ISSN: 0973-7510
E-ISSN: 2581-690X
Hospital-acquired infections (HAIs) are continuing to be a major risk in health care settings. World Health Organization (WHO) describes surgical site infections (SSIs) as one among the major health issue, causing enormous burden to both patients as well as doctors. Multidrug-resistant pathogens that cause SSIs continue to be an ongoing and increasing challenge to health care settings. The objective of the present study was to know the prevalence of extended-spectrum beta-lactamase (ESBL) producing gram-negative bacilli causing SSIs at a tertiary healthcare facility. The present cross-sectional observational study was done for a period of one year. Pus samples from clinically suspected cases of SSIs were collected and subjected to bacterial culture and sensitivity testing. From the total of 140 samples collected, a total of 138 bacterial isolates were isolated. Out of 138 isolates, 85 isolates (61.6%) were identified as gram-negative bacilli of which 33 isolates (38.8%) were identified to be ESBL phenotypes. Majority of the ESBL phenotypes were Escherichia coli (25.9%) followed by Klebsiella pneumoniae (7%), Acinetobacter species (2.4%), Pseudomonas aeruginosa (2.4%) and Proteus species (1.2%). Regular surveillance of antibiotic sensitivity pattern and screening for beta-lactamase production should be done which helps to know the trends of pathogenic bacteria causing SSI and guides in planning antibiotic therapy.
ESBL, Surgical site infection (SSI), Hospital-acquired infections (HAI), antimicrobial resistance
Hospital-acquired infections (HAI) are the infections occurring after 48 to 72 hours of hospital admission1. They account to significant part of mortality and morbidity in hospitals, and it is estimated that approximately more than 90,000 deaths have occurred following 1.7 million HAIs during 20192. Surgical site infection (SSI) is an infection that occurs after surgery in the part of the body where the surgery took place. SSI can be superficial involving only the skin or it could be more serious involving the tissues under the skin, organs, or implants3. Globally SSI is the third most common nosocomial infection after urinary tract infection and pneumonia4.
Surgical site infection was a major cause of death during 19th century with nearly 80% of the patients undergoing surgery were troubled with “hospital gangrene”. Introduction of antisepsis in the field of surgery by Sir Joseph Lister was a breakthrough in prevention of SSI and it turned out to be a boon to mankind5. In 20th century with the discovery of antibiotics, the surgical field was further reformed and made reconstructive, complicated and lifesaving surgeries possible. However, with the emergence of drug resistance in bacteria, SSI became one of the major challenges in healthcare system5,6. SSI infection is known to increase the burden, by increasing the duration of hospital stay, increasing mortality and adds to the financial burden of the patients7. Infections due to drug resistant gram-negative bacilli has narrowed the choice of antibiotics for treatment8.
Extended-spectrum beta-lactamase (ESBL) are the enzymes which inhibit the action of betalactam antibiotics by breaking the betalactam ring. Bacteria producing these enzymes possess resistant to betalactam antibiotics as well as to other class of antibiotics leading to therapeutic challenge to the surgeons9. Identification of the causative organism and its antibiotic sensitivity pattern if provided well in time can help in appropriate antibiotic therapy of SSIs. Objective of this study was to know the prevalence of ESBL producing gram negative bacilli causing SSI and to know their antibiotic sensitivity pattern.
The present cross-sectional study was done at department of Microbiology, JJM Medical College, Davanagere, Karnataka. Pus samples from clinically suspected patients with SSI were collected from various wards of hospital for a duration of one year (June 2015 to May 2016). Considering prevalence rate of surgical site infections, sample size was calculated by Cogent QC system and estimated sample size was 140. Ethical approval was obtained from Institutional ethical committee before the start of the study. (Ref. No.24/2014-15)
SSIs were considered according to the definition recommended by Centre for Disease Control (CDC) guidelines. 3 Samples were collected from patients after considering inclusion and exclusion criteria defined.
Inclusion criteria
Pus samples from clinically suspected surgical site infections.
Exclusion criteria
- Wounds which are primarily healed were excluded.
- Pus swabs from patients who have not undergone surgery.
- Wound infection formed from cases directly related to skin, subcutaneous tissue, stitch abscesses, infected sebaceous cyst, episiotomy, newborn circumcision site.
- Wounds without any clinical suspicion of surgical site infection.
Sample collection and transportation
Pus samples from the surgical wounds were collected using two sterile cotton swabs after cleaning the wound with sterile normal saline. Samples were collected preferably from depth of the wound under aseptic precaution and care was taken to avoid contamination from normal flora of skin. Samples collected were transferred immediately to the laboratory for further processing. One swab was used for Gram stain and another swab was used for bacterial culture10.
Microscopy, Culture and Sensitivity Testing
Using the first swab, smears were made on clean glass slides and Gram staining was done. Smears were screened for Gram reaction and morphology of bacteria was noted. Using the second swab, pus samples were inoculated on Blood agar, MacConkey agar and Thioglycolate broth and were incubated aerobically at 37°C for 18-24 hours. In case of no growth on plates after 24 hours, the respective Thioglycolate broth were examined for turbidity and subcultured if required11,12. The bacterial colonies obtained were further processed and identified conventionally, based on colony morphology on culture plates and standard biochemical tests. Gram negative bacilli were identified using motility test and biochemical reactions such as Indole test, Methyl red test, Voges Proskauer test, Triple sugar iron test and Citrate test. Antibiotic sensitivity testing was done on Muller Hinton agar by Kirby Bauer Disc Diffusion method and interpretation were done as per CLSI guidelines13,14. Following antibiotics were used for sensitivity testing of gram-negative bacilli: Ciprofloxacin (5µg), Gentamycin (15µg), Piperacillin-Tazobactam (100/10µg), Meropenem (10µg), Imipenem (10µg), Cefotaxime (30µg), Ceftazidime (30µg), Cefpodoxime (10µg), Ceftriaxone (30µg) and Cotrimoxazole (25µg).
Phenotypic detection of ESBL production
Phenotypic screening test- Phenotypic test for screening ESBL production was done by Disc diffusion test using Ceftazidime (30µg), Cefotaxime (30µg), Ceftriaxone (30µg), Cefpodoxime (10µg) and Aztreonam discs (50µg). A 0.5 McFarland standard suspension of the test organism was lawn cultured on Muller Hinton agar plate and above mentioned discs were placed approximately at a distance of 30 mm apart edge to edge and incubated at 37°C for 18 to 24 hrs. Zone of inhibition were measured carefully and interpretation was done according to CLSI guidelines. Accordingly, the zone diameters for the following antibiotics may indicate ESBL production: Aztreonam (AT 50µg) ≤ 27 mm; Ceftazidime (CAZ 30µg) ≤ 22 mm; Cefotaxime (CTX 30µg) ≤ 27 mm; Ceftriaxone (CTR 30µg) ≤ 25 mm and Cefpodoxime (CPD 10µg) ≤ 22 mm14.
Phenotypic confirmatory test
Those isolates positive for screening test were subjected to confirmatory test by combined disc diffusion test using Ceftazidime-30µg (CAZ), Ceftazidime clavulanic acid 30/10µg (CAC), Cefotaxime 30µg (CTX) and Cefotaxime clavulanic acid 30/10µg (CEC) discs. A 0.5 McFarland standard suspension of the test organism was prepared, and lawn cultured on Muller Hinton agar plate and above mentioned discs were placed at an approximate distance of 30 mm apart edge to edge. After which plates were incubated at 37°C for 18 to 24 hrs. Isolate showing an increase in the zone diameter of 5 mm with either antimicrobial agent tested in combination with clavulanate versus the zone diameter of the antimicrobial agent alone were considered to be phenotypic confirmatory test positive14,15.
Statistical analysis
Data was entered in Microsoft excel data sheet and was analysed using SPSS 22 version software (IBM SPSS Statistics, Somers NY, USA). Categorical data was represented in the form of frequencies and proportions.
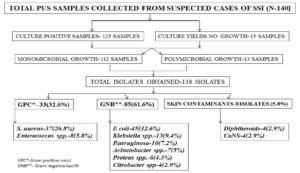
From total 140 patients, 76 (54.3%) were male patients and 64 (45.7%) were female patients with male: female ratio of 1.2:1. In the present study a total of 140 samples were collected and subjected for bacterial culture and sensitivity (Fig. 1). Out of 140 samples, 125 samples (89.3%) showed growth and 15 samples (10.7%) showed no growth. Out of 125 samples which showed growth, 13 samples yielded polymicrobial and 112 samples yielded monomicrobial growth, hence a total of 138 isolates were obtained. Out of 125 culture positive cases, 72 samples (57.6%) were collected from wounds over abdominal wall and 40 samples (32%) were collected from wounds over limbs and few samples from wounds over the groin, thorax, cheek, and neck. In our study 85 isolates (61.6%) were identified as gram-negative bacilli, 45 isolates (32.6%) were gram positive cocci, and 8 isolates (5.8%) were identified as skin contaminants. The most common bacteria causing SSI in our study is Escherichia coli 45 (32.6%) followed Staphylococcus aureus 36 (26%), Klebsiella pneumoniae 13 (9.4%) and Pseudomonas aeruginosa 10 (7.2%). Out of 85 gram negative bacilli causing SSI, 33 isolates (38.8%) were found to be ESBL producers and E. coli was the most common ESBL producer, followed by Klebsiella pneumoniae 6 (7%), Pseudomonas aeruginosa 2 (2.4%), Acinetobacter species 2 (2.4%) and Proteus species 1 (1.2%). Antibiotic-resistant pattern and distribution of ESBL producing gram negative bacilli is shown in Table 1 and Table 2, respectively.
Table (1):
Antibiotic resistant pattern of gram-negative bacilli.
| Organism Antibiotics | E. coli n=45 | Klebsiella pneumoniae n=13 | Proteus species n=6 | Citrobacter species n=4 | P aeruginosa n=10 | Acinetobacter species n=7 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | ||
| Ciprofloxacin (5µg) | 22 | 48.9 | 5 | 35.5 | 3 | 50 | 3 | 75 | 10 | 100 | 7 | 100 | |
| Gentamicin (15µg) | 15 | 33.3 | 4 | 30.8 | 1 | 16.6 | 3 | 75 | 6 | 60 | 6 | 85.7 | |
| Piperacillin-Tazobactam (100/10µg) | 8 | 17.7 | 3 | 23.1 | 2 | 33.3 | 1 | 25 | 1 | 10 | 3 | 42.9 | |
| Meropenem (10µg) | 2 | 4.4 | 1 | 7.7 | 0 | 0 | 1 | 25 | 2 | 20 | 2 | 28.6 | |
| Imipenem (10µg) | 2 | 4.4 | 1 | 7.7 | 1 | 16.6 | 1 | 25 | 2 | 20 | 1 | 14.3 | |
| Cefotaxime (30µg) | 21 | 46.6 | 3 | 23.1 | 0 | 0 | 3 | 75 | 2 | 20 | 6 | 85.7 | |
| Ceftazidime (30µg) | 23 | 51.1 | 6 | 46.2 | 2 | 33.3 | 2 | 50 | 4 | 40 | 2 | 28.6 | |
| Cefpodoxime (10µg) | 14 | 31.1 | 5 | 38.5 | 2 | 33.3 | 2 | 50 | 4 | 40 | 1 | 14.3 | |
| Ceftriaxone (30µg) | 18 | 40 | 4 | 30.8 | 1 | 16.6 | 3 | 75 | 2 | 20 | 2 | 28.6 | |
| Ceftazidime-clavulanic acid (30/10µg) | 22 | 48.8 | 5 | 38.5 | 1 | 16.6 | 1 | 25 | 4 | 40 | 2 | 28.6 | |
| Cefotaxime-clavulanic acid (30/10µg) | 21 | 46.6 | 5 | 38.5 | 1 | 16.6 | 1 | 25 | 2 | 20 | 1 | 14.3 | |
| Cotrimoxazole (25µg) | 13 | 28.8 | 7 | 53.8 | 3 | 50 | 2 | 50 | – | – | – | – | |
Table (2):
ESBL production among gram negative bacilli from surgical site infections.
| Gram Negative Bacteria | No. of isolates tested for ESBL production (n=85) | Phenotypic detection of ESBL production | ||||
|---|---|---|---|---|---|---|
| Detected by Screening test | Detected by Confirmatory test | |||||
| No. | % | No. | % | No. | % | |
| E. coli | 45 | 52.9 | 24 | 28.2 | 22 | 25.9 |
| K pneumoniae | 13 | 15.3 | 6 | 7 | 6 | 7 |
| Proteus species | 6 | 7 | 2 | 2.3 | 1 | 1.2 |
| Pseudomonas aeruginosa | 10 | 11.8 | 4 | 4.7 | 2 | 2.4 |
| Acinetobacter species | 7 | 8.2 | 2 | 2.4 | 2 | 2.4 |
| Citrobacter species | 4 | 4.7 | 0 | 0 | 0 | 0 |
| Total % of ESBL production | 38 | 44.7 | 33 | 38.8 | ||
In the present study out of 85 gram-negative bacilli, 33 (38.8%) were phenotypically identified as ESBL producers. Recent studies have shown that many healthcare setups are facing issues due to ESBL producing bacteria16,17. ESBL producing pathogens are difficult to treat and are serious public health concern. Infections due to such resistant pathogens can convert the non-fatal cases to fatal condition making it difficult to treat18,19. Genes coding for ESBLs are carried on bacterial chromosomes and can move among bacterial population either by inheritance or by plasmids20. Such drug-resistant pathogens are of great concern, as they cause fatal infections and increases the duration of hospitalization, thereby increasing the cost of treatment21. The prevalence of ESBL producing bacteria varies worldwide. Data from the Tigecycline Evaluation and Surveillance Trial (TEST) global surveillance database says that K. pneumoniae showed highest ESBL production rate which were collected in Latin America, followed by those of the Asia/Pacific rim, Europe, and North America (44%,22.4%,13.3% and 7.5%, respectively)22.
In the present study, prevalence of ESBL producing GNB causing SSI is 38.8%. E coli was the most common ESBL producer with 22 isolates (25.9%), followed by K. pneumonia 6 (7%), P aeruginosa 2 (2.4%), Acinetobacter species 2 (2.4%) and Proteus species 1 (1.2%). In a study done by Akhilesh P.S et al, E coli (36.9%) was the most common pathogen causing SSI, 11.66% of gram-negative bacilli were ESBL producers and it was also found that 71.13% of gram-negative bacilli were MDR strains (Multidrug-Resistant strains)23. In a study done by Kasukurthy L R et al gram-negative bacilli were the predominant pathogens of SSI, K pneumoniae (29%) was the most common isolate followed by E coli (22%) and prevalence of ESBL production was 44% of gram-negative bacilli were ESBL producers24. ESBL prevalence rates of 35% to 85% have been reported in different places of India, which could be due to different geographic locations, antimicrobial susceptibility pattern and different detection methods25.
Our study indicates that, tests targeting detection of resistant bacteria and strict hospital infection control programs, investigating ESBL phenotypes causing SSI should be considered with high priority.
Antibiotic resistant pattern of the gram-negative isolates is shown in Table 1. In the present study members of family Enterobacteriaceae showed significant resistance to Ciprofloxacin, third generation Cephalosporin and Cotrimoxazole. However, they were sensitive to Piperacillin-tazobactam, Imipenem and Meropenem. Among the non-fermenters (Acinetobacter species and P. aeruginosa), isolates showed significant resistance to Ciprofloxacin and Gentamycin, but were sensitive to Imipenem, Meropenem and Betalactam-betalactam inhibitor combination like Piperacillin-tazobactam, Cefotaxime-clavulanic acids. In the present study most of the gram-negative isolates causing surgical site infection were resistant to commonly used antibiotics. However, Carbapenems and Betalactam-betalactam inhibitor combinations remained sensitive which could be considered for treatment. Significant increase in the antimicrobial resistance is reported worldwide as per the third national summary of NHSN (National Healthcare Safety Network) and from a global systematic review26,27. Antibiotic resistant pattern may differ in each country, also in different region which could be due to genetic alterations and improper antibiotic usage2.
In the present study E. coli [45] isolates (32.6%) was the most common pathogen causing surgical site infection. Significant number of gram-negative bacilli were identified to be ESBL producers 33 (38.8%). These drug resistant strains can spread and cause cross infections in the hospitalized patients. Antibiotic resistance surveillance should be carried periodically to know the resistant trends and for appropriate antibiotic therapy.
Healthcare setups should protocolize strict infection control practices, follow best practices in management of SSI and educate the healthcare workers regarding rational use of antibiotics.
ACKNOWLEDGMENTS
Authors are grateful to all the patients who agreed to participate in the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
The study was approved by the institutional Ethics committee, JJM Medical College, Davangere, Karnataka, India (Ref. No.24/2014-15)
AVAILABILITY OF DATA
All the data sets generated and analyzed during this study are included in the manuscript.
- Sadeghi H, Khoei SG, Bakht M, et al. A retrospective cross-sectional survey on nosocomial bacterial infections and their antimicrobial susceptibility patterns in hospitalized patients in northwest of Iran. BMC Res Notes. 2021;14(1):88.
Crossref - Shadkam S, Goli HR, Mirzaei B, Gholami M, Ahanjan M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann Clin Microbiol Antimicrob. 2021;20(1):13.
Crossref - CDC healthcare-associated infections. 2010. Accessed: November 27, 2020. https://www.cdc.gov/hai/ssi/ssi.html.
- Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF. Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili national hospital, Tanzania. BMC Res Notes. 2014;7:500.
Crossref - Carroll E, Lewis A. Prevention of surgical site infections after brain surgery: the prehistoric period to the present. Neurosurgical Focus FOC. 2019;47(2):E2.
Crossref - Deka S, Kalita D, Mahanta P, Baruah D. High Prevalence of Antibiotic-Resistant Gram-Negative Bacteria Causing Surgical Site Infection in a Tertiary Care Hospital of Northeast India. Cureus. 2020;12(12):e12208.
Crossref - Takesue Y, Kusachi S, Mikamo H, et al. Antimicrobial susceptibility of pathogens isolated from surgical site infections in Japan: Comparison of data from nationwide surveillance studies conducted in 2010 and 2014-2015. J Infect Chemother. 2017;23(6):339-348.
Crossref - Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13(1):380.
Crossref - Oli AN, Eze DE, Gugu TH, Ezeobi I, Maduagwu UN, Ihekwereme CP. Multi-antibiotic resistant extended-spectrum beta-lactamase producing bacteria pose a challenge to the effective treatment of wound and skin infections. Pan Afr Med J. 2017;27:66.
Crossref - Murray PR, Baron E, Pfaller MA, Tenovar FC, Yolken RH: Manual of Clinical Microbiology. 7th edition. Washington, DC: American Society for Microbiology; 1999.
- Rajput RB, Telkar A, Chaudhary A, Chaudhary B. Bacteriological study of postoperative wound infections with special reference to MRSA and ESBL in a tertiary care hospital. Int J Adv Med 2019;6:1700-5.
Crossref - Collee JG, Duguid JP, Fraser AG, Marmion BP, Simmons A. Mackie and McCartney Practical Medical Microbiology. 14th edition, Churchill Livingstone Elsevier, New Delhi. 2007.
- Patrica MT, Bailey & Scott”s Diagnostic Microbiology; 13th Edition; Mosby Elsvier, China. 2014.
- Clinical and Laboratory Standards Institute. CLSI performance standards for antimicrobial susceptibility testing: 26th informational supplement. CLSI M 100-523.Clinical and Laboratory Standards Institute, Wayne, PA. 2016.
- Ranjan KP, Ranjan N, Bansal SK, Arora DR. Prevalence of Pseudomonas aeruginosa in Post-operative Wound Infection in a Referral Hospital in Haryana, India. J Lab Physicians. 2010;2(2):74-77.
Crossref - Mohammed I, Abass E. Phenotypic detection of Extended Spectrum β-Lactamases (ESBL) among gram negative uropathogens reveals highly susceptibility to imipenem. Pak J Med Sci. 2019;35(4):1104-1109.
Crossref - Tansarli GS, Athanasiou S, Falagas ME. Evaluation of antimicrobial susceptibility of Enterobacteriaceae causing urinary tract infections in Africa. Antimicrob Agents Chemother. 2013;57(8):3628-3639.
Crossref - Aruhomukama D. Review of phenotypic assays for detection of extended-spectrum β-lactamases and carbapenemases: a microbiology laboratory bench guide. Afri Health Sci. 2020;20(3):1090-1108.
Crossref - Shan S, Sajid S , Ahmad K. Detection of blaIMP Gene in Metallo-β-Lactamase Producing Isolates of Imipenem Resistant Pseudomonas aeruginosa; an Alarming Threat. J Microbiol Res. 2015;5(6):175-180.
Crossref - Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: A clinical update. Clin Microbiol Rev. 2005;18(4):657-686.
Crossref - Mirzaei B, Babaei R, Bazgir ZN, Goli HR, Keshavarzi S, Amiri E. Prevalence of Enterobacteriaceae spp. and its multidrug-resistant rates in clinical isolates: A two-center cross-sectional study. Molecular Biology Reports. 2021;48:665-675.
Crossref - Dhillon RHP, Clark J. ESBLs: A clear and present danger? Crit Care Res Pract. 2012;2012:625170.
Crossref - Tomar APS, Kushwah A, Shah H. Hospital acquired Infections due to multi-drug Resistant Organisms in a Tertiary Care Institute. International Journal of current Medical and Applied sciences. 2018;19(3):86-91.
- Kasukurthy LR, Bathala M, Bacteriological profile of Surgical Site Infections (SSIs) – a study in a tertiary care hospital. J Evid Based Med Healthc. 2020;7(32):1612 – 1616.
Crossref - Kaur J, Mahajan G, Chand K, Sheevani, Chopra S. Enhancing Phenotypic Detection Of ESBL In AmpC Co-Producers By Using Cefepime And Tazobactam. J Clin Diagn Res. 2016;10(1):Dc05-Dc08.
Crossref - Raouf M, Ghazal T, Kassem M, Agamya A, Amer A. Surveillance of surgical-site infections and antimicrobial resistance patterns in a tertiary hospital in Alexandria, Egypt. J Infect Dev Ctries. 2020;14(3):277-283.
Crossref - Weiner LM, Webb AK, Limbago B, et al. Antimicrobial resistant pathogens associated with healthcare-associated infections: Summary of data reported to the national healthcare safety network at the Centers for disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol. 2016;37(11):1288-1301.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.