ISSN: 0973-7510
E-ISSN: 2581-690X
Acinetobacter is an important nosocomial pathogen causing health care associated infections. It is highly antibiotic resistant gram-negative bacilli. The study was done to determine the prevalence of Acinetobacter species isolated from various clinical samples with their antibiotic susceptibility pattern. To determine the antimicrobial susceptibility pattern of the isolated Acinetobacter species and also the multidrug resistant mechanisms by phenotypic characterization. The retrospective study was carried out in patients diagnosed with Acinetobacter infection in the Microbiology Department, Krishna Institute of Medical Sciences, Krishna Vishwa Vidyapeeth, Deemed To Be University, Karad, a tertiary care hospital, including the clinical departments, during the period of two years from November 2020 till November 2022. Organism identification, biochemical test, antibiotic resistance pattern and phenotypic tests such as ESBL, MBL and Carbapenemase production were performed as per the Clinical and Laboratory Standards Institute (CLSI). The Modified Hodge test (MHT) was performed for Carbapenemase production detection in Acinetobacter species. A total of 150 Acinetobacter species were isolated from clinical samples. Acinetobacter baumannii was the most prevalent species 138 (92%), followed by Acinetobacter lwoffii 10 (7%) and Acinetobacter hemolyticus 2 (1%). The isolates showed highest resistance to Ampicillin 130 (87%) and sensitive to Colistin 113 (75.3%). Most of the isolates of of Acinetobacter baumannii showed maximum ESBL 21(14%) and MBL 75 (93%) production. Modified Hodge test showed positive results in Acinetobacter baumannii, only 11 (7%). The study showed that Acinetobacter baumannii was the most prevalent species showing drug resistance by phenotypic detection methods. At present Acinetobacter is a frequent pathogen in hospital acquired infections in critically ill patients admitted to ICU. The isolates of Acinetobacter species in our study showed resistant to most commonly used antibiotics. The study showed that ESBL production in Acinetobacter was 22 (15%) and MBL 80 (53%). Most Acinetobacter isolates were Multi Drug Resistant (MDR). MDR Acinetobacter is widely increasing due to inappropriate use of antibiotics in healthcare hospital. In our study, detection of carbapenemase by Modified Hodge test was positive in 11 (7%) isolates.
Acinetobacter, CLSI, ICU, ATCC, ESBL, MBL, MHT, MDR
Acinetobacter are ubiquitous, free livings saprophytes, small aerobic Gram negative cocco-bacilli that prefer moist environment and can be easily found in soil, water, food and sewage.1 They are also ubiquitous organisms in the hospital environment, where they play a significant role in the colonization and infection of patients admitted in hospitals.1 Acinetobacter causes wide spectrum of infections, including nosocomial pneumonia, secondary meningitis, surgical wound infections, skin and soft tissue infections, urinary tract infections (UTI) and bacteraemia.2 Acinetobacter spp. are generally considered a part of the normal flora of the skin,3 mucous membrane or the pharynx, and human respiratory secretions4 and are accountable for a wide variety of local and systemic infections, including pneumonia, septicaemia and wound infections.5 The main body areas populated by these microorganisms in hospitalized patients are the skin, oropharynx, and digestive tract. Acinetobacter spp. were isolated from various locations of the healthy individuals’ body including the forehead, nose, ear, throat, trachea, conjunctiva, hand, vagina and perineum, inhabiting humid areas, such as axillae, the groin and toe webs.6 Acinetobacter spp. are considered to be relatively low-grade bacteria.7 In nature, the bacteria belonging to the genus Acinetobacter, an environmental bacterium, is widely distributed and able to survive on environmental surfaces and linked to numerous nosocomial and opportunistic infections.
The sites in which Acinetobacter nosocomial infections predominantly occur are contingent on the duration and local epidemiological factors present. In the preliminary reports, it has been observed that urinary tract infections (UTIs) are prevalent in intensive care units. However, it is worth nothing that the incidence of UTIs has witnessed a decline, owing to the improved care of urinary catheters. On the contrary, there has been a considerable increase in the occurrence of nosocomial pneumonia.8
Of the Acinetobacter spp, A. baumannii is an important pathogen with a high morbidity and mortality, especially in the critically ill patients.9 Acinetobacter spp. is associated with a wide variety of infections – ventilator-associated pneumonia (VAP), blood stream infections (BIs), urinary tract infections, bacteraemia, meningitis, skin and wound diseases, ventriculitis, cholangitis, peritonitis, and infective endocarditis. Colonization of the skin and respiratory tract by the bacteria may occur without causing an infection. The survival of A. baumannii in harsh environmental conditions and its ability to develop multidrug resistance attributes to making infections caused this organism highly lethal, mainly in patients who have undergone major surgeries, the immunocompromised, those with malignancy, prolonged illness and in the extremes of age.9 Antimicrobials are chemical compounds either bactericidal or bacteriostatic, used in medical interventions to actively kill or inhibit the pathogens.
The emergence of antimicrobial-resistant Acinetobacter species is due both to the selective pressure exerted by the use of broad-spectrum antimicrobials and transmission of strains among patients, although the relative contributions of these mechanisms are not yet known.10
Infections caused by antibiotic-susceptible Acinetobacter isolates have usually been treated with broad-spectrum cephalosporins, β-lactam–β-lactamase inhibitor combinations (e.g., a combination that includes sulbactam, a drug marketed only in combination, in the United States), or carbapenems (e.g., imipenem or meropenem), used alone or in combination with an aminoglycoside.11 The primary goal for the control of Acinetobacter infection is recognizing its presence in a hospital or long-term care facility at an early stage, controlling its spread aggressively and preventing the establishment of endemic strains. Control measures are based almost entirely on experiences from outbreaks of Acinetobacter infection and generally address the organism’s major epidemic modes of transmission and the excessive use of broad-spectrum antibiotics.
Prevalence of Extended-spectrum beta-lactamase (ESBL), Metallo beta-lactamase (MBL) and Carbapenemase producing Acinetobacter species isolated from various clinical sample in tertiary care hospital’’ study was carried out in the Microbiology department at Krishna Institute of Medical Sciences and Krishna Hospital and Medical Research Centre, Karad over a 2 years period from November 2020 to November 2022.
Inclusion criteria
Isolates of Acinetobacter species, from clinical samples of patients admitted with clinical infection to the IPD/OPD. Patients with both sexes involved.
Exclusion criteria
To avoid duplication, isolates from the same patients and specimens were excluded.
Statistical analysis
Data were filled in the MS Excel Software. Then, analyzed results were expressed as percentage and p values, by Chi square test using Graph Pad Instant software. If the probability is less than 0.05, the association or difference is said to be significant.
Sample collection
The various clinical samples from which Acinetobacter species were isolated includes -Pus, Sputum, Urine, Blood, ETT secretions, Body fluids, Wound swab, CSF and others, from all age groups and both gender of patients. Appropriate sterile containers were used for collection of the samples and then transported to the laboratory.
Bacterial identification
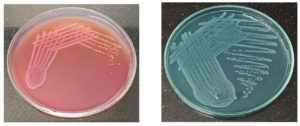
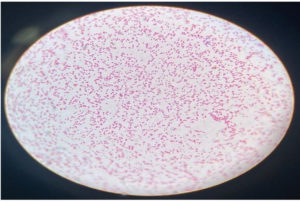
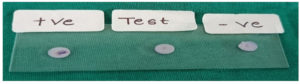
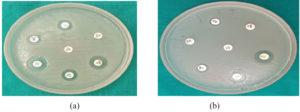
The clinical samples were cultured on appropriate culture media and the organisms isolated were identified using standard Microbiology procedures.12 Nutrient agar, MacConkey agar, (Figure 1a and 1b), Blood agar, Chocolate agar were used for inoculation of the clinical samples and incubated at 37°C for 24 hours. The isolates were then identified based on colony morphology on agar and Gram stain of the smear of colonies (Figure 2). Oxidase, Catalase reactions were performed (Figure 3). Further Biochemical reactions carried out for identification of the organisms (Figure 4a, 4b and Figure 5).12
Figure 3. Biochemical tests from left to right
TSI, Indole, MR, Nitrate reduction, Citrate, Urease, VP
Antibiotic susceptibility testing
Antimicrobial susceptibility testing by Disc diffusion method of Kirby-Bauer’s was carried out on Muller Hinton agar. The opacity adjusted to 0.5 McFarland standard. The antibiotic discs used were Amikacin (30 µg), Ciprofloxacin (5 µg), Cefepime (30 µg), Piperacillin (100 µg), Imipenem (10 µg), Meropenem (10 µg), Gentamicin (5 µg), Levofloxacin (5 µg), Tigecycline (15 µg), Colistin (10 µg), Co-trimoxazole (25 µg), Nalidixic acid (30 µg), Ampicillin (10 µg), Ceftazidime (30 µg) (Figure 6a and 6b). CLSI guidelines were used for the interpretation of the Zone diameter.13,14
Extended Spectrum of Beta Lactamase (ESBL) production testing
Double disc synergy test
Double disc synergy test (DDST) was performed for testing ESBL production, using Ceftazidime + Clavulanic acid along with Ceftazidime (Cephalosporin). Test and control organism inoculum was prepared and matched with 0.5 McFarland turbidity standard. The bacterial strains cultured on Mueller Hinton agar plates, as per CLSI guidelines. A disc containing Ceftazidime + Clavulanic acid (30 µg +10 µg), applied on the plate at a distance of 25mm from that of Ceftazidime (30 µg) and incubated for 18-24 hours. An increase ≥ 5 mm in the inhibition diameter of the ceftazidime disc applied after pre-diffusion of Ceftazidime-Clavulanic acid in comparison with ceftazidime disc, considered as positive for ESBL production (Figure 7a).15
Positive control
Klebsiella pneumoniae ATCC 700603.
Negative control
Escherichia coli ATCC 25922.
Metallo-β-lactamase detection (MBL) testing
Imipenem-EDTA combined disc diffusion test: The screening test for the detection and confirmation was tested by Imipenem-EDTA combined disc diffusion test. Test and control organism inoculum was prepared and matched with 0.5 McFarland turbidity standard. The bacterial strains cultured on Mueller Hinton agar plates, as per CLSI guidelines. Imipenem 10 µg disc, placed 20 mm apart, center from an Imipenem + EDTA disc containing 0.5 µl of 0.5 mg EDTA (750 µg per disk). Plates incubated at 37°C for 16-18 hours. On overnight incubation, an increase in zone size ≥ 7 mm around the Imipenem-EDTA disc compared to the Imipenem disc alone was recorded positive for MBL (Figure 7b).16
Control strains
Pseudomonas aeruginosa ATCC 27853.
Carbapenemase production detection testing
Modified Hodge Test (MHT)
Meropenem 10 µg disc placed in the center of the Muller Hinton agar plate and 10 µl of 50 mg Zinc sulfate solution added to the Meropenem disc and incubated at 37°C overnight. Zone around Meropenem disc with clover leaf-like indentation, was interpreted as positive for Carbapenemase detection by the Modified Hodge Test (Figure 7c).17
Figure 7. (a) ESBL production (DDST); (b) MBL production (CDDT); (c): Carbapenemase production (MHT)
Meropenem 10 µg disc placed in the center of Muller Hinton agar plate and 10 µl of 50 mg Zinc sulfate solution added to the Meropenem disc and the plates incubated at 37°C overnight. Zone around the Meropenem disc with clover leaf-like indentation, was interpreted as positive for Carbapenemase production by the Modified Hodge Test.17
Positive control
Klebsiella pneumoniae ATCC 1705.
Negative control
Klebsiella pneumoniae ATCC 17051706.
Table 1 shows age and gender wise distribution of Acinetobacter isolated. The isolates in the age group of 0-20 years were 14(9%), followed by age group of 21-40 years 46(31%), 41-60 years 57(38%), >60 years 33(22%) respectively.
In the present study, a total of 150 Acinetobacter was identified from 450 non-lactose fermenting bacteria. Our study is comparable with other studies,18 wherein maximum males were affected than females as given in the Table 1. The most commonly isolated species was Acinetobacter baumannii, followed by Acinetobacter lwoffii and Acinetobacter hemolyticus (Table 2). This finding can be corelated with the other study,19 wherein they have been reported maximum number of isolates from pus, followed by blood, endotracheal aspirate, urine, sputum, BAL (Bronchoalveolar lavage), swab (gluteal abscess), throat swab, CVP tip (Table 3). Similarly, other study20 reported that majority of isolates were from pus sample (Table 4). The other study21 has reported a similar observation of maximum isolates, from ICU (Figure 8 and 9). Other study22 reported maximum isolates from ICU.
Table (1):
Age and gender wise distribution of Acinetobacter
Age group |
Male n (%) |
Female n (%) |
Total n |
Percentage % |
|---|---|---|---|---|
0-20 |
8 (5) |
6 (4) |
14 |
9 |
21-40 |
28 (19) |
18 (12) |
46 |
31 |
41-60 |
42 (28) |
15 (10) |
57 |
38 |
˃60 |
22 (15) |
11 (7) |
33 |
22 |
Total (n) |
100 (67) |
50 (33) |
150 |
100 |
x2 = 0.5993, p value = 0.8966, Not significant
Table (2):
Speciation of Acinetobacter isolates
Species |
No. of Isolates |
Percentage (%) |
|---|---|---|
Acinetobacter baumannii |
138 |
92 |
Acinetobacter lwoffii |
10 |
7 |
Acinetobacter hemolyticus |
2 |
1 |
Total |
150 |
100 |
Table (3):
Acinetobacter species distribution in various IPD departments
| IPD clinical samples | Acinetobacter species | Total | ||
|---|---|---|---|---|
| Acinetobacter baumannii | Acinetobacter lwoffii | Acinetobacter Hemolyticus | ||
| Tracheal aspirates | 38 (27.3%) | 0 | 0 | 38 (27.3%) |
| Pus | 33 (23.7%) | 0 | 0 | 33 (25.3%) |
| Urine | 26 (18.7%) | 2 (1.4%) | 0 | 28 (19%) |
| Sputum | 12 (8.6%) | 1 (0.7%) | 0 | 13 (11%) |
| Body Fluids | 10 (7.1%) | 0 | 0 | 10 (8.7%) |
| Blood | 09 (6.4%) | 06 (4.3%) | 01 (0.7%) | 16 (11%) |
| CSF | 01 (0.7%) | 0 | 0 | 01(0.7%) |
x2= 46.436, p value = < 0.0001, significant.
Distribution of Acinetobacter species in various IPD departments.
Maximum isolates were from tracheal aspirates 38(27.3%), followed by pus 33(25.3%), urine 28(19%), sputum 13(11%), body fluids 10(8.7%), blood 16(11%), CSF 1(0.7%)
Table (4):
Acinetobacter species distribution in various OPD departments
| OPD clinical samples | Acinetobacter species | Total | ||
|---|---|---|---|---|
| Acinetobacter baumannii | Acinetobacter lwoffii | Acinetobacter Hemolyticus | ||
| Pus | 6 (54.5%) | 1 (09%) | 0 | 7 (64%) |
| Sputum | 2 (18.1%) | 0 | 1 (09%) | 3 (27%) |
| Urine | 1 (09%) | 0 | 0 | 1 (09%) |
x2= 3.376, p value = 0.4970, Not significant.
Of the Acinetobacter species isolated from various OPD departments, maximum isolates were from pus 7 (25.3%) followed by sputum 3 (11%), urine 1(19%)
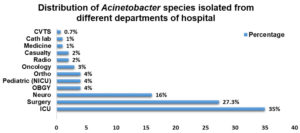
Figure 8. Distribution of Acinetobacter species isolated from different departments of hospital
Maximum isolates were from ICU 52(35%), followed by Surgery 41 (27.3%), Neuro 24 (16%), OBGY 6 (4%), Pediatric (NICU) 6 (4%), Ortho 6 (4%), Oncology 4 (3%), Radio 3 (2%), Casualty 3 (2%), Medicine 2 (1%), Cath lab 2 (1%), CVTS 1 (0.7%)
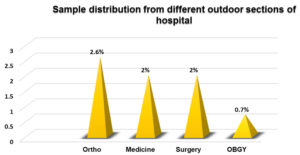
Figure 9. Sample distribution from different outdoor sections of hospital
Sample distribution from different outdoor sections of hospital. Maximum isolates were from Ortho 4 (2.6%), followed by Medicine 3 (2%), Surgery 3 (2%), OBGY 1 (0.7%).
The most commonly isolated species was Acinetobacter baumannii, followed by Acinetobacter lwoffii and Acinetobacter hemolyticus (Figure 3).
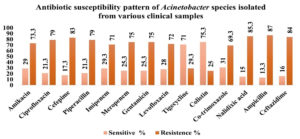
Similar findings have been observed from the other study23 reporting maximum isolate from Acinetobacter baumannii (Figure 10). In our study, we observed that the antibiotic sensitivity pattern showed maximum sensitivity to Colistin and Tigecycline (Table 5). Similarly, a study20 showed that most of Acinetobacter, were Colistin sensitive, and other study24 reported maximum susceptibility of Acinetobacter to Colistin. Acinetobacter showed extremely high degree of resistant to Ampicillin, Nalidixic acid, Ceftazidime, Cefepime, Ciprofloxacin, Piperacillin, Gentamicin, Amikacin, Meropenem, Imipenem correlating with the other studies by Guckan and Peymani et al.25,26 reported resistance to Ceftriaxone, Ceftazidime, Piperacillin + Tazobactam, Cefepime, Gentamicin, Ciprofloxacin, Ticarcillin + Clavulanic acid, Amikacin, Meropenem, Imipenem. It thus proves that extensive use of carbapenems has created a selective antibiotics pressure resulting in increased prevalence of carbapenems resistant Acinetobacter (CRA).27
Table (5):
Antibiotic susceptibility pattern of Acinetobacter species isolated from various clinical samples
| Antibiotic | Sensitive | Resistant | ||
|---|---|---|---|---|
| No. of isolates | Percentage % | No. of isolates | Percentage % | |
| Amikacin | 43 | 29 | 107 | 73.3 |
| Ciprofloxacin | 32 | 21.3 | 118 | 79 |
| Cefepime | 26 | 17.3 | 124 | 83 |
| Piperacillin | 32 | 21.3 | 118 | 79 |
| Imipenem | 44 | 29.3 | 106 | 71 |
| Meropenem | 38 | 25.3 | 112 | 75 |
| Gentamicin | 38 | 25.3 | 112 | 75 |
| Levofloxacin | 42 | 28 | 108 | 72 |
| Tigecycline | 106 | 71 | 44 | 29.3 |
| Colistin | 113 | 75.3 | 37 | 25 |
| Co-trimoxazole | 46 | 31 | 104 | 69.3 |
| Nalidixic acid | 22 | 15 | 128 | 85.3 |
| Ampicillin | 20 | 13.3 | 130 | 87 |
| Ceftazidime | 24 | 16 | 126 | 84 |
x2 =342.55, p value = < 0.0001, significant
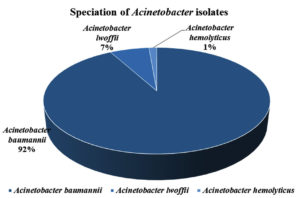
Figure 10. Speciation of Acinetobacter isolates
The most common species isolated was Acinetobacter baumannii 138 (92%), followed by Acinetobacter lwoffii 10 (7%) and Acinetobacter hemolyticus 2 (1%) respectively
Our study recorded that, the resistance towards Imipenem and Meropenem (Figure 4 and Figure 11). Other study23 reported high carbapenem resistance; resistant to Imipenem and Meropenem. The findings of our study showed ESBL production, comparable to the other study28 documenting same ESBL production (Table 6 and Figure 12).
Table (6):
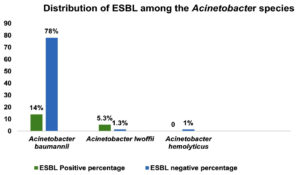
Distribution of ESBL among the Acinetobacter species
| Species | ESBL | Total | |
|---|---|---|---|
| Positive | Negative | ||
| A. baumannii | 21 (14%) | 117 (78%) | 138 (92%) |
| A. lwoffii | 8 (5.3%) | 2 (1.3%) | 10 (7%) |
| A. hemolyticus | 0 (0%) | 2 (1%) | 2 (1%) |
x2=25.578, p value = < 0.0001, significant
Figure 11. Antibiotic susceptibility pattern of Acinetobacter species isolated from various clinical samples
The different resistance pattern of bacterial isolates was observed against antimicrobial agents. Maximum sensitivity to Colistin 113(75.3%) was showed by Acinetobacter baumannii, followed by Tigecycline 106(71%), whereas, maximum resistance was to Ampicillin 130(87%), followed by Nalidixic acid 128(85.3%), Ceftazidime 126(84%)
Figure 12. Distribution of ESBL among the Acinetobacter species
Distribution of ESBL among the Acinetobacter species. Maximum ESBL production was observed in Acinetobacter baumannii 21(14%), followed by Acinetobacter lwoffii 8(5.3%). Non-ESBL production were observed in Acinetobacter baumannii 117(78%), Acinetobacter lwoffii 2(1.3%) and Acinetobacter hemolyticus 2(1%)
In the present study, MBL production was similar to the findings by other study29 documenting MBL production (Table 7 and Figure 13).
Table (7):
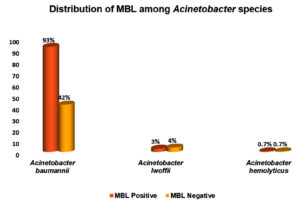
Distribution of MBL among the Acinetobacter species
| Species | MBL | Total | |
|---|---|---|---|
| Positive | Negative | ||
| A. baumannii | 75 (93%) | 63 (42%) | 138 (92%) |
| A. lwoffii | 4 (3%) | 6 (4%) | 10 (7%) |
| A. hemolyticus | 1 (0.7%) | 1 (0.7%) | 2 (1.3%) |
x2=0.7803, p value = 0.6770, Not significant
Figure 13. Distribution of MBL among the Acinetobacter species
Out of 150 isolates, maximum MBL positive isolates were observed in Acinetobacter baumannii 75(93%), followed by Acinetobacter lwoffii 4(3%), Acinetobacter hemolyticus 1(0.7%). MBL negative isolates were observed in Acinetobacter baumannii 64(43%), followed by Acinetobacter lwoffii 6(4%)
The present study showed Carbapenemase production in Acinetobacter baumannii isolates, by Modified Hodge Test (Table 8 and Figure 14). As compared to the other study,30 present study exhibited less positive percentage for Carbapenemase detection.
Table (8):
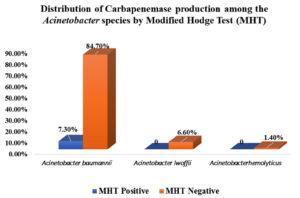
Distribution of Carbapenemase production among the Acinetobacter species by Modified Hodge Test (MHT)
| Species | MHT | |
|---|---|---|
| Positive | Negative | |
| A. baumannii | 11 (7.3%) | 127 (84.7%) |
| A. lwoffii | 0 | 10 (6.6%) |
| A. hemolyticus | 0 | 2 (1.4%) |
Figure 14. Distribution of Carbapenemase production among the Acinetobacter species by Modified Hodge Test (MHT). Modified Hodge test showed positive results in Acinetobacter baumannii only 11 (7%) of Acinetobacter baumannii were positive and 139 (93127 (84.67%)) were negative by Modified Hodge test.
Acinetobacter lwoffii and Acinetobacter hemolyticus, were negative for Carbapenemase production by Modified Hodge test
At present, Acinetobacter is common threat in health care associated infections particularly in critically ill ICU patients. Maximum percentage of Acinetobacter isolates were from pus sample followed by tracheal aspirates. Acinetobacter baumannii was the most common bacterial isolate among the Acinetobacter species. Our study showed that Acinetobacter species were resistant to most of the commonly used antibiotics such as Ampicillin, Nalidixic acid and Ceftazidime.
It thus proves that extensive use of carbapenems has created a selective antibiotics pressure resulting in increased prevalence of carbapenems resistant Acinetobacter (CRA). Acinetobacter, reported as MDR, showed susceptibility to Colistin and Tigecycline, which remains the drug of choice in the treatment for patients. All isolates were sensitive to Colistin 113 (75.3%) and resistant towards Ampicillin 130 (87%). Ceftazidime has been proposed as the indicator of ESBL production as compared to other antibiotics. The remaining isolates showed resistance to Ceftazidime.
Thus, Ceftazidime is a superior indicator for the detection of ESBL production. Imipenem has been suggested as the indicator of MBL production in comparison to others. So, Imipenem is a better indicator for the detection of MBL production. The study showed that ESBL production in Acinetobacter is 22(15%) and MBL 80(53%) and is on the ascent world over, in this way making these infections challenging to treat. ESBL and MBL production detection would be important for decreasing death rate and spread of multidrug resistant organisms. Acinetobacter species isolates were resistant to carbapenems such as imipenem 106(71%) and meropenem 112(75%). Modified Hodge test is a simple and easy test to be performed to identify carbapenems producing organisms. In our study, detection of Modified Hodge test by carbapenemase was positive in 11(7%) isolates.
ACKNOWLEDGMENTS
The authors are thankful to the Department of Microbiology, Krishna Institute of Medical Sciences, “Deemed to be University”, Karad, for providing the necessary research facilities to carry out the work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
The study was approved by the Institutional Ethical and Research Committee, Krishna Institute of Medical Sciences, Deemed University Karad, for conducting the present research, with protocol number (054/2021-22).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Lahiri KK, Mani NS, Purai SS. Acinetobacter spp as nosocomial pathogen: Clinical significance and antimicrobial sensitivity. Med J Armed Forces India. 2004;60(1):7-10.
Crossref - Ferdous J, Murshed M, Shahnaz S, Duza SS, Siddique PR. Isolation of Acinetobacter species and their antimicrobial resistance pattern in a tertiary care hospital in Dhaka, Bangladesh. Bangladesh J Med Microbiol. 2016;10(1):18-21.
Crossref - Tomaras AP, Dorsey CW, McQueary C, Actis LA. Molecular basis of Acinetobacter virulence and pathogenicity. Acinetobacter Molecular Biology. Norfolk, UK: Caistr Academic Pres. 2008:265-97.
- Munoz-Prise LS, Weinstein RA. Acinetobacter Infection. N Engl J Med. 2008;358(12):1271-1281.
Crossref - Beggs CB, Kerr KG, Snelling AM, Sleigh PA. Acinetobacter spp. and the clinical environment. Indoor and Built Environment. 2006;15(1):19-24.
Crossref - Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997;35(11):2819-2825.
Crossref - Pedersen MM, Marso E, Picket MJ. Non fermentative bacilli associated with man III pathogenecity and antibiotic susceptibility. Am J Clin Pathol. 1970;54(2):178-192.
Crossref - Bergogne-Berezin E. Guidelines on antimicrobial chemotherapy for prevention and treatment of infections in the intensive care unit. J Chemother. 2001;13(sup4):134-149.
Crossref - Doughari HJ, Ndakidemi PA, Human IS, Benade S. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ. 2011;26(2):101- 112.
Crossref - Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46(8):1254-1263.
Crossref - Lesho E, Wortmann G, Moran K, Craft D. Fatal Acinetobacter baumannii infection with discordant carbapenem susceptibility. Clin Infect Dis. 2005;41(5):758-759.
Crossref - Gupta S, Ramya TS. Acinetobacter Genus: Pathogenic and Non-pathogenic species. International Journal of Emerging Technologies and Innovative Research. 2021;8(5):a36-a48. http://www.jetir.org/papers/JETIR2105134.pdf.
- Bouvet PJM, Grimont PAD. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsoniisp nov., and Acinetobacter juniisp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Syst Bacteriol. 1986;36(2):228-240.
Crossref - Negi V, Pal S, Juyal D, Sharma MK. Bacteriological Profile of Surgical Site Infections and Their Antibiogram: A study From Resource Constrained Rural Setting of Uttarakhand State, India. J Clin Diagn Res. 2015;9(10):DC17-20.
Crossref - Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC. Diagnostic microbiology. The nonfermentative gram-negative bacilli. Philedelphia: Lippincott-Raven Publishers. 1997:253-320.
- Powell EA, Haslam D, Mortensen JE. Performance of the check-points check-MDR CT103XL assay utilizing the CDC/FDA antimicrobial resistance isolate bank. Diagn Microbiol Infect Dis. 2017;88(3):219-221.
Crossref - John S, Balagurunathan R. Metallo beta lactamase producing Pseudomonas aeruginosa and Acinetobacter baumannii. Indian Journal of Medical Microbiology. 2011 Jul 1;29(3):302-4.
- Lal A, Prasad R, Kiran D. Bacterio-Etiologic Profile of Surgical Site Infections: An Observational Study. European Journal of Molecular & Clinical Medicine. 2021 Jan 28;7(11):4770-4.
- Apoorva B, Mohan S, Sarwat T, Kakru DK. Antibiogram of Acinetobacter spp. isolates in a tertiary care hospital: a lead towards antibiotic stewardship. Acta Sci Microbiol. 2020;3(5):71-76.
Crossref - Islahi S, Ahmad F, Khare V, et al. Prevalence and Resistance Pattern of Acinetobacter Species in Hospitalized Patients in a Tertiary Care Centre. J Evol Med Dent Sci. 2014;3(17):4629-4635.
Crossref - Barai L, Fatema K, Haq JA, Faruq MO, Ahsan AA, Morshed MA, Hossain MB. Bacterial profile and their antimicrobial resistance pattern in an intensive care unit of a tertiary care hospital of Dhaka. Ibrahim Medical College Journal. 2010;4(2):66-9.
- Prashanth K, Badrinath S. Nosocomial infections due to Acinetobacter species: Clinical findings, Risk and Prognostic factors. Indian J Med Microbiol. 2006;24(1):39-44.
Crossref - Panjwani DM, Lakhani SJ, Lakhani JD, Khara R, Vasava S. Bacteriological profile and antimicrobial resistance pattern of Acinetobacter species isolated from patients of tertiary care hospital of Gujarat. Int Arch Integr Med. 2016;3(7):203-210.
- Lopez-Hernandez S, Alarcon T, Lopez-Brea M. Biochemical characterization of chromosomal cephalosporinases from isolates belonging to the Acinetobacter baumannii complex. Clin Microbiol Infect. 2002;7(4):218-226.
Crossref - Guckan R, Kolinc C, Demir ADB, Capraz A, Yanik K. Antimicrobial susceptibility of Acinetobacter baumannii complex isolated from different clinical samples in a tertiary care hospital. Journal of Antibiotics Research. 2015;1(1):103.
- Peymani A, Nahaei MR, Farajnia S, et al. High prevalence of metallo-betalactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn J Infect Dis. 2011;64(1):69-71.
Crossref - Jabeen F, Khan Z, Sohail M, Tahir A, Tipu I, Saleem HGM. Antibiotic resistance pattern of Acinetobacter baumannii isolated from bacteremia patients in Pakistan. J Ayub Med Coll Abbottabad. 2022;34(1):95- 100.
- Owlia P, Azimi L, Gholami A, Asghari B, Lari AR. ESBL- and MBL-mediated resistance in Acinetobacter baumannii: a global threat to burn patients. Infez Med. 2012;20(3):182-187.
- Talukdar A, Hodiwala AB, Sharma R. A microbiology study of Acinetobacter baumannii With Special reference to multi – drug resistance. Int J Curr Microbial App Sci. 2018;7(2):1176-1186.
Crossref - Thiriveni J, Umadevi U, Lakshmipriya N. Phenotypic and Molecular Characterization of Carbapenamase among Acinetobacter Isolates in a Tertiary Care Hospital, Tamil Nadu, India. Int J Curr Microbiol App Sci. 2018;7(12):2762-2771.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.