ISSN: 0973-7510
E-ISSN: 2581-690X
This survey aimed to study the occurrence of Listeria species in fish and meat samples and characterization of their virulence genes. Over all, Listeria spp. was found in 25.22% samples out of which 9.0% and 16.21% were L. monocytogenes and L. innocua, respectively. L. monocytogenes (n=10) belonged to 4b, 4d and 4e serovars. All the isolates revealed presence of virulence genes- plcA and iap, while plcB gene was also present in 90% of the isolates. The occurrence of L. monocytogenes in samples shows cogent evidence for their zoonotic potential and has public health significance.
Listeria spp., Prevalence, Serotypes, Virulence gene, Antimicrobials, Susceptibility
The genus Listeria is facultatively anaerobic, rod-shaped bacteria and dispersed in food, environmental and clinical samples1. L. monocytogenes is a known pathogen, whereas the pathogenicity of L. seeligeri, L. ivanovii and L. innocua have been less documented2. Listeria monocytogenes is disseminated from animals to humans through fecal-oral route. The fatality rate is higher (30 to 75%) especially in pregnant women, neonates, aged people and people with grievous underlying disease conditions like immune-suppression3. Ninety percent of cases of Listeria contamination due to use of contaminated food products are reported4.
Listeria monocytogenes was differentiated into 13 serotypes5. Only serotypes 1/2a, 1/2b, 1/2c and 4b are responsible for 95% of human illnesses6. In 2012 in the US, serotypes 4b (54%) and 1/2a (28%) were most commonly identified7. Many virulence determinants act as essential factors for the pathogenesis of L. monocytogenes, including hlyA, inlA, inlB, inlC, inlJ, actA, iap and plcA gene8. The hlyA gene is the main virulence determinant in L. monocytogenes; a hemolysin gene. The pore-forming cytolysin listeriolysin O is encoded by this hlyA gene. The product of hlyA is the first virulence factor which play a vital role in the L. monocytogenes pathogenesis, helps in intracellular parasitism9.
Little information is available on Listeria contamination in animal originated foods from North-East India. This survey aims to estimate the Listeria spp., identify the main serotypes and to screen the L. monocytogenes isolates for presence of different virulent genes.
Collection of Samples
One hundred and eleven (111) samples, including fish gill, fish intestine and beef (15 each), chicken and pork (24 each) and mutton (18) were collected from various markets in Aizawl, Mizoram, a hilly state in the North-Eastern region India. The samples were collected aseptically by maintaining cold chain and were processed for microbiological analysis within 24 h. This study was conducted for a period of one year from July 2018 to June 2019.
Isolation of Listeria
For isolation of Listeria, ISO 11290 method was used. Each sample was inoculated aseptically in Half Fraser Broth followed by incubation at 30°C. It was subcultured in Fraser broth, further streaked onto PALCAM agar and incubated at 37°C. The grey-green colonies on PALCAM agar with diffuse black-zone was plated on Tryptone Soya Yeast Extract agar. Gram staining and Biochemical tests comprising catalase test, fermentation and acid production from various sugars, followed by MR/VP tests were performed for identification of the isolates. All the Listeria isolates and the control strain were preserved in nutrient agar stab (with 1.5% agar agar) and 16% glycerol for subsequent analysis. L. monocytogenes ATCC 13832 was utilized as the standard strain.
Confirmation of Listeria by PCR
Listeria were cultured in Brain Heart Infusion Broth for overnight for extraction of genomic DNA using Bacterial DNA isolation kit (Genaid, Taiwan). To confirm the suspected isolates, PCR was performed by amplifying Listeria genus-specific prs gene using specific primer10 (1st BASE, Malaysia) (Table 1). To confirm L. monocytogenes isolates, primers specific for the virulence associated hlyA gene11 were self-designed using primer BLAST online tool. For detection of L. innocua, PCR amplification of in9 gene was targeted24. A total volume of 25 ml mixture comprising 12.5 ml of 2X Mastermix, 0.5 ml (10 pmol) each of bi-directional primers, DNA (1 ml) and nuclease-free water (10.5 ml) was prepared for PCR. The amplification of prs, hlyA and in9 genes was done by 5 min. denaturation at 95°C, accompanied by thirty cycles of 95°C (30 sec.), 53°C (30 sec.), 72°C (30 sec.) and final elongation at 72°C (5 min.).
Table (1):
Primers used for identification of Listeria species.
Species |
TargetGene |
Primers |
Sequences |
ProductSize (bp) |
References |
|---|---|---|---|---|---|
Genus Listeria |
prs |
prs-F prs-R |
5¢-GCTGAAGAGATTGCGAA-3¢ 5¢-AGAAGCAAAGAAACCTTGGATTTGCGG-3¢ |
370 |
Michel et al.10 |
L. monocytogenes |
hlyA |
hlyA-F hlyA-R |
5¢-GCTTTTGACGCTGCCGTAAG-3¢ 5¢-GCAACGTATCCTCCAGAGTGATCG-3¢ |
335 |
Designed. |
L. innocua |
in9 |
in9-F in9-R |
5¢-GGCTTCAGCGATTCTTCCG-3¢ 5¢-GCCCGATTTCCTCACTGTCTAA-3¢ |
421 |
Tingting et al.24 |
Identification of serogroup
A simplex PCR was used to determine the serogroup using group-specific primers (Table 2). Amplification of ORF2819 was used to identify 1/2b and 3b serovars and amplification of both ORF2819 and OFR2110 to identify 4b, 4d and 4e serovars. This PCR does not differentiate within serovars.
Table (2):
Primers used for identification of L. monocytogenes serogroups.
Target Gene |
Primer |
Sequences |
Product Size (bp) |
Serotype Specificity |
References |
|---|---|---|---|---|---|
ORF2819 |
ORF2819-F ORF2819-R |
5¢-AGCAAAATGCCAA AACTCGT-3¢ 5¢-CATCACTAAAGCC TCCCATTG-3¢ |
470 |
1/2b,3b,4b,4d,4e |
Michel et al.10 |
ORF2110 |
ORF2110-F ORF2110-R |
5¢-AGTGGACAATTGATTG GTGAA-3¢ 5¢-CATCCATCCCTTA CTTTGGAC-3¢ |
597 |
4b,4d,4e |
Michel et al.10 |
A 25 µl volume of reaction mixture was prepared for PCR same as prepared for Listeria identification. Listeria monocytogenes (ATCC 13832) serotype 4b was used as the positive control. PCR conditions for amplification were as previously described by Michel et al.10
Virulence associated gene detection
After confirming the isolates by hlyA gene, a simplex PCR was done to determine the three virulence genes, plcA, plcB and iap (Table 3). The composition of reaction mixture was the same as for the other genes used in this study. PCR conditions for amplification of all the three virulence genes were standardized at denaturation at 95°C (5 min.), followed by thirty cycles of 95°C (30 sec.), 52°C (30 sec.), 72°C (30 sec.) and a final elongation at 72°C (5 min.).
Table (3):
Primers used for detection of virulence-associated genes in L. monocytogenes isolates.
| Target Gene | Primer | Sequences | Product Size (bp) | References |
|---|---|---|---|---|
| plcA | plcA-F plcA-R |
5¢-TCCGCTCTACCTGACACAAC-3¢ 5¢-TCGTTGCTGTTTTGCTCGTC-3¢ |
317 | The primers were designed in this study. |
| plcB | plcB-F plcB-R |
5¢-TTGCCGGGTTTTG CTAATGC-3¢ 5¢-TAGTCCGCTTTCGC CCTTTT-3¢ |
323 | |
| iap | iap-F iap-R |
5¢-AGTGGCGCTGG TGTTGATAA-3¢ 5¢-GCTGTTTGTTGTT GCGTTGC-3¢ |
605 |
Prevalence of Listeria
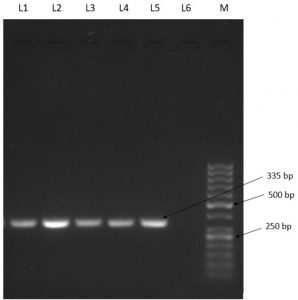
Occurrence of Listeria spp. collected from various markets in Aizawl is shown in Table 4. Out of 111 samples, 28 (25.22%) were positive for Listeria., 10 (9.0%) for L. monocytogenes and 18 (16.21%) for L. innocua. The hlyA gene was found in L. monocytogenes using PCR (Fig. 1). Incidence of L. monocytogenes rated higher in mutton and pork (16.6% each) followed by chicken (12.5%). The incidence of L. innocua was found in mutton (50%) followed by fish gill and pork (33.33% each), chicken (16.66%), fish intestine and beef (6.66%).
L1: Positive control (ATCC 13832), L2-L5: Listeria isolates, L6:Negative control, M:50bp Ladder
Fig.1. Identification of L. monocytogenes by PCR
Table (4):
Prevalence of Listeria spp. in various food samples.
| Type of Sample | No. of samples tested | No. of samples positive for Listeria spp. (%) | Total No. of Positive Samples (%) | |
|---|---|---|---|---|
| L. monocytogenes | L. innocua | |||
| Fish gill | 15 | 0 | 5 (33.3%) | 5 (33.33%) |
| Fish intestine | 15 | 0 | 1 (6.6%) | 1 (6.66%) |
| Pork | 24 | 4 (16.6%) | 4 (16.6%) | 8 (33.33%) |
| Mutton | 18 | 3 (16.6%) | 6 (33.3%) | 9 (50%) |
| Chicken | 24 | 3 (12.5%) | 1 (4.16%) | 4 (16.66%) |
| Beef | 15 | 0 | 1 (6.6%) | 1 (6.66%) |
| Total | 111 | 10 (9%) | 18 (16.21%) | 28 (25.22%) |
Nayak et al.12 recorded the overall occurrence of Listeria in 9% of foods and the maximum incidence was found in milk samples (16%) followed by meat and fish (8% each), and milk (4%). Barbuddhe13 and Nayak14 with their co-workers reported occurrence of Listeria from 10.17% and 6.7% meat samples, respectively. Murtiningsih and Sunarya15 reported L. innocua as the superior species isolated from fish and seafood samples (11.3%). Variation in occurrence of Listeria spp. reported by different workers might be owing to the difference in specimen, source and variation in hygienic condition maintained in different locations.
Serotyping
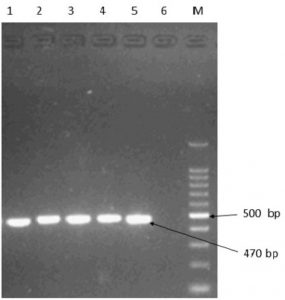
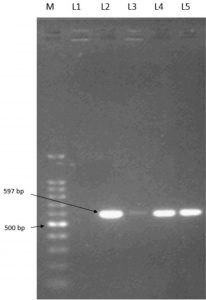
All L. monocytogenes showed PCR amplification of both ORF2819 and ORF2110, which identified them as serovars 4b, 4d or 4e (Fig.’s 2 and 3). Serovars 1/2a, 1/2b and 4b are linked with listeriosis16 and 4b was linked with major outbreaks of listeriosis17. Thus, the presence of serovar 4b in foods inflicts greater warnings on human health18. Serotype 4b was proclaimed from 37.2% of foods in Turkey19 as well in India and the United States of America with 60.4%20 and 16.4%21, respectively. The difference in rate of the prevalence might be due to contradiction in the types of foods selected for each investigation. According to Michel et al.10, the serotypes 3a, 4d, and 4e are infrequent in foods, therefore the serovars 4b, 4d and 4e were marked as 4b serogroup. In the present study, all the isolated L. monocytogenes were confirmed as 4b serogroup which is a major public health concern, since isolates of L. monocytogenes belonging to this serogroup are potential human pathogens.
L1: Positive control (ATCC 13832), L2-L5: Listeria isolates, L6: Negative control, M: 100 bp Ladder
Fig.2. Identification of L.monocytogenes serotypes (1/2b,3b,4b,4d,4e) isolates by PCR
M: 100 bp Ladder, L1: Negative control, L2: Positive control (ATCC 13832), L3-L5: Listeria isolates
Fig.3. Identification of L.monocytogenes serotypes (4b, 4d, 4e) isolates by PCR
Virulence-associated genes
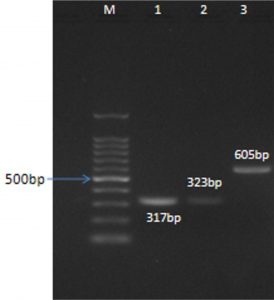
Virulence-associated gene profiling in L. monocytogenes was done by PCR amplification of three genes (Fig. 4). The plcA and iap genes showed presence in all the isolates, while plcB gene was present in 90% of the isolates.
M: 100 bp Ladder, L1: PCR Product (plcA gene), L2: PCR Product (plcB gene), L3: PCR Product (iap gene)
Fig.4. Virulent gene detection in L.monocytogenes by PCR
In a study in North-East India, Pegu et al.22 found virulence-associated genes, hlyA (40.7%), iap (29.6%), plcA (40.7%) and plcB (22.2%) of 27 isolates from fish. The mechanism of pathogenicity of L. monocytogenes is stated to be usually associated with production of haemolysin encoded by hlyA gene23. In our study, all the suspected isolates exhibited existence of hlyA, plcA and iap genes. This indicated that the isolates are potentially pathogenic. Occurrence of such pathogenic bacteria in meat has significant public health implications.
This seems to be the first study on isolation and serogrouping of L. monocytogenes in foods from Mizoram. The prevalence of serogroup 4b reported to be associated frequently with human listeriosis seems to be of serious public health concern. These virulent strains can contaminate other foods during processing, packaging or storage. Hence, large-scale screening of various food and food products sold in the markets of northeast India at regular intervals for the incidence of virulent strains is of utmost importance.
ACKNOWLEDGMENTS
The Authors thank the Advanced State Level Biotech Hub, Mizoram University, Aizawl, Mizoram funded by the Department of Biotechnology, New Delhi Government of India for the support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by Advanced State Level Biotech Hub, College of Veterinary Science, Khanapara, Assam funded by the Department of Biotechnology, New Delhi Government of India (Grant number: BT/04/NE/2009 Dtd.22.12.2010).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed in this research are included in the manuscript.
- Den Bakker HC, Warchocki S, Wright EM, et al. Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int J Syst Evol Microbiol. 2014;64:1882-1889.
Crossref - Perrin M, Bremer M, Delamare C. Fatal cases of Listeria innocua bacteria. J Clin Microbiol. 2003;41:5308-5309.
Crossref - Mead PS, Slutsker L, Dietz V, McCaig L, Bresee J . Food related illness and death in the United States. Emerg Infact Dis. 1999;5(5):607-625.
Crossref - Liu H, Lu L, Pan Y, et al. Rapid detection and differentiation of Listeria monocytogenes and Listeria species in deli meats by a new multiplex PCR method. Food Control. 2015;52:78-84.
Crossref - Jamali H, Radmehr B, Thong KL. Prevalence, characterization, and antimicrobial resistance of Listeria species and Listeria monocytogenes isolates from raw milk in farm bulk tanks. Food Control. 2013;34(1): 121-125.
Crossref - Huang B, Fang N, Dimovski K, Wang X, Hogg G, Bates J. Observation of a new pattern in serogroup-related PCR typing of Listeria monocytogenes 4b isolates. J Clin Microbiol. 2011;49(1):426-429.
Crossref - Centers for Disease Control and Prevention (CDC). 2014 National Listeria Surveillance annual summary 2012. Atlanta, Georgia: US Department of Health and Human Services, CDC.
- Sant’Ana AS, Igarashi MC, Landgraf M, Destro MT, Franco BD. Prevalence, populations and pheno-and genotypic characteristics of Listeria monocytogenes isolated from ready-to-eat vegetables marketed in Sao Paulo, Brazil. Int J Food Microbiol. 2012;155:1–9.
Crossref - Vazquez-Boland JA, Kuhn M, Berche P, et al. Listeria Pathogenesis and Molecular Virulence Determinants. Clin Microbiol Rev. 2001;14(3):584–640.
Crossref - Michel D, Carmen B, Philippe G, Christine J, Paul M. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J Clin Microbiol. 2004;42(8):3819–3822.
Crossref - Kocaman N, Sarimehmetoglu B. Isolation of Listeria monocytogenes in lamb meat and determination of the antibiotic resistance. Ankara Univ Vet Fak Derg. 2017;64:273-279.
Crossref - Nayak DN, Savalia CV, Kalyani IH, Kumar R, Kshirsagar DP. Isolation, identification, and characterization of Listeria spp. from various animal origin foods.Veterinary World. EISSN: 2231-0916,2015;8(6):695-701.
Crossref - Barbuddhe SB, Chaudhari SP, Malik SVS. The occurrence of pathogenic Listeria monocytogenes and antibodies against Listeriolysin-O in buffaloes. J Vet Med B 2002;49:181-184.
Crossref - Nayak JB, Brahmbhatt MN, Savalia CV, et al. Detection and characterization of Listeria species from buffalo meat. Buffalo Bull. 2010;29:83-87.
- Murtiningsih RE, Sunarya HN. Incidence of Listeria in Fish and Seafood in Indonesia In: Fish utilization in Asia and the Pacific. Proceedings of the APFIC Symposium, Beijing, China, RAP Publication (FAO). Regional Office for Asia and the Pacific, FAO, Bangkok, Thailand. Asia-Pacific Fishery Commission. 1998;270-276.
- Hossein J, Kwai LThong. Genotypic characterization and antimicrobial resistance of Listeria monocytogenes from ready-to-eat foods. Food Control. 2014;44:1-6.
Crossref - Buchrieser C, Brosch R, Catimel B, Rocourt J.Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can J Microbiol. 1993;39:395–401.
Crossref - Ward TJ, Usgaard T, Evans P. A targeted multilocus genotyping assay for lineage, serogroup and epidemic clone typing of Listeria monocytogenes. Appl Environ Microb. 2010;76:6680–6684.
Crossref - Ayaz ND, Erol I. Relation between serotype distribution and antibiotic resistance profiles of Listeria monocytogenes isolated from ground Turkey. J Food Prot. 2010;73(5):967-972.
Crossref - Shen J, Rump L, Zhang Y, Chen Y, Wang X, Meng J. Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiology,2013;35(1):58-64.
Crossref - Barbuddhe SB, Doijad SP, Goesmann A, Hilker R, Poharkar KV, Rawool DB, et al. Presence of a widely disseminated Listeria monocytogenes serotype 4b clone in India. Emerging Microbes and Infections. 2016;5:55.
Crossref - Pegu RK, Sen A, I Shakuntala, Kumar A, Ahmed G. Prevalence of Listeria Monocytogenes in Freshwater Fish of Northeast India and Their Molecular Characterization by PCR. Indian Journal of Hill Farming. 2017;30(2):220-226.
- Gellin BG, Broome CV, Bibb WF, Weaver RE, Gaventa S, L Mascola. The Epidemiology of Listeriosis in United States-1986. Listeriosis Study Group. American j Epidemiol. 1991;133: 392-401.
Crossref - Tingting T, Qiming C, Xiaomei B, Fengxia L, Zhaoxin Lu. Investigation on prevalence of Listeria spp. and Listeria monocytogenes in animal derived foods by multiplex PCR assay targeting novel genes. Food Control. 2017;73:704-711.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.