resistant Staphylococcus aureus in and around Bangalore, southern India. Rev Soc Bras Med Trop. 2011;44(3):309-12.
Crossref - Best N, Fraser JD, Rainey PB, Roberts SA, Thomas MG, Ritchie SR. Nasal carriage of Staphylococcus aureus in healthy Aucklanders. Journal of The New Zealand Medical Association. 2011;124:1332.
- Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36(2):131-139.
Crossref - Aouati H, Hadajadi L, Aouati F, et al. Emergence of Methicillin Resistant Staphylococcus aureus ST239/241 SCCmec-III merury in Eastern Algeria. Pathogens. 2021;10(11):1503.
Crossref - George K, Abdulkader JK, Sugumar M, Rajagopal GK. Prevalence of MRSA Nasal Carriage in Patients Admitted to a Tertiary Care Hospital in Southern India. J Clin Diagn Res. 2016; 10(2):DC11-DC13.
Crossref - Edslev SM, Westh H, Andersen PS, et al. Identification of a PVL-negative SCCmec-IVa sublineage of the methicillin-resistant Staphylococcus aureus CC80 lineage: understanding the clonal origin of CA-MRSA. Clin Microbiol Infect. 2018;24(3):273-278.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.