ISSN: 0973-7510
E-ISSN: 2581-690X
Multidrug resistance patterns of Acinetobacter spp. have led to their emergence as an important source of nosocomial infections. This study investigated the prevalence and clinical characteristics of Acinetobacter spp. in hospital-acquired wound and urinary tract infections. A total of 432 samples [wound swabs (210) and urine samples (222)] were analyzed for the presence of Acinetobacter spp. through selective culturing on MacConkey and Leeds Acinetobacter medium followed by identification with API 20E strips and Vitek 2 compact system. Antimicrobial susceptibility was assessed by adopting the disk diffusion method on Muller-Hinton agar, whereas the minimum inhibitory concentration procedure was carried out by using a ComASP™ Colistin test kit. Biofilm formation was examined using microtiter plates and following the crystal violet staining method. PCR was performed to amplify virulence (lasB, bap, and plcN) and antimicrobial resistance (blaOXA-51like) genes. The results revealed a low prevalence of Acinetobacter spp. (1.85 %) where Acinetobacter baumannii was the predominant species. Acinetobacter baumannii isolates harbored blaOXA-51-like gene to exert extensive or pan-drug resistance. Most Acinetobacter baumannii isolates demonstrated weaker to moderate biofilm-forming capabilities and carried the bap gene. Acinetobacter baumannii isolates lacked the combination of virulence factors encoding lasB and plcN genes. Acinetobacter baumannii infections are rising in Saudi Arabia. The results of this study highlight the epidemiology of virulent and antibiotic-resistant Acinetobacter spp., particularly A. baumannii, in Saudi Arabia. The detailed elaboration on the diversity, virulence, and antimicrobial susceptibility patterns of Acinetobacter spp. in Saudi Arabia requires further in-depth molecular investigations.
Acinetobacter baumannii, Wound Infections, Antimicrobial Resistance, Virulence Factors, Biofilm
Acinetobacter genus is characterized by gram-negative, non-fermentative, and strictly aerobic bacteria, which are categorized into 38 species. Acinetobacter lwoffii and A. baumannii are the most common human pathogens. A. haemolyticus, A. nosocomialis, and A. pitti are newly emerging human pathogens of the genus Acinetobacter.1-3 Acinetobacter species are ubiquitously found in nature with more frequent occurrence in water and soil. They are capable of surviving on dry and moist surfaces and resisting common disinfectants, which allows the growth of some Acinetobacter species in the hospital environment.1,2
Acinetobacter species are often detected in nosocomial infections, particularly in ICUs (intensive care units) where sporadic, endemic, and epidemic cases are commonly treated. A. baumannii is frequently implicated in various hospital-acquired infections such as secondary meningitis, bacteremia, wound and burn infections, UTIs (urinary tract infections), and infective endocarditis.1,2,4 The bacteria enters the bloodstream in some cases leading to the occurrence of bacteremia with significantly high mortality rates of 32% to 52%. A. baumannii-related UTIs require continuous catheterization and antibiotic therapy. It also infects soft tissues in traumatic injuries, postsurgical wounds, and skin. A. baumannii infection in burn wounds complicates its treatment therapies.2-4
Antibiotic resistance in A. baumannii enhances its clinical significance and its resistance to broad-spectrum β-lactam antibiotics, cephalosporins, quinolones, aminoglycosides, and carbapenems is steadily increasing.2,3 Therefore, the World Health Organization (WHO) has categorized A. baumannii as a “critical priority pathogen” to human health that necessitates urgent development of novel antibiotics.4
Because of the intrinsic oxacillinase (blaOXA-51-like) enzyme, which has over 40 sequence variants, Acinetobacter spp. are resistant to carbapenems. In Acinetobacter baumannii, the blaOXA-51-like gene is widely distributed and is thought to be an essential genetic marker for identifying Acinetobacter to the species level. Other types of bla genes in Acinetobacter may also include blaOXA-23, blaOXA-40, and blaOXA-58. The blaOXA-51-like may hydrolyze imipenem and meropenem (carbapenems) as well as benzylpenicillin, ampicillin, ticarcillin, and piperacillin (penicillins).1,2,4
Multiple virulence factors contribute to A. baumannii pathogenesis and associated high mortality rates, including protein secretion systems, outer membrane proteins, biofilm production, lipopolysaccharide, quorum sensing, capsule, nutrient-acquisition systems, efflux pumps, phospholipase, and elastase. These factors facilitate pathogen survival under stressed conditions and promote antimicrobial resistance.2-4
Little is still known about Acinetobacter true pathogenic potential or virulence repertory, despite a great deal of research being done on its virulence potential. OmpA, a member of the outer membrane proteins (OMPs), has been found to considerably contribute to the disease-causing capability of A. baumannii, even though it is thought that other factors may contribute to the pathogen’s virulence potential. OmpA, the pathogen’s most prevalent surface protein, is also implicated in complement resistance and biofilm development. Because A. baumannii may form biofilms, it can thrive continuously in unfavorable settings and conditions. In fact, it has been demonstrated that A. baumannii forms biofilms on abiotic surfaces, such as glass and tools used in intensive care units and/or on biological surfaces like epithelial cells. After A. baumannii connects to specific surfaces, the onset of biofilm development and maturation is facilitated by both pili assembly and the creation of biofilm-associated protein (BAP). Bacterial cell surfaces have BAP, which stabilizes the mature biofilm on biotic or abiotic surfaces, aiding in the growth and maturity of biofilms. Phospholipase D and C are two more important proteins that have been demonstrated to support the virulence of A. baumannii. Phospholipase C increases toxicity to epithelial cells, whereas phospholipase D is crucial for pathogenesis, resistance to human serum, and epithelial cell evasion.1,2,4
Antibiotic-resistant Acinetobacter baumannii infections are rising in Saudi Arabia, however, only a few studies have documented their prevalence in various cities along with antimicrobial susceptibility patterns.5-9 Thus, the elucidation of Acinetobacter baumannii epidemiology in Saudi Arabia requires further in-depth investigation. Therefore, the current study investigated Acinetobacter spp. prevalence, antibiotic susceptibility profiles, and virulence factors associated with wound and UTI infections in hospitalized patients in Makkah, western Saudi Arabia.
Sample collection
A total of 432 clinical samples [wound swabs (210) and urine samples (222)] of hospitalized patients were collected (October 2021 to January 2022) from four hospitals in Makkah, Saudi Arabia. The wound swab samples were collected using sterile swabs with Amies and charcoal transport media (Zhejiang Runlab Technology Co., Ltd., China) whereas urine samples were collected in sterile urine sample bottles. An ice box was used to transfer the samples to the laboratory without direct exposure to the sunlight. Microbiological examinations were initiated on the sampling day.
Isolation and identification of Acinetobacter spp.
MacConkey agar (Oxoid, UK) and Leeds Acinetobacter selective agar plates (HiMedia, India) were used to culture the samples (wound swabs and urine) for 24 h at 37°C.10-13 Acinetobacter spp. were identified by using API 20E strips (bioMerieux, France), and further confirmed by Vitek 2 Compact System (bioMerieux).14,15
Antimicrobial susceptibility profiles of Acinetobacter spp.
Kirby-Bauer disk diffusion method and Clinical and Laboratory Standard Institute’s (CLSI) guidelines were followed to perform antimicrobial susceptibility tests of Acinetobacter spp.16,17 The choice of antimicrobial agent tested in this study was based on the recommendations and guidelines of CLSI clinical breakpoints for Acinetobacter spp.17 Briefly, McFarland standard suspension (0.5) of each confirmed Acinetobacter spp. isolate was prepared followed by spreading onto Mueller-Hinton agar (HiMedia) plates and incubation (18-24 h) at 37°C along with antibiotic disks. Antimicrobial susceptibility test involved 14 antibiotics (Oxoid, Basingstoke, UK) belonging to 8 antimicrobial classes17 including Piperacillin (100 µg), Ampicillin-Sulbactam (10/10 µg) [penicillins], Ciprofloxacin (5 µg), Levofloxacin (5 µg) [fluoroquinolones], Ceftazidime (30 µg), Cefepime (30 µg) [cephalosporins], Gentamicin (10 µg), Tobramycin (10 µg), Amikacin (30 µg) [aminoglycosides], Imipenem (10 µg), Meropenem (10 µg) [carbapenemes], Trimethoprim-Sulfamethoxazole (1.25/23.75 µg) [folate pathway antagonists], and Tetracycline (10 µg) [tetracyclines]. Colistin [lipopeptides] susceptibility was assessed by adopting the broth dilution method and calculating minimum inhibitory concentration (MIC) according to the CLSI recommendations.17 ComASP™ Colistin test kit (Liofilchem Diagnostics, Italy) was used for the assay by following manufacturers’ guidelines.18,19 Briefly, the McFarland (0.5) standard was prepared, and sterile saline was used to dilute the suspension (1:2 (v/v)) for the preparation of solution (A). Solution B was prepared by adding solution A (0.4 ml) in a tube containing Mueller Hinton II broth. Solution B aliquot (100 µl) was dispensed into each well of a microtiter plate containing different concentrations of antibiotics. The inoculated microtiter plates were incubated (16-20 h) at 36 ± 2°C. CHROMagar ESBL (Saudi Laboratory Prepared Media – SPLM, Saudi Arabia) was used to detect extended-spectrum beta-lactamase (ESBL) Acinetobacter spp. phenotypes after the incubation of plates (24 h) at 37°C.20
Calculation of multiple antibiotic resistance index (MARI)
The number of antibiotics facing isolate resistance was divided by the total number of tested antibiotics to calculate the multiple antibiotic resistance index (MARI).21 MARI ≥ 0.2 depicts the origin of isolates from frequent antibiotic usage areas whereas MARI ≤ 0.2 presents the bacterial origin from less frequent antibiotic usage areas.22
Biofilm-forming capability of Acinetobacter spp.
A modified 96-well microtiter plate method was followed to assess the biofilm formation capability of Acinetobacter spp.23,24 Briefly, overnight nutrient broth (HiMedia) cultures were diluted in a fresh medium (108 CFU, equal inoculum level with 0.5 McFarland standard). Inoculum aliquots (20 µl) were separately transferred to the wells of a flat uncoated 96-well microtiter plate (Corning incorporated, life sciences, USA) along with fresh broth (180 µl) followed by incubation (24 h) at 37°C. Distilled water was used to wash microtiter plates thrice before staining with crystal violet solution (1%, 200 µl). The plates were again rinsed with distilled water thrice to remove unbound cells. Then, plates were dried and ethanol (95%, 150 µl) was added before measuring the absorbance (590 nm) in a microplate reader (BioTek Synergy 2 Multimode Plate Reader, USA). The assay was performed in triplicate to obtain consistent results. Biofilm formation capabilities of Acinetobacter spp. were calculated and categorized as strong (S), moderate (M), and weak (W) according to the formula mentioned in Table 1.23,24
Table (1):
Biofilm formation in Acinetobacter spp.
Biofilm formation |
Strong |
Moderate |
Weak |
|---|---|---|---|
BF= AB – CW |
0.200-0.299 |
0.100-0.199 |
<0.100 |
BF= AB ÷ CW |
4.00-5.99 |
2.00-3.99 |
<2.00 |
BF: biofilm, AB: stained attached bacteria, CW: stained control wells
Molecular detection of virulence factors-encoding genes and antimicrobial resistance in Acinetobacter spp.
AllPrep Bacteria kit (Qiagen, USA) was used to extract Acinetobacter spp. DNA. DNA quantity and quality were measured Spectrophotometrically (Denovix DS-11 Spectrophotometer, USA). The PCR amplification of genes [plcN (466 bp), blaOXA-51-like (353 bp), bap (560 bp), and lasB (300 bp)] was carried out using DreamTaq Green PCR Master Mix (2X) (Thermo Fisher Scientific, USA). Primer sequences, product sizes, and annealing temperatures of the genes are listed in Table 2.25 A Veriti Thermal Cycler (Thermo Fisher Scientific) was used for the PCR amplification, and the reaction mix (25 µl) consisted of 2X master mix (12.5 µl), forward and reverse primers (10 µM, 2.0 µl), DNA (10 ng/µl, 1.0 µl), and nuclease-free water (9.5 µl). PCR conditions were separately adjusted for each gene as blaOAX-51-like [initial denaturation (95°C, 3 min), 35 cycles of denaturation (95°C, 30 sec), annealing (55°C, 30 sec), and extension (72°C, 1 min) followed by a final extension (72°C, 7 min)], bap [initial denaturation (95°C, 3 min), 35 cycles of denaturation (95°C, 30 sec), annealing (58°C, 30 sec), and extension (72°C, 1 min) followed by a final extension (72°C, 7 min)], plcN [initial denaturation (95°C, 3 min), 35 cycles of denaturation (95°C, 30 sec), annealing (55°C, 30 sec), and extension (72°C, 1 min) followed by a final extension (72°C, 7 min)], and lasB [initial denaturation (95°C, 3 min), 35 cycles of denaturation (95°C, 30 sec), annealing (55°C, 30 sec), and extension (72°C, 1 min) followed by a final extension (72°C, 7 min)]. Agarose gel electrophoresis verified the quality and size of PCR amplification products. The gel (2%) was prepared using ultrapure Agarose (Cleaver Scientific, UK) and TBE buffer (1X) followed by the addition of SYBR-safe DNA stain (Invitrogen, USA). PCR products (4 μl) were loaded into the wells of the gel. A DNA ladder of 100-1000 bp (Thermo Fisher) served as a reference to estimate the size of PCR products. The gel was run at 100 mV for 30 minutes and DNA fragments were observed in a UV Trans-illuminator (Gel Documentation and imaging system).25
Table (2):
Primers for antimicrobial resistance and virulence factors encoding genes in Acinetobacter spp.
Gene primer |
Sequence 5′→3′ |
Product size (bp) |
Annealing temperature |
|---|---|---|---|
blaOXA-51-like |
5′-TAATGCTTTGATCGGCCTTG-3′ 5′-TGGATTGCACTTCATCTTGG-3′ |
353 |
55 °C |
bap |
5′-ATGCCTGAGATACAAATTATTGCCAAGGATAATC-3′ 5′-AGGTGCTGAAGAATCATCATCATTAC-3′ |
560 |
58°C |
plcN |
5′-GTTATCGCAACCAGCCCTAC-3′ 5′-AGGTCGAACACCTGGAACAC-3′ |
466 |
55 °C |
lasB |
5′-GGAATGAACGAAGCGTTCTC-3′ 5′-GGTCCAGTAGTAGCGGTTGG-3′ |
300 |
55 °C |
Statistical analysis
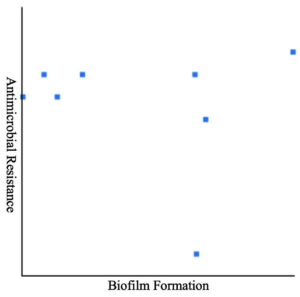
SPSS Statistics (SPSS version 21.0) was used for the statistical analysis. Pearson correlation coefficient revealed the linear association between Acinetobacter spp. biofilm forming capability and multidrug resistance.
Control strains
The controls were comprised of Acinetobacter baumannii BAA-747, a clinical reference isolate obtained from Al-Borg Medical Diagnostic Laboratories, Jeddah, Saudi Arabia, and Escherichia coli ATCC® 25922™.
A total of 432 clinical samples[wound swabs (210) and urine (222)] were examined, which revealed the presence of Acinetobacter spp. in only seven wound swabs (3.33%), and one urine sample (0.45%). The overall Acinetobacter spp. incidence was noted as 1.85% among 432 clinical samples. The bacterial growth on MacConkey plates was mostly characterized by lactose-fermenting bacteria (Klebsiella and E. coli), and a few non-lactose fermenting bacteria (Pseudomonas and/or Acinetobacter). The Leeds Acinetobacter medium differentiated Acinetobacter spp. and Pseudomonas spp., which were further confirmed by API 20E strips and the Vitek 2 compact system. API 20E strips identified all isolates as Acinetobacter spp. The vitek 2 compact system recognized six Acinetobacter baumannii isolates (AB1, AB2, AB3, AB6, AB7, and AB8) from wound swabs whereas one isolate of wound origin was identified as Acinetobacter haemolyticus (AB5). The only isolate from a urine sample was identified as Acinetobacter junii (AB4).
Antimicrobial susceptibility profiles of Acinetobacter spp.
Seven out of eight wound swab isolates (88%) exhibited resistance to three or more antimicrobial agents (Table 3). The urine isolate was susceptible to all tested antimicrobial agents except colistin. Overall, the highest resistance rates (88%, n = 8) were noted against ciprofloxacin, tetracycline, and trimethoprim/sulfamethoxazole followed by 75% (n = 8) resistance to ceftazidime, imipenem, meropenem, ampicillin/sulbactam, piperacillin, and levofloxacin (Table 3). Approximately, 63% of Acinetobacter spp. demonstrated resistance to cefepime, gentamicin, and amikacin. The lowest Acinetobacter spp. resistance (38%) was noted against colistin (Table 3).
Table (3):
Antimicrobial resistance profiles of Acinetobacter spp.
| Isolates | Sample | Clear zone diameter (mm) | MIC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | FEP | IPM | MEM | SAM | PRL | CN | SXT | AK | CIP | LEV | TOB | TE | CL† | ||
| AB6 | WS | 13 (R) | 12 (R) | 12 (R) | 10 (R) | 14 (I) | 8 (R) | 19 (S) | 0 (R) | 21 (S) | 0 (R) | 18 (S) | 20 (S) | 0 (R) | 0.5 (S) |
| AB7 | WS | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 1 (I) |
| AB5 | WS | 25 (S) | 21 (S) | 14 (R) | 0 (R) | 0 (R) | 0 (R) | 17 (S) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 22 (S) | 0 (R) | 16 (R) |
| AB3 | WS | 0 (R) | 0 (R) | 12 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 2 (I) |
| AB2 | WS | 0 (R) | 16 (I) | 27 (S) | 29 (S) | 0 (R) | 19 (I) | 0 (R) | 0 (R) | 14 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 16 (R) |
| AB1 | WS | 0 (R) | 0 (R) | 12 (R) | 0 (R) | 11 (R) | 0 (R) | 12 (R) | 0 (R) | 21 (S) | 0 (R) | 0 (R) | 20 (S) | 0 (R) | 0.5 (S) |
| AB4 | UR | 24 (S) | 27 (S) | 35 (S) | 31 (S) | 30 (S) | 29 (S) | 25 (S) | 25 (S) | 22 (S) | 27 (S) | 27 (S) | 30 (S) | 26 (S) | 16 (R) |
| AB8 | WS | 0 (R) | 0 (R) | 9 (R) | 7 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0 (R) | 0.5 (S) |
| Total resistance (%) | 6 (75) | 5 (63) | 6 (75) | 6 (75) | 6 (75) | 6 (75) | 5 (63) | 7 (88) | 5 (63) | 7 (88) | 6 (75) | 4 (50) | 7 (88) | 3 (38) | |
CAZ: Ceftazidime, FEP: Cefepime, IPM: Imipenem, MEM: Meropenem, SAM: Ampicillin-Sulbactam, PRL: Piperacillin, CN: Gentamicin, SXT: Trimethoprim-Sulfamethoxazole, AK: Amikacin, CIP: Ciprofloxacin, LEV: Levofloxacin, TOB: Tobramycin, TE: Tetracycline, CL: Colistin
†: MIC-based antimicrobial susceptibility testing against colistin using ComASP™ Colistin test kit. All other AST was carried out by the Kerby-Baur disc diffusion method and measured in millimeters (mm)
S: susceptible, I: intermediate, R: resistant
WS: Wound swabs, UR: Urine samples
Interpretations of antimicrobial susceptibility testing were based on the CLSI clinical breakpoints 17 for Acinetobacter spp.
Seven wound-swab Acinetobacter spp. isolates (88%, n = 8) exhibited multidrug-resistance (MDR) patterns (resistance to three or more antimicrobials) (Table 4). Notably, Acinetobacter baumannii (AB3) and Acinetobacter baumannii (AB7) exerted resistance against all the 14 tested antibiotics of eight different antimicrobial classes including colistin, which suggests them as pan-drug-resistant (PDR) isolates. It was followed by Acinetobacter baumannii (AB1 and AB2) and Acinetobacter haemolyticus (AB5), which were resistant to 11, 12, and 10 antibiotics of seven classes, respectively. These results categorized them as extensively drug-resistant (XDR) isolates (Table 4).
Table (4):
Correlation between antimicrobial resistance and biofilm formation in Acinetobacter spp.
| Isolate | Species | Resistant pattern | Number of classes | Biofilm formation | blaoxa-51-like | MARI |
|---|---|---|---|---|---|---|
| AB6 | A. baumannii | CAZ, FEP, IPM, MEM, SAM, PRL, SXT, CIP, TE | 6 | Weak | + | 0.43 |
| AB7 | A. baumannii | CAZ, FEP, IPM, MEM, SAM, PRL, CN, SXT, AK, CIP, LEV, Tob, TE, CL | 8 | Weak | + | 0.6 |
| AB5 | A. haemolyticus | IPM, MEM, SAM, PRL, SXT, AK, CIP, LEV, TE, CL | 7 | Weak | – | 0.5 |
| AB3 | A. baumannii | CAZ, FEP, IPM, MEM, SAM, PRL, CN, SXT, AK, CIP, LEV, Tob, TE, CL | 8 | Strong | + | 0.6 |
| AB2 | A. baumannii | CAZ, FEP, SAM, PRL, CN, SXT, AK, CIP, LEV, Tob, TE, CL | 7 | Weak | + | 0.5 |
| AB1 | A. baumannii | CAZ, FEP, IPM, MEM, SAM, PRL, CN, SXT, CIP, LEV, TE | 7 | Moderate | + | 0.5 |
| AB4 | A. junii | CL | 1 | Moderate | + | 0.072 |
| AB8 | A. baumannii | CAZ, FEP, IPM, MEM, SAM, PRL, CN, SXT, AK, CIP, LEV, Tob, TE | 7 | Weak | + | 0.5 |
| Pearson correlation | r = -0.1351 (no significance) | |||||
CAZ: Ceftazidime, FEP: Cefepime, IPM: Imipenem, MEM: Meropenem, SAM: Ampicillin-Sulbactam, PRL: Piperacillin,
CN: Gentamicin, SXT: Trimethoprim-Sulfamethoxazole, AK: Amikacin, CIP: Ciprofloxacin, LEV: Levofloxacin, Tob: Tobramycin,
TE: Tetracycline, CL: Colistin, MARI: Multiple Antibiotic Resistance Index
Pearson correlation examined the association between biofilm formation and multidrug resistance in Acinetobacter spp.
Five ESBL-positive Acinetobacter spp. (63%) isolates from wound swabs grew on CHROMagar ESBL and harbored the blaOXA-51-like gene as well. blaOXA-51-like gene-containing Acinetobacter junii of urine sample was susceptible to all the antibiotics except colistin and remained ESBL-negative on CHROMagar ESBL (Table 4). MARI of all wound swabs Acinetobacter baumannii and A. haemolyticus ranged between 0.4 and 0.6 depicting their origin from frequent antibiotic usage areas. Contrarily, 0.07 MARI of A. junii of UTI revealed their origin from areas with lesser usage of antibiotics.
Table (5):
Biofilm Formation by Acinetobacter spp.
Isolate Code |
Species |
Origin |
Strong |
Moderate |
Weak |
|---|---|---|---|---|---|
AB 1 |
Acinetobacter baumannii |
Wound |
0.162 |
||
AB 2 |
Acinetobacter baumannii |
Wound |
0.058 |
||
AB 3 |
Acinetobacter baumannii |
Wound |
0.236 |
||
AB 4 |
Acinetobacter junii |
Urine |
0.163 |
||
AB 5 |
Acinetobacter haemolyticus |
Wound |
0.032 |
||
AB 6 |
Acinetobacter baumannii |
Wound |
0.17 |
||
AB 7 |
Acinetobacter baumannii |
Wound |
0.048 |
||
AB 8 |
Acinetobacter baumannii |
Wound |
0.077 |
||
Total (%) |
1 (12.5 %) |
2 (25 %) |
5 (62.5 %) |
Biofilm-forming capability of Acinetobacter spp. and its correlation with multidrug-resistance
Biofilm formation of all eight Acinetobacter spp. was examined using microtiter plates with crystal violet staining. Table 5 depicts that a substantial number of Acinetobacter spp. (87.5 %, n = 8) demonstrated either moderate (25%, n = 8) or a weak (62.5%, n = 8) biofilm-forming capability. Pearson correlation coefficients revealed a non-significant (r = -0.1351) relationship between MDR and biofilm formation (Table 4 and Figure 1). Notably, there was no association between biofilm formation and resistance to a particular drug. Seven Acinetobacter spp. of wound swabs contained biofilm-association protein (bap)-encoding genes (Table 6).
Table (6):
Prevalence of virulence factors and antimicrobial resistance encoding genes in Acinetobacter spp.
Isolate Code |
Species |
Source |
blaOXA-51-like |
bap |
plcN |
lasB |
|---|---|---|---|---|---|---|
AB1 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
AB2 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
AB3 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
AB4 |
Acinetobacter junii |
UR |
+ |
+ |
– |
– |
AB5 |
Acinetobacter haemolyticus |
WS |
– |
– |
+ |
+ |
AB6 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
AB7 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
AB8 |
Acinetobacter baumannii |
WS |
+ |
+ |
– |
– |
WS: wound swabs, UR: Urine samples, bap: Biofilm-associated protein gene, plcN: Phospholipase C gene, lasB: Elastase gene, blaOXA 51-like: Class D β-lactamase resistance gene
Figure 1. Pearson correlation between biofilm formation in Acinetobacter and multidrug resistance, no significance was observed
Prevalence of virulence factors-encoding genes in Acinetobacter spp.
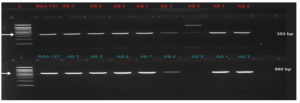
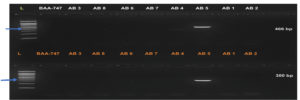
PCR amplification of virulence factors-encoding genes revealed that seven out of eight (87.5 %) Acinetobacter baumannii (wound swabs) and Acinetobacter junii (UTI) harbored biofilm-association protein (bap) and blaOXA-51-like (class D oxacillinase) genes (Table 6 and Figure 2 and 3). Elastase (lasB) and phospholipase C (plcN)-encoding genes were detected in only one isolate (12.5 %) of wound swab origin that was identified as Acinetobacter haemolyticus (AB5). This isolate did not contain blaOXA-51-like and bap genes (Table 6 and Figure 2 and 3).
Figure 2. Upper row: blaOXA-51-like gene (253 bp) in Acinetobacter spp. Lane (L) represents DNA marker (Merck KGaA, Darmstadt, Germany), Lane (BAA-747) represents positive control, and Lanes AB1-AB8 represent Acinetobacter spp. Bottom row: bap virulence gene (560 bp) in Acinetobacter spp. Lane (L) represents DNA marker (Merck KGaA, Darmstadt, Germany), Lane (BAA-747) represents positive control, and Lanes AB1-AB8 represent Acinetobacter spp.
Figure 3. Upper row: plcN gene (466 bp) in Acinetobacter spp. Lane (L) represents DNA marker (Merck KGaA, Darmstadt, Germany), Lane (BAA-747) represents positive control, and Lanes AB1-AB8 represents Acinetobacter spp. Bottom row: lasB gene (300 bp) in Acinetobacter spp. Lane (L) represents DNA marker (Merck KGaA, Darmstadt, Germany), Lane (BAA-747) represents positive control, and Lanes AB1-AB8 represents Acinetobacter spp.
Immunocompromised hospitalized patients are more prone to Acinetobacter spp. infections. This study investigated Acinetobacter spp. incidence in wound swabs and urine samples of hospitalized patients in multiple centers. Overall Acinetobacter prevalence was quite low (1.85%, n = 432) in both types of samples. Most Acinetobacter spp. isolates (87.5%, n = 8) were recovered from wound swabs whereas only one isolate was identified in urine samples. Acinetobacter baumannii was the most prevalent (75%) species in wound swabs. Acinetobacter haemolyticus was also recovered from the wound swabs whereas Acinetobacter junii was the only identified isolate in urine samples. The clinical prevalence of Acinetobacter spp., particularly A. baumannii, varies among countries, cities, and regions of the same country. For example, A. baumannii clinical isolation rates range between <1% to < 30% in Europe, whereas an average clinical isolation rate of only 0.7% has been reported in North America. Recently, an average clinical Acinetobacter baumannii isolation rate of 4.6% has been reported in the Middle East.26 However, a study investigated Acinetobacter spp. incidence in 11 hospitals in five Saudi Arabian cities (Makkah, Medina, Tabuk, Jeddah, and Riyadh) and reported its comparatively low prevalence
(3.9%, n = 124).8 Thus, the low clinical prevalence of Acinetobacter baumannii during this study is in line with the previous global and Saudi Arabian reports.8,26,27
The low incidence of Acinetobacter spp., particularly Acinetobacter baumannii, could be attributed to its opportunistic nature as it mainly infects immunocompromised patients.4 Moreover, a recent study has linked clinical Acinetobacter baumannii prevalence with the overall antibiotics applications in clinical settings.26 Pearson correlation revealed that lower antibiotic resistance rates in a particular city or region lead to lower Acinetobacter spp. clinical isolation rates in that area. However, Acinetobacter spp. prevalence rises with the overall increase in antibiotic resistance, which highlights a high risk of more Acinetobacter-linked nosocomial infections in the future.26
During this study, eight Acinetobacter spp. isolates were recovered from clinical samples, and Acinetobacter baumannii was the most prevalent species. All six Acinetobacter baumannii (75%, n = 8) isolates and one Acinetobacter haemolyticus (12.5%, n = 8) isolate were recovered from the wound swabs whereas only one Acinetobacter junii (12.5%, n = 8) isolate was detected in urine samples. Aedh et al.28 have also reported the highest prevalence of Acinetobacter baumannii (98.4%, n = 124) among samples of 12 Saudi Arabian hospitals.8 Similar findings have been reported in other parts of the world.1,27 Gupta et al.27 also reported the predominance of Acinetobacter baumannii (72%) among 111 isolates of Acinetobacter spp. Acinetobacter baumannii is known to cause 80% of total Acinetobacter-related infections.29
The predominant prevalence of Acinetobacter baumannii than other Acinetobacter spp. could be attributed to its virulence determinants such as biofilm formation, proteinase and gelatinase secretion to damage host tissues, phospholipase C secretion to enhance the toxicity to epithelial cells, and the presence of intrinsic class D oxacillinase.1,29,30 The presence of Acinetobacter haemolyticus and Acinetobacter junii was quite low, and both have never been reported in Saudi Arabia in wound infections (A. haemolyticus) and UTIs (A. junii). However, a similar low incidence of A. haemolyticus and A. junii-associated infections has been reported in other studies.27,30,31 The occurrence of A. haemolyticus-associated wound infection is less frequent, whereas A. junii is considered a rare pathogen of UTIs.27,31 Thus, the results of this are in accordance with the previous reports.
The overuse of clinical antibiotics in medical, agriculture, and veterinary applications contributes to the global rise in antimicrobial resistance (AMR).32 The detection of multidrug, extensively, and pan-drug-resistant Acinetobacter baumannii in nosocomial infections has significantly increased.1 During the current study, seven out of eight Acinetobacter spp. demonstrated multidrug-resistance patterns, which included six Acinetobacter baumannii isolates and one Acinetobacter haemolyticus isolate. Furthermore, two of the six Acinetobacter baumannii were resistant to all the tested antibiotics of eight antimicrobial classes, which established these isolates as pan-drug-resistant (PDR). Three Acinetobacter baumannii were resistant to 10 or more antibiotics of seven antimicrobial classes, which confirmed their extensively drug-resistant (XDR) nature. Multidrug-resistant Acinetobacter baumannii has been reported in Saudi Arabia with the rising incidence of XDR and MDR Acinetobacter baumannii isolates as well.7,28,33-35
A study in Makkah City revealed MDR patterns in 90% of the isolated Acinetobacter baumannii.36 An investigation in Jeddah reported a rise in MDR Acinetobacter baumannii (55% to 67%) and PDR Acinetobacter baumannii (20% to 33%) from 2010 to 2013.37 Similarly, an increasing trend of PDR and XDR Acinetobacter baumannii occurrence has been observed in Europe, the USA, and other regions.38-40 MDR patterns of Acinetobacter species could be attributed to their intrinsic chromosomal acquisition of genes encoding resistance to carbapenems and cephalosporins, and point mutations for resisting colistin, quinolones, aminoglycosides, and tertracyclines. Moreover, the acquisition of transposons, plasmids, and other mobile genetic elements facilitates the transfer of more resistance genes to Acinetobacter spp., particularly A. baumannii.41
Broad-spectrum beta-lactam combinations (ampicillin-sulbactam), and cephalosporins (cefepime and ceftazidime) are considered the first line of drugs for treating Acinetobacter baumannii. However, Acinetobacter baumannii presented high resistance to these antibiotics during this study, which is in line with the previous reports in Saudi Arabia7,8,28,33 and other countries.4,26,38,40 Therefore, fluoroquinolones, carbapenems, and aminoglycoside have become the first choice for MDR-Acinetobacter baumannii infection treatment.4,26 Acinetobacter spp. (Acinetobacter baumannii and A. haemolyticus) of wound swabs exhibited significant resistance (75%, n=8) to carbapenems (imipenem and meropenem) during this study. Acinetobacter baumannii isolates were found to have intrinsic blaOXA-51-like genes.42 The incidence of carbapenem-resistant Acinetobacter baumannii is increasing in Saudi Arabia and other countries. Acinetobacter baumannii also exerted higher resistance against fluoroquinolones followed by aminoglycosides and cephalosporins, which is similar to the previous reports in Saudi Arabia and other countries.4,7,8,26,28,33,38,40 Therefore, polymyxins (colistin) tetracyclines have become the second line of choice for countering MDR Acinetobacter baumannii infections.4,26 Acinetobacter baumannii isolates detected in this study were also significantly resistant to colistin, which is considered a last treatment option despite its hazards to kidneys and other side effects.1 Therefore, treatment of PDR and XDR Acinetobacter spp. infected wounds are becoming a challenging task, which necessitates the development of novel therapeutic options and the implementation of strict infection control strategies.
Multiple studies have investigated the virulence potential of Acinetobacter baumannii, but still further in-depth studies are required. Several factors might contribute to the Acinetobacter baumannii pathogenicity including biofilm synthesis, efflux pumps, protein secretion systems, and phospholipases.43 This study investigated various virulence determinates of Acinetobacter baumannii, A. haemolyticus, and A. junii such as biofilm-forming capability, bap-encoding genes, phospholipase C (plcN), and elastase (lasB). The biofilm formation was noted in all the Acinetobacter isolates. However, most isolates demonstrated weak biofilm-forming capability except one isolate of Acinetobacter baumannii from wound infections, which exhibited strong biofilm-forming efficiency. Bala et al.44 have also reported the inability or weaker biofilm-forming capability of 75 Acinetobacter baumannii isolates. Acinetobacter spp. biofilms on abiotic and biotic surfaces are associated with persistent and chronic infections, antibiotic resistance, and survivability in hospital environments.44
Biofilm formation could be crucial in the rising emergence of MDR pathogens. However, Pearson correlation (r = -0.1351) analysis during this study did not present any correlation between biofilm-forming capability and MDR Acinetobacter spp. Avila-Novoa et al.45 have also reported that there is no relationship between biofilm-forming capability and MDR development in Acinetobacter baumannii that was revealed by genotypic and phenotypic methods. Contrarily, Rao et al.46 have associated a higher tendency of biofilm formation in A. baumannii with MDR. Moreover, they noted higher antibiotic resistance patterns in biofilm-producing isolates than the isolates that do not produce biofilm. Similarly, another study has also established an association between the biofilm-forming capability of Acinetobacter baumannii and MDR profiles.47 Therefore, it is difficult to draw definitive conclusions regarding the biofilm formation and its association with MDR development in Acinetobacter spp.
The majority of Acinetobacter spp. isolates presented a weak biofilm-forming capability. However, the bap gene was amplified from these isolates, which has been linked to biofilm formation and maturation.43 However, all Acinetobacter spp. do not express the bap gene and their biofilm-forming capability does not solely rely on this gene. Outer membrane protein A (ompA) and pili assembly system (csu) are also known to contribute to biofilm formation.48 Aliramezani et al.25 have also noted that all bap gene-harboring Acinetobacter baumannii isolates did not exhibit strong biofilm-forming capabilities.
Elastase and phospholipases are well-documented Acinetobacter baumannii virulence factors.49 plcN gene-encoded Phospholipase C hydrolyzes phospholipids to spread infection in host tissues whereas lasB gene-encoded elastase degrades elastin to damage the host’s defense mechanisms and tissues.25 During this study, Acinetobacter baumannii isolates did not contain both of these genes (lasB and plcN) except Acinetobacter haemolyticus. Different studies have reported varying carriage of lasB and plcN genes in clinical isolates of Acinetobacter baumannii. Aliramezani et al.25 demonstrated the presence of the plcN gene in 20% (n = 100) Acinetobacter baumannii isolates, whereas the lasB gene was noted only in 4% of isolates in Iran. Kareem et al.50 reported the detection of the plcN gene in 23.3% (n = 30) isolates, whereas the lasB gene was found in 53.33% of Acinetobacter baumannii isolates in Iraq. The combined presence of both genes (plcN and lasB) might enhance the Acinetobacter baumannii virulence.50
This study demonstrates lower Acinetobacter baumannii incidence in nosocomial wound and UTI cases in western Saudi Arabia. Nonetheless, some isolates exhibited PDR, XDR, and MDR against tested antibiotics. However, a weak biofilm-forming capability was noted in most Acinetobacter baumannii isolates even in the presence of the bap gene. Acinetobacter baumannii mostly lacked the combined presence of the plcN and lasB genes except for one isolate. This study first time reports the incidence of a virulent and MDR Acinetobacter haemolyticus that is associated with wound infections. Acinetobacter junii was isolated from urine samples, which exhibited colistin resistance. Acinetobacter baumannii infections are increasing in Saudi Arabia. Thus, the results of this study shed more light on the epidemiology of virulent and antibiotic-resistant Acinetobacter spp., particularly A. baumannii, in Saudi Arabia. Further molecular-level studies could reveal more details about the diversity, virulence, and antimicrobial susceptibility patterns of Acinetobacter spp. in Saudi Arabia.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
HHA, HAM and LAN conceptualized the study. HHA, NAO, KE, and HAM applied methodology. HSAI-G, SA and MMSA investigated the study. HSAI-G, LAN, NAO performed data curation. HSAI-G, LAN and HAM performed formal analysis. KE and IA collected the resources. HHA and HSAI-G wrote original draft. HHA and IA wrote, reviewed and edited the manuscript. HAM, LAN and HHA performed supervision. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was reviewed and approved by the Department of Biology Postgraduate and Research Ethics Committee and also approved by the Faculty of Applied Science Postgraduate and Research Ethics Committee, approval number (342140314434/52962) on 27 October 2021. All urine and wound samples analysed in this study were anonymous, and only the gender of the sample provider was disclosed. Personal, clinical and epidemiological data related to these samples were not provided or disclosed during the study.
- Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii. Virulence. 2012;3(3):243-250.
Crossref - Asif M, Alvi IA, Rehman SU. Insight into Acinetobacter baumannii:pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist. 2018;11:1249-1260.
Crossref - Dehbanipour R, Ghalavand Z. Acinetobacter baumannii:pathogenesis, virulence factors, novel therapeutic options and mechanisms of resistance to antimicrobial agents with emphasis on tigecycline. J Clin Pharm Therapeut. 2022;47(11):1875-1884.
Crossref - Moubareck CA, Halat DH. Insights into Acinetobacter baumannii:a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics. 2020;9(3):119.
Crossref - Almaghrabi MK, Joseph MRP, Assiry MM, Hamid ME. Multidrug-resistant Acinetobacter baumannii: an emerging health threat in Asser region, Kingdom of Saudi Arabia. Can J Med Microbiol Infect Dis. 2018;9182747.
Crossref - Ibrahim ME. Prevalence of Acinetobacter baumannii in Saudi Arabia:risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob. 2019;18:1.
Crossref - Kharaba A, Abdelaziz HMA, Al-Hameed FM, et al. Acinetobacter baumannii in Saudi Arabia:the new growing threat. Saudi Crit Care J. 2019;3(1):54-57.
Crossref - Kharaba A, Algethamy H, Hussein M, et al. Incidence, outcomes, and predictors of Acinetobacter infection in Saudi Arabian critical care units. J Crit Care. 2021;66:109-116.
Crossref - Aldali JA. Acinetobacter baumannii:a multidrug-resistant pathogen, has emerged in Saudi Arabia. Saudi Med J. 2023;44(8):732-744.
Crossref - Jawad A, Hawkey PM, Heritage J, Snelling AM. Description of Leeds Acinetobacter medium, a new selective and differential medium for isolation of clinically important Acinetobacter spp., and comparison with Herellea agar and Holton’s agar. J Clin Microbiol. 1994;32(10):2353-2358.
Crossref - Alados JC, Serrano J, Garcia JA, Miranda C, Orellana G, de la Rosa M. Usefulness of Leeds Acinetobacter medium for recovery of Acinetobacter species from respiratory specimens collected in intensive care unit. Euro J Clin Microbiol Infect Dis. 1997;16(6):474-476.
Crossref - Ajao AO, Robinson G, Lee MS, et al. Comparison of culture media for detection of Acinetobacter baumannii in surveillance cultures of critically-ill patients. Euro J Clin Microbiol Infect Dis. 2011;30(11):1425-1430.
Crossref - Yusuf I, Skiebe E, Wilharm G. Evaluation of CHROMagar Acinetobacter and MacConkey media for the recovery of Acinetobacter baumannii from soil samples. Lett Appl Microbiol. 2023;76(2):ovac051.
Crossref - Shayegani M, Maupin PS, McGlynn DM. Evaluation of the API 20E system for identification of nonfermentative Gram-negative bacteria. J Clin Microbiol. 1978;7(6):539-545.
Crossref - Joyanes P, del Carmen Conejo M, Martinez-Martines L, Perea EJ. Evaluation of the vitek 2 system for the identification and susceptibility testing of three species of non-fermentative Gram-negative rods frequently isolated from clinical samples. J Clin Microbiol. 2001;39(9):3247-3253.
Crossref - Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-496.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed. CLSI supplement M100, March 2023. Clinical and Laboratory Standards Institute, USA.
- Galani I, Adamou P, Karaiskos I, Gimarellou H, Souli M. Evaluation of ComASP™ colistin (formerly SensiTest™ colistin), a commercial broth microdilution-based method to evaluate the colistin minimum inhibitory concentration for carbapenem-resistant Klebsiella pneumoniae isolate. J Glob Antimicrob Resist. 2018;15:123-126.
Crossref - Shams N, AlHiraky H, Moulana N, et al. Comparing quantitative and qualitative methods for detecting the in vitro activity of colistin against different Gram-negative bacilli. J Bacteriol Mycol. 2021;8(5):1181.
Crossref - Roberts LW, Hoi LT, Khokhar FA, et al. Genomic characterization of multidrug-resistant Escherichia coli, Klebsiella pneumoniae and, Acinetobacter baumannii in two intensive care units in Hanoi, Viet Nam:a prospective observational cohort study. Lancet Microbe. 2022;3(11):e857-e866.
Crossref - Apun K, Chong YL, Abdullah M, Micky V. Antimicrobial susceptibilities of Escherichia coli isolates from food animals and wildlife animals in Sarawak, Malaysia. Asian J Animal Vet Adv. 2008;3(6):409-416.
Crossref - Thenmozhi S, Rajeswari P, Kumar BS, Saipiryanga V, Kalpana M. Multi-drug resistance patterns of biofilm-forming Aeromonas hydrophila from urine samples. Int J Pharmaceut Sci Res. 2014;5(7):2908-2918.
- Naves P, del Prado G, Huelves L, et al. Measurements of biofilm formation by clinical isolates of Escherichia coli is a method-dependent. J Appl Microbiol. 2008;105(2):585-590.
Crossref - Arafa SH, Alshehri WA, Organji SR, et al. Antimicrobial resistance, virulence factors-encoding genes, and biofilm-forming ability of community associated uropathogenic Escherichia coli in western Saudi Arabia. Polish J Microbiol. 2022;71(3):325-339.
Crossref - Aliramezani A, Soleimani M, Fard RMN, Nojoomi F. Virulence determinants and biofilm formation of Acinetobacter baumannii isolated from hospitalized patients. Germs. 2019;9(3):148-153.
Crossref - Ma C, McClean S. Mapping global prevalence of Acinetobacter baumannii and recent vaccine development to tackle it. Vaccines. 2021;9(6):570.
Crossref - Gupta N, Gandham N, Jadhav S, Mishra RN. Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J Nat Sci Biol Med. 2015;6(1):159-162.
Crossref - Aedh AI, Al-Swedan AD, Mohammed AA, et al. Occurrence of multidrug-resistant strains of Acinetobacter spp.:an emerging threat for nosocomial-borne infection in Najran region, KSA. Trop Med Infect Dis. 2023;8(2):108.
Crossref - Camp C, Tatum OL. A review of Acinetobacter baumannii as a highly successful pathogen in times of war. Lab Med. 2010;41(11):649-657.
Crossref - Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and physiological overview of Acinetobacter infections:a century of challenges. Clin Microbiol Rev. 2017;30(1):409-447.
Crossref - Abo-Zed A, Yassin M, Phan T. Acinetobacter junii as a rare pathogen of urinary tract infection. Urol Case Rep. 2020;32:101209.
Crossref - Samreen, Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers:a potential threat to public health. J Glob Antimicrob Resist. 2021;27:101-111.
Crossref - Al-Obeid S, Jabri L, Al-Agamy M, Al-Omari A, Shibl A. Epidemiology of extensive drug resistant Acinetobacter baumannii (XDRAB) at security forces hospital (SFH) in Kingdom of Saudi Arabia (KSA). J Chemother. 2015;27(3):156-162.
Crossref - Aballah E, Ahamed F, Al-Omari AS. Antibiotic susceptibility patterns of some clinical isolates from Al-Rass General hospital. Int J Biosci. 2015;6:47-54.
Crossref - Ahmed NJ, Alahmari AK, Alshehri AM, Yusufoglu HS, Al-Tamammi FK. The resistance of Acinetobacter baumannii at different healthcare institutions in Saudi Arabia. Latin Am J Pharm. 2022;41(5):1002-1006.
- Haseeb A, Faidah HS, Bakhsh AR, et al. Antimicrobial resistance among pilgrims:a retrospective study from two hospitals in Makkah, Saudi Arabia. Int J Infect Dis. 2016;47:92-94.
Crossref - Al Mobarak MF, Matbuli RM, Meir H, et al. Antimicrobial resistance patterns among Acinetobacter baumannii isolated from King Abdulaziz hospital, Jeddah, Saudi Arabia, four-years surveillance study (2010-2013). Egypt J Med Microbiol. 2014;23(4):53-60.
Crossref - Shi J, Sun T, Cui Y, et al. Multidrug resistant and extensively drug resistant Acinetobacter baumannii hospital infection associated with high mortality:a retrospective study in the pediatric intensive care unit. BMC Infect Dis. 2020(1);20:597.
Crossref - Karakonstantis S, Gikas A, Astrinaki E, Kritsotakis EI. Excess mortality due to pandrug-resistant Acinetobacter baumannii infections in hospitalized patients. J Hospital Infect. 2020;106(3):447-453.
Crossref - Sobouti B, Mirshekar M, Fallah S, Tabaei A, Mehrabadi JF, Darbandi A. Pan drug-resistant Acinetobacter baumannii causing nosocomial infections among burnt children. Med J Islamic Rep Iran. 2020;34:24.
Crossref - Manchanda V, Sanchaita S, Singh NP. Multidrug resistant Acinetobacter. J Glob Infect Dis. 2010;2(3):291-304.
Crossref - Turton JF, Woodford N, Glover J, Yard S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carapabenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-2976.
Crossref - Zhou JX, Feng DY, Li X, Zhu JX, Wu ZB, Zhang TT. Advances in research on virulence Factors of Acinetobacter baumannii and their potential as novel therapeutic targets. J Appl Microbiol. 2023;134(2):lxac089.
Crossref - Bala M, Gupte S, Aggarwal P, Kaur M, Manhas A. Biofilm producing multidrug resistant Acinetobacter species from a tertiary care hospital:a therapeutic challenge. Int J Res Med Sci. 2016;4(7):3024-3026.
Crossref - Avila-Novoa MG, Solis-Velazquez OA, Rangel-Lopez DE, Gonzalez-Gomez JP, Guerreto-Medina PJ, Gutierrez-Lomeli M. Biofilm formation and detection of fluoroquinolone-and carbapenem-resistant genes in multidrug-resistant Acinetobacter baumannii. Can J Infect Dis Med Microbiol. 2019;e3454907.
Crossref - Rao RS, Karthika RU, Singh SP, et al. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol. 2008;26(4):333-337.
Crossref - Yang CH, Su PW, Moi SH, Chuang LY. Biofilm formation in Acinetobacter baumannii:genotype-phenotype correlation. Molecules. 2019;24(10):1849.
Crossref - Gedefie A, Demsis W, Ashagrie M, et al. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis:a review. Infect Drug Resist. 2021;14:3711-3719.
Crossref - Morris FC, Dexter C, Kostoulias X, Ikhtear Uddin Peleg AY. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol. 2019;10:1601.
Crossref - Kareem SM, Al-Kadmy IMS, Al-Kaabi MH, Aziz SN, Ahmad M. Acinetobacter baumannii virulence is enhanced by the combined presence of virulence factors gene phospholipase C (plcN) and elastase (lasB). Microb Pathogen. 2017;110:568-572.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.