ISSN: 0973-7510
E-ISSN: 2581-690X
Extensive and unregulated use of antibiotics favours the acquisition of resistance to antimicrobial agents. Hospitalized patients are at usually high risk of infections, and therefore the hospital surroundings favours the acquisition of resistance to antimicrobial agents, complicating the treatment of health facility infections as a result of drug resistant pathogens. The pattern of hospital infection has modified over the years, reflective advances in drugs and development of antimicrobial agents. Hospital effluent could contain multi drug resistant (MDR) coliforms and enteric pathogens which could pose a great problem for communities. There also exists a relationship between the metal tolerance and the anti-microbial resistance in microorganisms. Bacteria have continued to adapt to ecology of humans and their antimicrobial agents by diversifying the systems that they have evolved to counter those agents. As a result, unexpected and new problem of resistance have appeared and some resistance has become widely spread. So our aim is to determine the prevalence of antibiotic and heavy metals resistance in coliforms isolated from hospital wastewater. The present study was conducted at Department of (Ag.) Microbiology, Faculty of Agricultural Sciences; Aligarh Muslim University Aligarh.
Isolation, Waste water, Antibiotic, Coliform, Interaction with Metals.
Coliforms are Gram negative, rod shaped, non-spore-forming and aerobic or facultative anaerobic bacteria. The coliform group is not a particular substantial taxonomic group, it belongs to family Enterobacteriaceae. They ferment lactose with both gas and acid production at 35 °C. The coliform members include Citrobacter, Enterobacter, Escherichia, Hafnia and Proteus. Some pathogens like Salmonella, Shigella Yersinia and Klebsiella added to the coliform group.
The antibiotics that we intake are not all processed by our bodies and some of them are expelled as waste and reach our wastewater treatment plants. Sewage from hospitals and pharmaceuticals plants has been shown to contribute to antibiotic resistance in treatment plants4.
Hospital discharge gives significant volumes of wastewater loaded with microorganisms, heavy metals, toxic chemicals and radioactive elements, the majority of which are pathogenic. Humans are mainly exposed to these chemicals and infectious agents via the consumption of water from surface or sub-surface water bodies15.
Bacterial resistance to antibiotics and heavy metals is an expanding issue in today’s general public. Resistance to antibiotics is acquired by an adjustment in the genetic makeup of bacterium, which can happen by either transfer of antibiotic resistance genes between microorganisms or due to the mutations taking place in the environment. The indiscriminate utilization of antibiotics in medicinal services, in agribusiness and animal farming is thus adding to the developing issue of antibiotic resistance in microorganisms7. In addition to the antibiotics, heavy metals utilized as a part of industry and in family unit items are also mounting, specific pressure in the environment. As a result, the microorganisms have to make changes for better survival and duplication3.
Drug-resistant pathogenic bacteria are continuously identified in both clinical and community settings whose phenotypic resistance can be attributed to genes acquired from another organism through horizontal gene transfer (HGT) via conjugation, transformation, or transduction. The high concentrations of antibiotics used in clinical and agricultural settings likely provide strong selection pressure favouring such RG exchange between pathogens and microbes from virtually any habitat5.
Metals are critically dangerous factor to biota in nature, for instance, heavy metals may diminish metabolic movement and assorted qualities and in addition influence the qualitative and quantitative structure of microbial groups10. Resistance mechanisms to toxic metals are known to exist18. Metal resistance in microbes is controlled by resistance genes that are found on chromosomes, plasmids and transposons.
There also exists a relationship between the metal tolerance and the anti-microbial resistance in microorganisms due to the probability that resistance qualities to both the antibiotics and the heavy metals might be present on the same mobile genetic element (MGEs) for example, plasmids, transposons and integrons1. These numerous quality MGEs are exchanged between the microorganisms living in areas defiled by heavy metals.
Lawrence’s (2000) discussion of the Selfish Operon theory, clustering of genes on a plasmid, if all the genes clustered are useful to the organism interns of survival of its species, those genes may be transferred together in the conjugation process. In an environment with antibiotics and heavy metals stress, it would be more ecologically favorable for a bacterium to acquire resistance to both stresses. Plasmid mediated resistance genes passes to other bacteria, and those bacteria would then have a better chance of survival.
Relationship between resistance to antibiotics and heavy metals has been accounted for by the few workers where genes encoding resistance to metals were located on transmissible plasmids6, 20. Genes that code for antibiotics resistance attributes and genes that code for metal resistance are regularly carried on the same plasmids or mobile genetic elements. Transferable plasmids encoding resistance to different heavy metals and antibiotics in Salmonella have been accounted for9.
Collection of samples
Wastewater samples were collected from a drain of J. N. Medical College, Aligarh Muslim University Aligarh, during the months of January-February 2016. Samples were collected in sterile glass bottle and transported to the laboratory immediately for initial processing, so that elapsed time between the sample collection and initial processing did not exceed 3 h.
Isolation and characterization of Coliform bacteria
Wastewater samples were serially diluted in sterile NSS. For the determination of the total coliforms present in the wastewater samples the presumptive test was used. For this double and single strength lactose broth was prepared. Durham tubes were inverted in the broth. Single strength lactose broth was inoculated with 1 ml of the water sample. After incubation at 37 0C for 48 h, in the Durham’s tube presence of gas as well as acid production with colour change to yellow was the presumptive evidence of the presence of coliform bacteria in the water sample.
A sterile inoculating loop was used to streak Eosin Methylene Blue (EMB) agar plates from the positive lactose broth tubes. After incubation for 24 h at 37 0C, the presence of pink mucoid colonies and green metallic sheen colonies was recorded as a suspected indicative result for coliform bacteria. Well-isolated colonies were further inoculated into brilliant green lactose bile broth for the complete test. Bacterial isolates were tentatively identified on the basis of morphological, cultural and biochemical characteristics.
Antibiotic sensitivity of the bacterial isolates
Antibiotic sensitivity behaviour of the isolated strains was determined by disc diffusion method as described by Bauer, et al., (1996). The antibiotic discs were obtained from Hi-Media Lab. Pvt. Ltd., Mumbai, India. The potency of disc were checked against a sensitive strain i.e., Escherichia coli B.
Muller Hinton agar plates were prepared. 0.1 ml culture from broth was spread evenly onto Muller Hinton agar plates. Plates were kept at room temperature for 10-15 min. Antibiotic discs were mounted and plates were incubated at 37 oC overnight. The plates were scored for resistance or sensitive after 18-24 hours by comparing the chart based on the inhibitory zone diameter as given by the disc manufacturer
Determination of minimum inhibitory concentration (MIC) of heavy metals
MIC of heavy metals was worked out by Agar dilution method against test strains. Different concentrations of freshly prepared heavy metals solutions were added to sterilize nutrient agar medium of pH 7.2, mixed well and poured into petriplates. The recommendations of WHO (1961) were followed in conducting the test as adapted by Malik et al., 2002.
Each plate was divided into 36 equal sectors. A loopful (5mm diameter) of 18 h broth culture was spot inoculated in duplicate, in each sector. All the test strains were also inoculated in duplicate, in each sector. Inoculated plates were allowed to absorb inoculum and the plates were incubated at 37 °C for overnight. The plates were examined for the presence or absence of growth on the spotted area. The presence of growth was indicated by (++) and absence of growth by (—). Concentration of heavy metals at which there was no growth was considered as their respective MICs.
Isolation of plasmid DNA
Cells were grown in 5 ml broth for overnight at 37 oC and pelleted by centrifugation at 10,000 rpm, at 4oC for 5 minute. The supernatant was discarded and the pellet washed with STE buffer. The cell pellet was thoroughly suspended in 100 µl of solution A, mixed properly followed by the addition of 200 µl of solution B (freshly prepared), the suspension was inverted several times and kept in ice for 5 min. ice cold solution C (150 µl) was added to the suspension. The suspension was centrifuged at 10,000 rpm for 5 min. and the supernatant thus obtained was transferred to fresh microfuge tubes, equal volume of phenol: chloroform (1:1) was added to the solution. The solution was centrifuged at 10,000 rpm for 10 min. The aqueous layer was transferred to fresh microfuge tubes followed by precipitation with 2 volume of absolute ethanol; the solution was kept at room temperature for 15 min and centrifuged for 5 min at maximum speed remove supernatant completely. To remove the residual salts, the pellet was washed with 1 ml of 70% ethanol. The final pellet obtained was allowed to air dry and dissolved in 30 µl TE buffer.
Agarose gel electrophoresis
Agarose gel electrophoresis was performed as described by Kado and Liu (1981). 0.7% agarose was used, which was prepared in 1x TAE buffer (49 ml distilled water added to 1 ml 50x TAE), the suspension was boiled till agarose dissolves completely. The solution was allowed to cool and ethidium bromide dyes (0.1µg/mL), was added to the solution. Electrophoresis was performed on a horizontal apparatus. Pellet of precipitated plasmid DNA was dissolved in 30 µL of TE buffer. To this 5 µL of marker dye (Bromothymol blue) and 5 µL double distilled water was added. This sample was loaded into wells of agarose gel slab. Electrophoresis was done at 50V/cm for 2-3 h. After running the gel, it was visualized under Gel documentation system (Bio RAD, USA).
In the present study, a total of 69 coliform bacteria were isolated from hospital wastewater. The isolates were characterized on the basis of morphological, cultural and biochemical characteristics in accordance with the Bergey‘s manual of Determinative Bacteriology.
Antibiotics sensitivity of the isolates against 10 antibiotics was conducted by disc diffusion method. The antibiotics / drugs used were Amoxycillin, Ampicillin, Ofloxacin, Kanamycin, Chloramphenicol, Methicillin, Streptomycin, Rifampicin, Norfloxacin and Nitrofurantoin. On the basis of sensitivity behaviour, incidence of antibiotic resistance was determined. All isolates demonstrated maximum resistance to Amoxycillin and Ampicillin, followed by Methicillin and least resistance to Ofloxacin, Chloramphenicol, Norfloxacin, Nitrofurantoin, and Kanamycin. The most effective antibiotic in vitro seems to be Streptomycin and Rifampicin shows high antibacterial activity towards all isolates (Table 1).
Table (1):
Percent resistance of the bacterial isolates from hospital wastewater against antibiotics
Antibiotics |
Concentration (µg/disc) |
No. of resistant isolates |
Percentage |
|---|---|---|---|
Amoxycillin |
30 |
69 |
100.00 |
Ampicillin |
10 |
69 |
100.00 |
Ofloxacin |
5 |
23 |
33.33 |
Kanamycin |
30 |
16 |
23.18 |
Chloramphenicol |
30 |
23 |
33.33 |
Methicillin |
30 |
68 |
98.55 |
Nitrofurantoin |
30 |
21 |
23.43 |
Streptomycin |
25 |
02 |
02.89 |
Rifampicin |
30 |
04 |
05.79 |
Norfloxacin |
10 |
21 |
23.43 |
Total number of isolates = 69
Table (2):
Antibiotic resistance pattern among isolated bacteria
No. of antibiotics |
Resistance pattern (No. of isolates) |
Percentage |
|---|---|---|
2 |
MET, AMX (02) AMP, AMX (01) |
2.89 1.44 |
3 |
MET, AMP, AMX (16) |
23.18 |
4 |
MET, AMP, RIF, AMX (01) MET, AMP, K, AMX (05) |
1.44 7.24 |
5 |
MET, OF, AMP, NX, AMX (01) MET, AMP, NX, C, AMX (03) MET, AMP, K, AMX, NIT (03) MET, OF, AMP, AMX, NIT (02) |
1.44 4.34 4.34 2.89 |
6 |
S, MET, OF, AMP, NX, AMP (01) MET, OF, AMP, NX, K, C (02) MET, OF, AMP, NX, RIF, AMX (02) |
1.44 2.89 2.89 |
7 |
MET, OF, AMP, NX, RIF, K, AMP (01) MET, AMP, NX, K, C, AMX, NIT (01) |
1.44 1.44 |
All the bacterial isolates (69) were also tested for their resistance against 6 different heavy metals i.e. Cr3+, Cu, Zn, Ni, Hg, and Cr+6. Present study indicated that 100% of the isolates were resistance to Cr3+ and Ni, whereas for Cu, Zn, Hg, Cr+6, exhibited 30.4%, 12.9%, 28.7%, 5.7% respectively (Table 3). In the present study the bacteria isolated from hospital sewage exhibited a maximum MIC of 200 µl for Hg. Further MIC values up to 3200 µl for other metals were recorded.
Table (3):
Incidence of metal resistance in bacterial isolates from hospital wastewater
| Metals | Sensitive range | Resistance range | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CrCl3 | MIC(µg/ml) No. of isolates |
200* (-) |
800 (01) (1.4) |
1600 (47) (68.1) |
3200 (21) (30.4) |
100% | ||||
| ZnSo4 | MIC(µg/ml) No. of isolates |
25 (1) (1.4) |
50 (-) |
100 (56) (81.1) |
200* (03) (4.3) |
400 (03) (4.3) |
800 (06) (8.6) |
12.9% | ||
| CuSO4 | MIC(µg/ml) No. of isolates |
100 (09) (13.0) |
200* (39) (56.5) |
400 (21) (30.4) |
30.4% | |||||
| K2Cr2O7 | MIC(µg/ml) No. of isolates |
25 (01) (1.4) |
200* (64) (92.7) |
400 (04) (5.7) |
5.7% | |||||
| NiCl2 | MIC(µg/ml) No. of isolates |
100* (-) |
400 (35) (50.7) |
800 (29) (42.0) |
1600 (05) (7.2) |
100% | ||||
| HgCl2 | MIC(µg/ml) No. of isolates |
12.5* (45) (65.2) |
25 (11) (15.9) |
50 (02) (2.8) |
100 (03) (4.3) |
200 (04) (5.7) |
28.7% | |||
*MIC of the standard strains
Total no. of isolates= 69
Values in parentheses indicates the percentage of total isolates:
Table (4):
Metal resistance pattern among bacteria from hospital wastewater
No. of metals |
Resistance pattern (No. of isolates) |
Percentage resistance (%) |
|---|---|---|
2 |
Cr3+, Ni2+ (32) |
46.37 |
3 |
Cr3+, Cu2+, Ni2+ (07) Cr3+, Ni2+, Hg2+ (09) |
10.14 13.04 |
4 |
Cr3+, Zn2+, Cr6+, Ni2+ (05) Cr3+, Cu2+, Ni2+, Hg2+ (13) |
07.24 18.84 |
5 |
Cr3+, Zn2+, Cr6+, Cu2+, Ni2+ (01) |
01.44 |
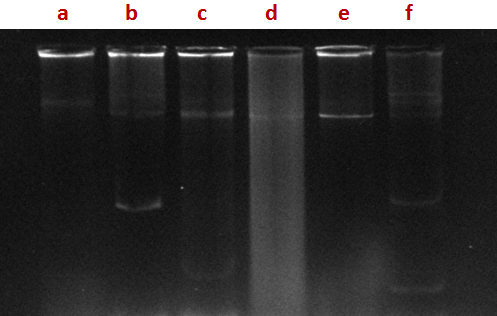
Fig. 1. Agarose gel electrophoresis profile of plasmid DNA, isolated from bacterial isolates. Lane a: SHA 10, lane b: SHA 31, lane c: SHA 03, lane d: SHA 35, lane: e SHA 65, lane f: SHA 66)
Several investigations on antibiotic resistance coliforms in wastewater have led to contradictory conclusions. Some investigators reported that antibiotics resistance bacteria were removed less efficiently than antibiotic sensitive organisms16.
The relative high level of resistance to antimicrobial agents is a reflection of misuse or abuse of these agents in the environment12. As pointed out by Hsu et al., (1992) differences in percentage of bacterial resistance to various antibiotics may reflect the history of antibiotics application and hence there is possibility of using bacterial resistance as an indicator of antibiotic application.
Majority of isolates from wastewater were found to have resistance to multiple drugs / antibiotics, ranging from resistance against 2 to 7 antibiotics. The combination of MET, AMP, AMX was the most prevalent (23.18%) followed by combination of MET, AMP, AMX, K (7.24%) (Table 2). In bacterial isolates resistance to antibiotics is highly prevalent, particularly in developing countries. This is due to the intensive use of antibiotics in both medicine and agriculture. Resistance to antibiotics is often plasmid mediated and several microcosm studies have shown plasmid transfer to occur in wastewater environment. We have found that antibiotic resistance bacteria were widespread in wastewater of India2. This finding, in and of itself, is not surprising since the intrinsic resistance of many organisms to antibiotics is well documented13. Studies have shown that the release of wastewater from hospitals was associated with an increase in the prevalence of antibiotic resistance8. Antibiotic resistance in bacteria is a serious threat to society today, and one of the reasons responsible for this problem is over use of antibiotics in humans14, 21.
A high frequency of resistance among isolates from hospital wastewater to antibiotics was observed in the present study. Isolates showed a high resistance against Ampicillin and Amoxacillin (100%), Methicillin (98.55%) and lower against Rifampicin (05.79%), Nalidixic acid (23.43%), chloramphenicol (33.33%) and least to Streptomycin (02.89%) (Table 1). Khan et al., 2006 also shows similar results to this study. They have also reported on transferable plasmids encoding resistance to various heavy metals and antibiotics in gram negative bacteria. Most of the isolates showed their MIC range 100-3200 µg/ml against Cr6+ while MIC 400-1600 µg/ml was recorded against Ni. Similar findings were also reported previously by Allen et al., 2010. In this study the prevalence of antibiotic and metal resistance among isolates was investigated. Hospital wastewater contained a higher number of single and multiple antibiotic resistances among coliform species.
In most of the strains of coliforms isolated from hospital sewage, plasmids of different molecular size were found (Fig.1). In the present study a few isolates were found to carry plasmids; however some tolerated high concentrations of the metals tested. There is the possibility that metal tolerance genes are carried on chromosomes or transposons which were not investigated during this study. Previous studies have showed the role of plasmids in conferring resistance to both antibiotics and metals. Mc Hugh et al. (1975) have shown that in Salmonella typhimurium isolates from human burn wounds treated with silver nitrate solution, plasmids conferring antibiotic and metal resistance.
Hospital effluent could contain multi drug resistant (MDR) coliforms and enteric pathogens. MDR strains are more and this posses a potential health hazard and require intervention measures. The pattern of antibiotic resistance in different bacterial isolates is more or less the same which implies that different bacterial strains carry similar antibiotic resistance determinants. Exposure to varying concentrations of these heavy metals (chromium, nickel, zinc, copper and mercury) to the native environmental bacteria has led to evolution of metal tolerant strains.
ACKNOWLEDGMENTS
All authors are thankful to Department of Agricultural Microbiology, Faculty of Agricultural Sciences, Aligarh Muslim University, Aligarh- India 202002.
- Aminov, R.I., Mackie, R.I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiology Letters; 2007; 271: 147–161.
- Ansari, MI., Malik, A. Seasonal variation of different microorganisms with nickel and cadmium in the Industrial wastewater and agricultural soils. Environment Monitoring Assessment; 2010; 167: 151-163.
- Calomiris, J., Armstrong, L., Seidler, J. Association of metal tolerance with multiple antibiotic resistances of bacteria isolated from drinking water. Appl. Environmental Microbiology; 1994; 47: 1238-1242.
- Chitnis, V., Chitnis, D., Patil, S., Kant, R. Hospital effluent a source of multiple drug resistance bacteria; 2000; pp. 989-991. Current science. 79.
- Dantas, GM., Sommer, OA. The complex interplay between resistome genotypes and resistance phenotypes; 2012.
- Dhakephalker, PK., Chopade, BA. High levels of multiple metal resistance and its correlation to antibiotic resistance in environmental isolates of Acinetobacter. Biometals; 1994; 7: 67-74.
- Dhanorkar, DV., Tambekar, DH. Studies on multidrug resistance pattern of clinical isolates. 45th Annual Conference of Association of Microbiologist of India; 2004.
- Elmanama, A. Journal of Al-Aqsa University (natural sciences series); 2006; 10:108.
- Ghosh, A., Singh, A., Ramteke, P., Singh, V. Characterization of large plasmids encoding resistance to toxic heavy metals in Salmonella abortusequi. Biochemical and biophysical research communications; 2000; 272(1): 6-11
- Giller, K., Witter, E., McGrath, SP. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils. A review, Soil Biology & Biochemistry; 1998; 30: 1389-1414.
- Kado, CI., Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol; 1981; 145: 1365-1373.
- Khan, MKR., Malik, A. Antibiotic resistance and detection of ²- lactamase in bacterial strains of Staphylococci and Escherichia coli isolated from foodstuffs. World Journal of Microbiology & Biotechnology; 2001; 17: 863-868.
- Malik, A., Aleem, A. Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environmental Monitoring Assessment; 2011; 178: 293-308.
- Okeke. Emerg Infect Dis; 2007; 13(11): 1640-1646.
- Pathak, SP., Gopal, k. Antibiotics resistance and metal tolerance among coliform sp. From drinking water in a hilly area. Journal of Environmental Biology; 1994; 15: 139-147.
- Rachel, E., Rosenberg, G., Shirley, A., Shawn G., Johnnie, A., Davis, Xin He., Ashish, G., Lara, M. Methicillin-Resistant Staphylococcus aureus(MRSA) Detected at Four U.S. Wastewater Treatment Plants. Environ Health Perspect; 2001
- Rouch, DA., Lee, BT., Morby, AP. Understanding cellular responses to toxic agents. A model for mechanism choice in bacterial metal resistance. Journal of Indian Microbiology; 1995; 14: 132-141.
- Scottel, L., Mandal, A., Clerk, D., Silver, S., Hedges, RW. Volatilization of mercury and organomercurials determined by F factor system in enteric bacilli. Nature; 1974; 251: 335 – 337.
- Vishal, D., Ashok, J., Rakesh, K. Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India; 2010.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


