ISSN: 0973-7510
E-ISSN: 2581-690X
Acinetobacter baumannii is an important opportunistic pathogen that mainly infects critically patients in intensive care units (ICU). The production of plasmid-mediated extended-spectrum b-lactamases (ESBLs) is one of the most important mechanisms of resistance against b-lactam antibiotics. This study aimed to evaluate the prevalence of ESBLs in A. baumannii isolated from ICU of Ghaem hospital, Mashhad, Iran. A total of 140 A. baumannii isolates recovered from hospitalized patients in ICU of Ghaem hospital in Mashhad city from December 2014 to March 2015. Identification of A. baumannii isolates carried out using biochemical laboratory methods and then confirmed by OXA-51 PCR screening. Susceptibility testing performed using disk diffusion (Kirby-Bauer) method as recommended by CLSI guidelines. A. baumannii isolates screened for production of ESBLs using combination disk test. blaPER, blaGES, blaTEM, blaSHV, blaCTX, blaVEB and blaOXA-10 beta-lactamase genes detected using conventional PCR. The most antibacterial resistance was against cefuroxime (99.3%) and colistin was the most effective antibiotic. None of the isolates were ESBL producer by combination disk test. However, results of PCR revealed that the prevalence of blaPER, blaGES and blaTEM genes were 7.1%, 4.3% and 27.1%, respectively. blaCTX, blaVEB, and blaOXA-10 were not found in any of isolates. According to the results, the high resistance was seen against selected antibiotics and the phenotypic tests are not sufficient alone for determination of ESBLs producer of A. baumannii isolates. So, molecular tests are also necessary for detection of these enzymes.
A. baumannii, ESBLs, ICU, Iran
Bacteria which constitute the Acinetobacter genus were originally identified in the first decade of the 20th century. Acinetobacter is a genus of gram-negative bacteria belonging to the gammaproteobacterial1. Acinetobacter are rode-shape during rapid growth and coco-bacillary in the stationary phase. They are generally encapsulated, nonmotile, aerobic, gram-negative organisms with tendency to retain crystal violet and therefore to be incorrectly identified as gram-positive cocci2. Frequent misidentification of Acinetobacter as Neisseria or Moraxella on gram staining is readily clarified by the negative oxidase reaction of Acinetobacter. Additionally, Acinetobacter are catalase-positive. Hemolysis of red blood cells, acidification of glucose, growth at 44°C, and variability in carbon source uptake are few of the phenotypic characteristics applied to distinguish Acinetobacter strains3, A. baumannii isolates are more likely caused disease in patients with immunosuppression, serious underlying disease and people who are exposed to invasive procedures accompanying treatment with broad-spectrum antibiotics. Therefore, the spread of these species in ICU and burn wards is more. A. baumannii is an important cause of nosocomial infection, such as ventilator associated pneumonia(VAP), urinary tract infections, wound infections, and septicemia4. A. baumannii is a significant opportunistic pathogen that mainly infects critically ill patients in ICU5.
As known the ability of A. baumannii to achieve different mechanisms of resistance, also, resistance to all available common antibiotics as well as lack of new effective antimicrobial drugs are the most important causes of risk about this organism. A. baumannii isolates which are resistant to three or more classes of antibiotics are called multi-drug resistant strains (MDR). Increasing antibiotic resistance in Acinetobacter inhibits from appropriate management in antibiotic therapy6.
A. baumannii has several innate resistance mechanisms to a number of antibiotics, such as aminopenicillins, first-and second- generation cephalosporins and chloramphenicol. Besides this, it has a considerable capacity to acquire mechanisms conferring resistance to broad-spectrum b-lactams, carbapenems, aminoglycosides and fluoroquinolones. Beta lactam antibiotics (mainly carbapenems) are now the first drug of choice to treat this microorganism; however, in the last decade, resistance to carbapenems has appeared in hospitals worldwide owing to the production of beta-lactamase, change in permeability, increase in efflux, and modification of the affinity of penicillin-binding proteins (PBPS) in these bacteria7. Production of plasmid-mediated extended-spectrum b-lactamases (ESBLs) is one of the most important mechanisms of resistance against beta-lactam antibiotics. Many of these enzymes have evolved from TEM and SHV -lactamases, but recently a large number of ESBLs are related to TEM and SHV, such as GES and VEB, have been described8. Plasmid is accounted for distribution of the most beta lactamases; however, the gene encoding these enzymes may also be on the chromosome or transposable elements and integrons.
ESBLs are also able to hydrolyze three and four generation cephalosporins and monobactams. ESBLs producer isolates are inhibited by b-lactamase inhibitors (clavulanic acid, sulbactam and tazobactam), At present, there are more than 300 different ESBL variants, and these have been clustered into nine different structures and these evolutionary families based on amino acid sequence9. TEM, CTX, SHV, GES, VEB, OXA-10 and PER are the major types.
In according to the information on the prevalence of these enzymes and antibacterial resistance pattern, control, prevention and treatment of this bacterium is important, thus, this study aimed to evaluate the prevalence of ESBLs in A. baumannii isolated from ICU of Ghaem hospital, Mashhad, Iran.
Bacterial sources
A total of 140 A. baumannii isolates were recovered from hospitalized patients in ICU of Ghaem hospital in Mashhad city from December 2014 to March 2015. All nonlactose fermenting members were subjected to microbiologic and biochemical tests such as; gram staining, oxidase, catalase, O/F, and growth at 42°C on nutrient agar medium. For confirmation of A. baumannii isolates, API20NE kit (version 6.0, bio-Merieux, Marcy L’Etoile, France) was applied. Then until use, clinical isolates were stored in nutrient broth containing 20% glycerol at -80°C.
Antibiotic Susceptibility Testing
Antibiotic Susceptibility Testing performed using modified Kirby-Bauer disk diffusion method based on CLSI guidelines6, 10. The potency of antibiotics disks was checked by reference strains Pseudomonas aeruginosa ATCC 27853. After incubation for a period of 24 h, results were reported as sensitive, intermediate, or resistant according to the zone diameters. The antibiotics used were imipenem (10 µg), meropenem (10 µg), colistin (25 µg), amikacin (30 µg), ceftazidime (30 µg), cefotaxime (30 µg), cefuroxime (30 µg), ceftriaxone (30 µg), cefepime (30 µg), ertapenem (10 µg) and ampicillin/sulbactam (10 µg).
Phenotypic Detection of Beta-Lactamase
In phenotypic confirmation of ESBLs producers on Muller Hinton agar, the combination disc test (CDT) was applied as previously defined11. Cefotaxime (30 µ) or ceftazidime disks (30 µ) with or without clavulanate (10 µ) were used. After incubation of plates for 24 h at 37°C, if the diameter of inhibition zone for each of these antibiotics in combination with clavulanic acid compared to antibiotics alone, increased by more than 5 mm, they defined as the ESBL-producing isolates, if no, isolates were reported as ESBL negative. P. aeruginosa ATCC 27853 was applied for quality control of isolates.
DNA extraction and PCR
For DNA extraction and template preparation, the boiling method was used as previously described12 and lastly samples stored at -20°C, till use. The primer pair sequences designed by primer premier software for detection of ESBL genes in clinical isolates of A. baumannii using PCR technique are shown in Table 1. Of note, PCR of blaOXA-51-like gene was also used for confirmation of isolates identification. The PCR program for blaGES, blaCTX and blaPER genes was composed of an initial denaturation step (94°C, 5 min) followed by 30 cycles of denaturation step (94°C, 1 min), annealing step (60°C, 1 min), and extension step (72°C, 1 min) with final extension (72°C, 7 min). The DNA amplification program for blaoxa-10, blaSHV, blaVEB and blaTEM genes was similar to previous genes except that the annealing temperature was 51°C. Components of PCR master mix (Amplicon, Denmark) were as follows; 1.5 mM Mgcl2, 10 pmol/µl of each primer, 0.2 mM dNTPs, 1U Taq DNA polymerase, 1X PCR buffer and 50 ng/µl DNA. PCR products were analyzed using 2% agarose gel electrophoresis (Cinaagen, IRAN). And 50bp DNA ladder (Fermentas company product) was used to detect the specific PCR products related to the bla genes. Then, results were observed under UV light gel documentation system.
Table (1):
Primer sequences of ESBL genes amplified by PCR
Primer Name |
5- primer sequence – 3′ |
Size, bp |
|---|---|---|
F- GES R- GES |
5′- CGCTTCATTCACGCACTATTACTG-3′ 5′-TCTCTCCAACAACCCAATCTTTAGG-3′ |
682 |
F- VEB R- VEB |
5′-GCCAGAATAGGAGTAGCAAT-3′ 5′-GGTTACTTCCTGTTGTTGTT-3′ |
547 |
F-TEM R-TEM |
5′-CAGTGCTGCCATAACCAT-3′ 5′-CGCCTCCATCCAGTCTAT-3′ |
271 |
F- SHV R-SHV |
5′-TATTCGCCTGTGTATTATCTCC-3′ 5′-CTGTTATCGCTCATGGTAATG-3′ |
378 |
F-OXA R-OXA |
5′-GCGGCACCTGAATATCTA-3′ 5′-TCTTAGCGGCAACTTACTT-3′ |
165 |
F-PER R-PER |
5′-GCAATACTCGGTCTCGCACAG-3′ 5′-TTCGGCTTGACTCGGCTGAG-3′ |
461 |
F-CTX R-CTX |
5′- GCGACAATACTGCCATGAATAAGC-3′ 5′-ATATCGTTGGTGGTGCCATAATCTC-3′ |
349 |
Sequencing of PCR products
The PCR products of three samples for each mentioned gene were subjected to direct sequencing and the nucleotide sequences were evaluated and analyzed with CLUSTAL W2 and BLAST software’s.
Data analysis
SPSS software (version 22, Chicago, IL, USA) was used for performing the statistical analysis using chi-squire and Fisher’s exact tests. Also, P-value<0.05 was considered as significant statistically.
In this study, a total of 140 isolates A. baumannii collected from ICU of Ghaem hospital in Mashhad, Iran, from December 2014 to March 2015. The sources of isolates were as follows; Trashes 71 (50.7%), Urine 50 (35.7%), Wound 10 (7.2%) and Blood Culture 9 (6.4%). Overall, 51.4%, 48.6% of the hospitalized patients were female and male, respectively. The most frequent isolates of A. baumannii (with prevalence of 40%) were in the age group 31-50 years. Also, the most rates of isolates (with prevalence of 51.4%) were seen in female than male.
As shown in Table 2, results of antibacterial susceptibility pattern revealed that in A. baumannii the high resistance was to all antibiotics except colistin, as resistance rates to imipenem, meropenem, ceftazidime, cefotaxime, cefuroxime, ceftriaxone, Cefepime, ertapenem and ampicillin/sulbactam were 97.9%, 98.1%, 96.4%, 97.9%, 99.3%, 97.9%, 97.9%, 98.6% and 97.1%, respectively. The most effective antibiotic against A. baumannii was colistin with susceptibility 97.9% followed by amikacin with sensitivity 27.1% (Table 2).
Table (2):
Antimicrobial susceptibility pattern of A. baumannii isolates
Antimicrobials |
Resistance, No. (%) |
Intermediate, No. (%) |
Sensitivity, No. (%) |
|---|---|---|---|
imipenem |
137 (97.9) |
0 |
3 (2.1) |
meropenem |
138 (98.6) |
0 |
2 (1.4) |
colistin |
3 (2.1) |
0 |
137 (97.9) |
amikacin |
94 (67.1) |
8 (5.7) |
38 (27.1) |
ceftazidime |
135 (96.4) |
0 |
5 (3.6) |
cefotaxime |
137 (97.9) |
1 (0.7) |
2 (1.4) |
cefuroxime |
139 (99.3) |
0 |
1 (0.7) |
ceftriaxone |
137 (97.9) |
1 (0.7) |
2 (1.4) |
cefepime |
137 (97.9) |
0 |
3 (2.1) |
ertapenem |
138 (98.6) |
0 |
2 (1.4) |
ampicillin/sulbactam |
136 (97.1) |
2 (1.4) |
2 (1.4) |
It was also presented that none of the isolates were ESBL producers by Combination disk method. Although A. baumannii isolates exhibited a high degree of resistance to third-generation cephalosporins but they did not produce ESBL.
PCR revealed that the prevalence of blaPER, blaGES, blaTEM genes were 7.1%, 4.3% and 27.1%, respectively. blaCTX, blaVEB, and blaOXA-10 were not found in any of isolates (Table 3 and Figures 1-3).
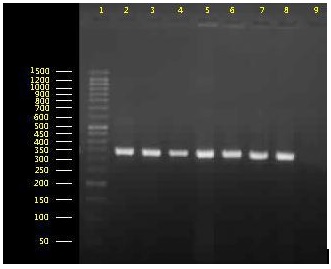
Fig. 1. Agarose gel electrophoresis for analyzing of blaoxA-51 gene amplification. Lane 1: 50bp DNA Ladder, lanes 2-8: Isolates containing the fragment of 353bp blaoxA-51 gene, lane 9: negative control.
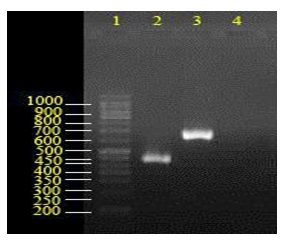
Fig. 2. Agarose gel electrophoresis for blaPER and blaGES genes amplification. Lane 1: 50bp DNA ladder, lane 2: isolate harboring blaPER (461 bp) and lane 3: isolate with 682 bpblaGES gene. 4: negative control.
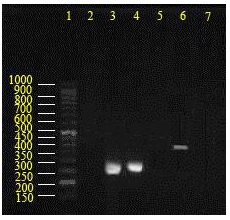
Fig. 3. Agarose gel electrophoresis for amplification analysis of blaTEM and blaSHV genes. Lane 1: 50bp DNA ladder, lanes 2: isolate without blaTEM and blaSHV genes, lanes 3-4: isolates with blaTEM (271 bp), lane 5: negative control for reaction with blaTEM primer, lane 6: isolate with 378 bpblaSHVgene, lane 7: negative control for reaction withblaSHVprimer.
Table (3):
Frequency distribution of ESBLs genes in clinical A. baumannii isolates
| Genes | Number | Genes | Number (%) |
|---|---|---|---|
| blaTEM | 38(27.1%) | blaCTX | 000 |
| blaSHV | 9(6.4%) | blaOXA-10 | |
| blaPER | 10(7.1%) | blaVEB | |
| blaGES | 6(4.2%) |
A. baumannii isolates which at one time had two ESBL genes were: blaPER/blaGES 2 (1.4%), blaSHV/blaGES 2 (1.4%), blaSHV/blaTEM 2 (1.4%), blaGES/blaTEM 1 (0.7%).
In addition, results showed that there no correlation was found between prevalence of ESBLs genes and types of clinical samples (p>0.05), as per Table 4.
Table (4):
Frequency distribution of ESBLs genes in A. baumannii isolates based on types of clinical samples
Genes |
Trashes, N=71 |
Urine, N=50 |
Wound, N=10 |
Blood culture, N=9 |
P- value |
|---|---|---|---|---|---|
blaTEM |
18(25.4%) |
15(30%) |
2(20%) |
3(33.3%) |
0.86 |
blaSHV |
5(7%) |
3(6%) |
0 |
1(11.1%) |
0.78 |
blaPER |
3(4.2%) |
6(12%) |
1(10%) |
0 |
0.32 |
blaGES |
2(2.8%) |
4(8%) |
0 |
0 |
0.40 |
blaCTX |
0 |
0 |
0 |
0 |
0.00 |
blaOXA-10 |
0 |
0 |
0 |
0 |
0.00 |
blaVEB |
0 |
0 |
0 |
0 |
0.00 |
A. baumannii is mostly found in hospital settings and is nowadays noticed more than ever due to its survival ability in such environments and causing nosocomial infections. In this study, among the aminoglycosides, amikacin was resistant in 67.1% of cases and in other groups; cephalosporins, carbapenems, and penicillin were almost 100% resistant, indicating multiple drug resistance in these isolates. Colistin was the most effective antibiotic (97.9%). Our results demonstrated that the rate of resistance was significantly lower in colistin compared to other antibiotics. The possibly reason for low resistance to this antibiotic may be owing to its infrequent prescription during the recent period. In a study conducted by Shahcheraghi et al. (2009) on A. baumannii isolated from patients hospitalized in Tehran hospitals showed a large proportion of isolates were resistant to ceftriaxone (96.8%), cefotaxime (95.7%), ceftazidime (78.9%), and cefexime (100%), while 95.8% of isolates were susceptible to colistin, which their findings were consistent with current study. This study, like the studies carried out by Tseng et al. in China in 2007 13 and Smolyakov et al. in 200014, confirmed that most isolates were resistant to ceftazidime and cefepime. Regarding the present results and similar studies, due to over-administration of the third generation cephalosporins and lack of observing the hygienic principles in the community, a considerable resistance has been developed against this generation of cephalosporins. So, based these findings, the third generation cephalosporins are not good choice for treatment of infections caused by MDR A. baumannii isolates.
The study conducted by Srinivasan and colleagues in Ohio, USA15, more than 80% of the isolates were resistant to a wide range of cephalosporins and 20% to imipenem, while in our study, resistance to imipenem was more than 90%, which this dissimilarity might be due to unnecessary overuse of antibiotics. An another study conducted by Akan et al. in 2002, in Turkey on 277 A. baumannii isolates revealed that the resistance rate to imipenem and amikacin was 53.6% and 59.8%, respectively16, in contrast, this rate was much higher in our study and also in comparable above mentioned ones. All of studies stated here used disk diffusion agar method to evaluate antibiotic susceptibility similar to our study. Therefore, the difference in results could be attributed to diversity in types of isolates, variety in antibiotic disks used, and difference in geographical regions of the studies and policies of infection control17. Thus, the regional determination of antibiotic susceptibility of A. baumannii can act as a suitable guide for effective use of routine antibiotics. Since in this study and similar ones, the most A. baumannii isolates (50.7%) were obtained from pulmonary secretions, it appears that the respiratory tract is the most involved in infections caused by A. baumannii. So, disinfection and sterilization of equipment and respiratory devices like respiratory is one of the ways for prevention of infection dissemination. Based on the studies conducted in our country (Iran) and through study reported by Sharif et al. in 2013, 51% of A. baumannii isolates had the wide-range beta-lactamase-producing phenotype18. Also, Owlia et al. in 2012, in Tehran reported that 21% of A. baumannii isolates had the wide-range beta-lactamase-producing phenotype19. Similarly, this rate was reported 28% in a study conducted by Sinha et al. in India in 20078. Shakibaie et al. in 2012 identified only 3 ESBL-producing among 100 A. baumannii isolates20. The study by Jazani et al. (2010) reported only 1 ESBL-producing isolate from among 48 A. baumannii isolates recovered from clinical samples of patients hospitalized in burn hospital, Tehran21. In present study, we used the combination disk method similar to the method used in the mentioned studies, there was no positive test regarding phenotype. One probable reason for lack of production of extended b-lactamase-producing phenotype in the present study, compared to other studies, may be increased expression of AmpC genes and also beta-lactamases and metallo beta-lactamase enzymes. It is also possible that mechanisms other than extended b-lactamase like secretary pumps and variations in porins induce resistance in this organism. Indeed, resistance in A. baumannii is associated with a combination of various mechanisms including acquisition of b-lactamases, stable induction of AmpC, reduced permeability, changes in penicillin binding proteins, and somewhat, with an increase in Efflux pumps21. Performing of phenotypic tests alone is not able to determine the ESBL-producing isolates in A. baumannii. Some molecular tests need be performed to determine the presence of ESBL enzymes.
The blaOXA-51 gene is considered as a chromosol component of A. baumannii isolates which can used to identify this organism,22for this reason in present study we used blaOXA-51 gene for confirmation of the A. baumannii isolates.
Azhari and et al. in 2010 in Tabriz indicated that among 100 isolates of A. baumannii from different clinical samples, PER gene was not found in any of samples23. The first report of presence of PER gene was detected in a study conducted by Farajnia and et al. in 2013 in Tabriz, wherein its prevalence was 51.7% 24 which was higher than over results. But another in 2007 in Argentina presented that among 1 out of 6 isolates was positive for PER gene25.
Shahcheraghi et al. in 2009 in Tehran revealed that of 95 isolates, 12.8% were reported positive for TEM26 while this rate in study carried out by Sharif and et al. in 2012 in Tehran was 56%18. In another one in America in 2010, occurrence of TEM was 37%27, which is partly in line with present study.
GES was first reported in the study by Shahcheraghi et al. in 2011 in Tehran, Iran, in which 2 out of 100 A. baumannii isolates were resistant to imipenem26. Furthermore, in the study carried out by Bogaerts et al. in 2009 in Belgium, 9 out of 125 isolates were reported positive for GES gene which is relatively similar to our study with 6 positive isolates28. Also VEB was first reported in the study performed by Poirel et al. in 2003 in France, in which 7 cases (58.3%) out of 12 A. baumannii isolates were positive for VEB gene29. In the study conducted by Pasteran and colleagues in 2006 in USA, 47.6% of isolates possessed the VEB gene[30]. Moreover, in the study carried out by Farajnia et al. in 2009 in Tabriz, Iran, 10% out of 100 A. baumannii isolates were reported positive for VEB gene24, in contrast none of isolates were positive for VEB gene in our study.
Vafaei and et al. presented of 130 A. baumannii isolates, 19% of those were positive for CTX gene31. But in the study conducted by Ramoul and colleagues in 2013, CTX was not found in any of the isolates32, which is close to our results.
Several studies suggested varying distribution of resistant genes in different geographical regions. Geographical distance and also pattern of antibiotic usage can predispose to emergence of resistant genes in different geographical areas. As Beta-lactamase-producing isolates constitute a lower percentage compared to Beta-lactam-resistant isolates, it seems that in addition to the production of Beta-lactamases, other factors like the presence of Efflux pumps and cellular wall canals or purines also contribute to the creation of resistance. Due to the capacity of these isolates for transmitting resistance genes to other clinical isolates, the exact identification of Beta-lactamases genes contributing resistance is of most importance for care, treatment, and epidemiologic studies on transmission methods in hospitals33.
The high resistance of isolates to third and fourth generation of cephalosporins compared to the low number of ESBL producing isolates, proposed another resistance mechanisms such as secretory pump, purines, biofilm information involved in development of resistance.
Hence, the development in policies of antibiotic prescription and infection control are more critical to prevent the spreading of such resistant infectious organisms.
According to the results, the high resistance was seen against selected antibiotics and the phenotypic tests are not sufficient alone for determination of ESBLs producer of A. baumannii isolates. So, molecular tests are also necessary for detection of these enzymes.
ACKNOWLEDGMENTS
We would like thank our colleagues in Laboratory of Medical Microbiology of Ghaem hospital for Clinical Samples Collection.
- P. E. Fournier, H. Richet, and R. A. Weinstein, “The epidemiology and control of Acinetobacter baumannii in health care facilities,” Clinical infectious diseases, 2006; 42(5): pp. 692-699.
- R. F. Kheltabadi et al., “Antimicrobial Susceptibility patterns and the distribution of resistance genes among Acinetobacter species isolated from patients in shahid Beheshti hospital, Kashan,” Feyz Journals of Kashan University of Medical Sciences, 2009; 12(4).
- Y.-W. Tang, N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing, “Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli,” Journal of clinical microbiology, 1998; 36(12): pp. 3674-3679.
- M. L. Joly Guillou, “Clinical impact and pathogenicity of Acinetobacter,” Clinical microbiology and infection, 2005; 11(11), pp. 868-873.
- H. Saghi, A. Bahador, A. Khaledi, R. A. Kachoei, F. A. Dastjerdi, and D. Esmaeili, “Antibacterial effects of Origanum vulgare essence against multidrug-resistant Acinetobacter baumannii isolated from selected hospitals of Tehran, Iran,” Avicenna Journal of Clinical Microbiology and Infection, 2015; 2(1).
- G. M. Eliopoulos, L. L. Maragakis, and T. M. Perl, “Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options,” Clinical infectious diseases, 2008; 46(8): pp. 1254-1263.
- K. M. Hujer et al., “Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center,” Antimicrobial agents and chemotherapy, 2006; 50(12): pp. 4114-4123.
- M. Sinha, H. Srinivasa, and R. Macaden, “Antibiotic resistance profile & extended spectrum beta-lactamase (ESBL) production in Acinetobacter species,” Indian journal of medical research, 2007; 126(1): p. 63.
- D. L. Paterson and R. A. Bonomo, “Extended-spectrum b-lactamases: a clinical update,” Clinical microbiology reviews, 2005; 18(4): pp. 657-686.
- M. Moghadamshakib et al., “ISSN 0975-413X CODEN (USA): PCHHAX.”
- F. H. M’Zali, A. Chanawong, K. G. Kerr, D. Birkenhead, and P. M. Hawkey, “Detection of extended-spectrum â-lactamases in members of the family Enterobacteriaceae: comparison of the MAST DD test, the double disc and the Etest ESBL,” Journal of Antimicrobial Chemotherapy, 2000; 45(6), pp. 881-885.
- Y.-S. Yu, Q. Yang, X.-W. Xu, H.-S. Kong, G.-Y. Xu, and B.-Y. Zhong, “Typing and characterization of carbapenem-resistant Acinetobacter calcoaceticus–baumannii complex in a Chinese hospital,” Journal of medical microbiology, 2004; 53(7): pp. 653-656.
- Y.-C. Tseng, J.-T. Wang, F.-L. L. Wu, Y.-C. Chen, W.-C. Chie, and S.-C. Chang, “Prognosis of adult patients with bacteremia caused by extensively resistant Acinetobacter baumannii,” Diagnostic microbiology and infectious disease, 2007; 59(2): pp. 181-190.
- R. Smolyakov et al., “Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment,” Journal of Hospital Infection, 2003; 54(1): pp. 32-38.
- V. B. Srinivasan et al., “Genetic relatedness and molecular characterization of multidrug resistant Acinetobacter baumannii isolated in central Ohio, USA,” Annals of clinical microbiology and antimicrobials, 2009; 8(1): p. 21.
- O. Akan, “Antibiotic resistance of Acinetobacter baumannii isolates: data from Ibni Sina Hospital for the year 2002,” Mikrobiyoloji bulteni, 2003; 37(4): pp. 241-246.
- A. Bahador et al., “The prevalence of ISAba1 and ISAba4 in Acinetobacter baumannii species of different international clone lineages among patients with burning in Tehran, Iran,” Jundishapur journal of microbiology, 2015; 8(7).
- M. Sharif, R. Mirnejad, and N. Amirmozafari, “Molecular identification of TEM and SHV extended spectrum â-lactamase in clinical isolates of Acinetobacter baumannii from Tehran hospitals,” J Gen Microb Immun, 2014; 2: pp. 1-9.
- P. Owlia, L. Azimi, A. Gholami, B. Asghari, and A. R. Lari, “ESBL-and MBL-mediated resistance in Acinetobacter baumannii: a global threat to burn patients,” Infez Med, 2012; 20(3): pp. 182-7.
- M. R. Shakibaie, S. Adeli, and M. H. Salehi, “Antibiotic resistance patterns and extended-spectrum b-lactamase production among Acinetobacter spp. isolated from an intensive care Unit of a hospital in Kerman, Iran,” Antimicrobial resistance and infection control, 2012; 1(1), p. 1.
- N. Jazani, H. Babazadeh, M. Sohrabpour, M. Zartoshti, and M. Ghasemi-Rad, “The prevalence of extended spectrum beta-lactamases in Acinetobacter baumannii isolates from burn wounds in Iran,” The Internet Journal of Microbiology, 2011; 9(2): pp. 1-7.
- J. F. Turton, N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt, “Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species,” Journal of clinical microbiology, 2006; 44(8): pp. 2974-2976.
- F. Azhari, S. Farajnia, and M. Rahnema, “Investigation Of Prevalence Of Esbl Types Veb-1 And Per-2 Genes And Int-1 In Acinetobacter Bumanii Strains Isolated From Patients Of Imam Reza Hospital Intabriz,” 2010.
- S. Farajnia, F. Azhari, M. Y. Alikhani, M. K. Hosseini, A. Peymani, and N. Sohrabi, “Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant Acinetobacter baumannii isolates in North-West of Iran,” Iranian journal of basic medical sciences, 2013; 16(6): pp. 751-755.
- L. Poirel et al., “Identification of the novel narrow-spectrum â-lactamase SCO-1 in Acinetobacter spp. from Argentina,” Antimicrobial agents and chemotherapy, 2007; 51(6): pp. 2179-2184.
- F. Shahcheraghi, M. Abbasalipour, M. Feizabadi, G. Ebrahimipour, and N. Akbari, “Isolation and genetic characterization of metallo-â-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals,” Iranian journal of microbiology, 2011; 3(2): pp. 68-74.
- N. Dai et al., “Drug-resistant genes carried by Acinetobacter baumanii isolated from patients with lower respiratory tract infection,” Chinese medical journal, 2010; 123(18): pp. 2571-2575.
- P. Bogaerts et al., “GES extended-spectrum â-lactamases in Acinetobacter baumannii isolates in Belgium,” Antimicrobial agents and chemotherapy, 2010; 54(11), pp. 4872-4878.
- L. Poirel, O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann, “Outbreak of extended-spectrum â-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital,” Journal of Clinical Microbiology, 2003; 41(8): pp. 3542-3547.
- F. Pasterán et al., “Emergence of PER-2 and VEB-1a in Acinetobacter baumannii strains in the Americas,” Antimicrobial agents and chemotherapy, 2006; 50(9): pp. 3222-3224.
- S. Vafaei, R. Mirnejad, and N. Amirmozafari, “Determining the Patterns of Antimicrobial Susceptibility and the Distribution of blaCTX-M Genes in Strains of Acinetobacter Baumannii Isolated from Clinical Samples,” Journal of Isfahan Medical School, 2013; 31(252).
- A. Ramoul, S. Hammami, M. Dekhil, S. Aimiri, A. Slim, and I. B.-B. Boubaker, “Phenotypic and genotypic characterization of clinical multidrug resistant Acinetobacter baumannii from Algerian intensive care units,” African Journal of Microbiology Research, 2013; 7(10): pp. 868-874.
- F. Shahcheraghi, N. Akbari Shahmirzadi, H. Jabbari, and N. Amirmozafari, “Detection of blaCTX, blaTEMbeta-lactamase genes in clinical isolates of Acinetobacterspp. from selected Tehran hospitals,” Iranian Journal of Medical Microbiology, 2009; 3(1): pp. 1-9.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


