ISSN: 0973-7510
E-ISSN: 2581-690X
The relative high burden of morbidity and mortality caused by Staphylococcus aureus (SA) in healthcare and community settings is a major concern worldwide. It can cause invasive infections, sepsis and deaths. Despite progress in methicillin-resistant S. aureus (MRSA) prevention in healthcare settings, there is a critical need for assessment of the problem in both healthcare and community settings. This study was conducted for examining the prevalence, risk factors and antimicrobial susceptibility of MRSA in Mansoura University Hospitals (MUHs), Egypt. Samples were collected from patients in MUHs with clinically suspected nosocomial infections. MRSA isolates were identified by the standard bacteriological methods, biochemical reactions and disc diffusion method as recommended by the Clinical & Laboratory Standards Institute (CLSI), then confirmed by MecA gene PCR. A total of 2006 isolates was obtained. SA (32%) was the most frequently isolated pathogen. MRSA (130 isolates) represented 20% of SA and 6.48 % of all isolates. The mecA PCR identified SA as MRSA in 99.2% of cases. MRSA was isolated with another organism (mostly Gram-negative bacilli) from 40.8% of cases while 59.2% of MRSA was isolated alone. The most important reported risk factors for MRSA infections were prolonged hospital stays, recent antibiotic therapy, ICU admission, indwelling devices and presence of surgical sutures. MRSA was resistant to many antibiotics but sensitive to vancomycin in 99.2% of cases. Minimizing exposure to the risk factors with rapid diagnosis of MRSA infections are essential for early initiation of appropriate antibiotic treatment and limitation of the non-optimal use of glycopeptides and deaths.
Antibiotic resistance, b-lactamase, b-lactams, MRSA, Staphylococcus aureus.
SA is a leading cause of human bacterial infections in healthcare facilities and in the community all over the world. The severity of these infections varies widely from minor skin infections to sepsis, fatal necrotizing pneumonia and deaths. In the United States, an estimated 19,832 deaths from 119,247 SA bloodstream infections occurred in 20171.
About 20–30% of the global population is persistently colonized with SA in the anterior nares, with 60–100% of persons suspected to be transiently colonized at some time during their lives. SA infection frequently follows its nasal carriage2.
Antibiotic resistance in SA is becoming increasingly significant3. The b-lactamase-resistant penicillins (oxacillin, methicillin, flucloxacillin and cloxacillin) were developed as a treatment for penicillin-resistant SA. In 1959, methicillin, a semi-synthetic penicillin, was introduced to treat SA infections, but only 2 years later, the 1st case of MRSA was reported in England. Despite this, MRSA generally remained an uncommon finding even in hospitals till the 1990s when the occurrence of MRSA has rapidly increased as nosocomial and community-acquired infections resulting in a serious problem worldwide. Children, old ages, pregnant females and immunocompromised patients, for example those suffering from cancer, blood dialysis and transplantation, frequently become infected by SA, mostly the multi-antibiotic-resistant MRSA strains leading to longer hospitalization duration, more health care costs, treatment failures and deaths4.
The prevalence of MRSA infections shows considerable worldwide geographical variation, that has been related to efforts to decrease colonization and spread of these highly adaptive organisms. Multidrug resistant epidemic can occur from the endemic organisms. MRSA prevalence was reported to be 62.5% in Pakistan and ranged from 12% to 49.4% in six different hospitals of Saudi Arabia. In European countries, MRSA rates varied from 45% in Ireland, Italy, Belgium, Greece and United Kingdom down to 0.6% in Sweden and most nearby countries. The low MRSA prevalence in Sweden, Denmark, Norway, Finland, Iceland and the Netherlands has been suspected to be due to proper infection control measures in these countries. In the Mediterranean area, the highest overall proportions of MRSA were reported in Egypt, Jordan and Cyprus, where more than 50% of the SA blood culture isolates were resistant to methicillin5.
The commonest effective therapeutic option against the multi-antibiotic-resistant MRSA is VA or linezolid6-7. VA is a glycopeptide antibiotic that is active against Gram-positive bacteria including Staphylococci and Enterococci, however, Gram-negative bacteria are naturally resistant to it, mainly because of its outer membrane that acts as a penetration barrier8.
The non-optimal use of VA in treatment of methicillin-resistant Staphylococcal infections (both coagulase-positive &-negative), preceded the development of VA-resistant Staphylococci. Clinically, the first reported Staphylococcal resistance to VA was in a strain of Staphylococcus haemolyticus. In 1997, the 1st reported VA intermediate-resistant SA (VISA) was in Japan, and then subsequent cases were reported in other countries. The VISA isolates were all MRSA9. The 1st reported VRSA cases were in the USA, Jordan and Brazil in the 200210.
A critical assessment of the prevalence and the recent trends of MRSA is central to formulating a framework of approaches and informing public health policy to further prevent SA infections.
Objective
Screening for the prevalence, risk factors and antimicrobial susceptibility of MRSA in Mansoura University Hospitals (MUHs), Egypt.
Bacterial strains
The study was conducted over a period of 12 months starting from January till December, 2015. During the 1st 6 months, 130 MRSA isolates were isolated from patients with clinically suspected nosocomial infections. The isolates were from different medical and surgical departments of the MUHs dealing with the Microbiology diagnostics and infection control unit (MDICU) in the Microbiology and Immunology department, Faculty of Medicine, Mansoura University, Egypt.
Materials used
- Equipments for samples collection and processing: Sterile cotton swabs, wide-necked leak-proof containers, syringes, Eppendorf tubes and pipettes.
- Blood culture bottles (Egyptian Diagnostic Media) (EDM): The Composition: Trypticase (1.5 gm%), Dextrose (0.5 gm%), Yeast extract (0.25 gm%),Agar (0.075 gm%), L-cystine (0.05 gm%) Sodium thioglycolate (0.05 gm%), Resazurin (0.0001 gm%)
- Media: Stuart’s transport medium (STM) (Oxoid), Nutrient agar (Oxoid), Blood agar (Oxoid), Chocolate agar (Oxoid), MSA (Oxoid), Cysteine Lysine Electrolyte Deficient (CLED) medium (Becton-Dickinson), Mueller– Hinton (MH) agar (Oxoid), DNase agar (Oxoid), Nutrient broth (Oxoid).
- Chemicals: Hydrogen peroxide (3% H2O2), Hydrochloric acid (3.6% HCl).
- EDTA-anticoagulated human plasma.
- Antibiotics: Antibiotic disks (Oxoid).
- McFarland 0.5 standards that were used as references to adjust the bacterial suspensions’ turbidity as recommended by the CLSI 2014 11.
Culture conditions and Antibiotics’ sensitivity testing
The isolates were obtained as pure growth from the clinical samples and identified by the standard bacteriological methods and biochemical reactions12-13. Antibiotics’ sensitivity testing, including the identification of methicillin resistance by the cefoxitin-based method, was conducted by the disc diffusion method as recommended by the CLSI 201411.
Confirmation of MRSA isolates by MecA gene amplification PCR
Chromosomal DNA extraction was done according to Aushbel et al., 199014 to get the DNA templates. MecA gene amplification by PCR was done using a pair of primers (Sigma) selected according to Bignardi et al,199615 and the sequence of the primer used was:
F: 5′-CTCAGGTACTGCTATCCACC-3′
R: 5′-CACTTGGTATATCTTCACC-3′
The thermal cycler program was adjusted and proceeded as the following; initial denaturation for 5 minutes at 95°C, thirty cycles of 30 seconds denaturation then 30 seconds annealing then 30 seconds extension at 94°C, 42°C, 72°C respectively, followed by final extension for 10 minutes at 72°C. Agarose gel (1.5%) electrophoresis of the amplified MecA gene was done according to Davis et al, 198616 using the DNA molecular marker (100 bp DNA Ladder; Lonza Rockland. Inc, USA) to detect the expected (448 bp) bands visualized by staining with ethidium bromide (EB).
Determination of the MIC of VA on MRSA
This was done according to Sarker et al, 2007 using a microtitre plate incorporating resazurin as an indicator of bacterial growth17.
Statistical analysis
The c2 was done using a computer programme embedded in Microsoft Excel. Tests were considered non-significant if the probability of error is equal or more than 5% (p ³ 0.05), significant if (p < 0.05), highly significant if (p < 0.001)18.
Different samples were received from 2856 patients with clinically suspected nosocomial infections, from different medical and surgical departments of the MUHs dealing with the MDICU in the Microbiology and Immunology department, Faculty of Medicine, Mansoura University, Egypt. A total of 3047 clinical samples were collected. Samples were examined in MDICU laboratory after being processed and cultured on appropriate media under appropriate incubation conditions. Infections were detected in 1881 samples. Single pathogen was detected in 1756 samples whereas 125 samples yielded 2 pathogens. Consequently, the total number of the isolated nosocomial pathogens was 2006.
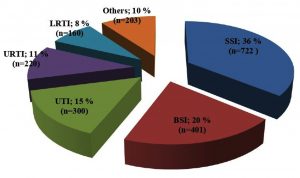
Surgical site infection (SSI) [722 isolates] represented the commonest nosocomial infection during the study period (36%) (Fig. 1). SA [642 isolates] was the most frequently isolated pathogen representing 32% of all isolates while Klebsiella species was the most common Gram-negative organism accounting for 22.1%.
Fig. 1. Different types of nosocomial infections in MUHs: A total of 2006 nosocomial isolates was obtained. Surgical site infection (SSI) represented the commonest nosocomial infection (36%) followed by blood stream infection (BSI), urinary tract infection (UTI), upper respiratory tract infection (URTI) then lower respiratory tract infection (LRTI).
One hundred & thirty MRSA isolates were detected by phenotypic methods representing 20% of SA. The pattern of resistance to methicillin was homogenous in 66 cases (50.8%) wile heterogenous in 64 cases (49.2%). MRSA represented 6.48% of all nosocomial isolates. One hundred and twenty-nine isolates (99.2% of cases) were positive for the mecA gene by PCR.
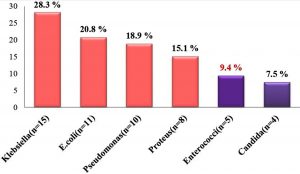
MRSA was isolated alone from 59.2% of cases (77 cultures) while 40.8% of MRSA isolates (53 cultures) were isolated with another organism. Gram negative bacilli (44 isolates;) were common co-pathogens as the following; Klebsiella (15 isolates), E. coli (11 isolates), Pseudomonas (10 isolates) and Proteus (8 isolates) (Fig. 2). All the associated Enterococci (5 isolates) were VA sensitive.
Fig. 2. Associated co-pathogens with nosocomial MRSA isolates: Gram negative bacilli were common co-pathogens. All the associated Enterococci were VA sensitive.
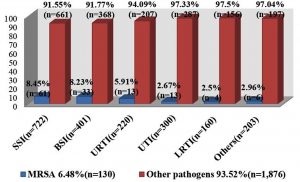
MRSA causing SSI (61 isolates) was the most predominant type for MRSA nosocomial infections representing (8.45%) “Fig. 3”. MRSA was isolated from surgical patients in 58.46% of cases (76 cases) and was from medical patients in 41.54% of cases (54 cases).
Fig. 3. Distribution of MRSA isolates according to the type of nosocomial infection in MUHs: Surgical site infection (SSI) was the most prevalent (8.45 %) followed by blood stream infection (BSI), upper respiratory tract infection (URTI) urinary tract infection (UTI), then lower respiratory tract infection (LRTI).
The most important reported risk factors for the development of MRSA infections were recent antibiotic therapy (especially b–lactams), prolonged hospital stays (especially if more than 7 days), indwelling devices (especially if more than 2 devices simultaneously), ICU admission and presence of surgical sutures (Table 1).
Table (1):
The risk factors for methicillin resistance among Staphylococcus aureus isolates.
| Risk factor | 130 MRSA | 512 MSSA(control) | Odds ratio (95%CI) | P value | |
|---|---|---|---|---|---|
| Antibiotic therapy | 117(90.0%) | 235(45.9%) | 10.61 (5.66-20.27) | <0.001** | |
| Hospitalization > 1 week | 110(84.6%) | 214(41.8%) | 7.66 (4.50-13.16) | <0.001** | |
| Indwelling devices | IV catheter | 97(74.6 %) | 125(24.4 %) | 9.10 (5.72-14.54) | <0.001** |
| Urinary catheter | 64(49.2%) | 201(39.3%) | 1.50 (1.0-2.25) | 0.049* | |
| Wound drains | 48(36.9 %) | 111(21.7 %) | 2.11 (1.37-3.26) | 0.005* | |
| Orthopedic prosthesis | 10(7.7%) | 33(6.4%) | 1.21 (0.54-2.64) | 0.75 | |
| Central venous line | 7(5.4%) | 20(3.9%) | 1.40 (0.52-3.59) | 0.61 | |
| ETT (Ventilator) | 7(5.4%) | 17(3.3%) | 1.66 (0.61-4.36) | 0.39 | |
| Old age (≥ 50) | 32(24.6 %) | 110(21.5 %) | 1.19 (0.74-1.92) | 0.51 | |
| ICU patients | 33(25.4 %) | 87(17.0 %) | 1.66 (1.02-2.69) | 0.039* | |
| Previous hospital admission | 26(20.0%) | 76(14.8%) | 1.43 (0.85-2.41) | 0.19 | |
| Diabetes mellitus | 32(24.6%) | 166(32.4%) | 0.76 (0.47-1.21) | 0.27 | |
| Surgical sutures | 76(58.5%) | 247(48.2%) | 1.51 (1.00-2.27) | 0.047* | |
| Pressure ulcers | 12(9.2%) | 44(8.6%) | 1.08 (0.52-2.20) | 0.96 | |
| Burn | 13(10.0%) | 39(7.6%) | 1.35 (0.66-2.71) | 0.48 | |
| Malignancy | 19(14.6%) | 53(10.4%) | 1.48 (0.81-2.69) | 0.22 | |
This table illustrates the reported risk factors for infection with MRSA. Recent antibiotic therapy, prolonged hospital stays, indwelling devices (such as IV lines, urinary catheters and wound drains), ICU admission & presence of surgical sutures were the most important reported risk factors for the development of MRSA infections. * Significant (p < 0.05); **Highly significant (p < 0.001).
Regarding b-lactams as a risk factors for methicillin resistance, they were prescribed alone in 73.1% of cases and in combination with other antibiotics in 7.7% of cases (Table 2).
Table (2):
Antibiotic therapy of the 130 MRSA infected patients during their hospital stay
| Antibiotic therapy | Patients infected | ||
|---|---|---|---|
| No. | % | ||
| No antibiotic therapy | 13 | 10.0 | |
| Single antibiotic type | β-lactam antibiotics | 95 | 73.1 |
| Others | 12 | 9.2 | |
| More than one antibiotic type (including β-lactams) | 10 | 7.7 | |
| Total | 130 | 100 | |
This table shows the antibiotics prescribed for the 130 MRSA infected patients during their hospital stay. The β-lactam antibiotics were the commonest.
The highest overall age incidence was in the age group 40-50 years and nearly the prevalence of MRSA infections among males is equal to the prevalence of MRSA infections among females (Table 3).
Table (3):
Age and gender distribution of the 130 MRSA infections.
| Age group (years) | Test group130 MRSA | |
|---|---|---|
| Males | Females | |
| <10 | 5(62.5%) | 3(37.5%) |
| 10- | 3(30.0%) | 7(70.0%) |
| 20- | 4(57.1%) | 3(42.9%) |
| 30- | 10(37.0%) | 17(63.0%) |
| 40- | 19(41.0%) | 27(59.0%) |
| 50- | 13(54.2%) | 11(45.8%) |
| 60- | 3(37.5%) | 5(62.5%) |
| Total | 57(43.8%) | 73(56.2%) |
This table shows the age and the gender distribution of MRSA patients. The highest overall age incidence was in the age group 40-50 years.
All isolates were resistant to penicillin, ampicillin and cephradine antibiotics. MRSA isolates were resistance to rifampicin (10%), fusidic acid (64%), ciprofloxacin (65%), trimethoprim-sulfamethoxazole (71%), gentamicin (80%), erythromycin (88%), clindamycin (89%) and tetracycline (92%). Fortunately, one hundred and twenty-nine MRSA isolates (99.2%) in the current study, were sensitive to VA (Table 4).
Table (4):
Antibiotic sensitivity of the MRSA isolates by the disc diffusion method.
| Antibiotic | Result | MRSA (No. = 130) | |
|---|---|---|---|
| No. | % | ||
| Cefoxitin (FOX) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Methicillin (MET) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Oxacillin (OX) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Penicillin G (P) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Ampicillin (AMP) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Amoxycillin/K+ clavulanate (AMC) | Sensitive | 1 | 0.8 |
| Resistant | 129 | 99.2 | |
| Cephradine (CE) | Sensitive | 0 | 0 |
| Resistant | 130 | 100 | |
| Cefuroxime (CXM) | Sensitive | 4 | 3 |
| Resistant | 126 | 97 | |
| Clindamycin (DA) | Sensitive | 14 | 11 |
| Resistant | 116 | 89 | |
| Erythromycin (E) | Sensitive | 16 | 12 |
| Resistant | 114 | 88 | |
| Gentamicin (CN) | Sensitive | 26 | 20 |
| Resistant | 104 | 80 | |
| Tetracycline (TE) | Sensitive | 10 | 8 |
| Resistant | 120 | 92 | |
| Sulphamethoxazole/Trimethoprim (SXT) | Sensitive | 38 | 29 |
| Resistant | 92 | 71 | |
| Ciprofloxacin (CIP) | Sensitive | 45 | 35 |
| Resistant | 85 | 65 | |
| Fusidic acid (FD) | Sensitive | 47 | 36 |
| Resistant | 83 | 64 | |
| Imipenem (IPM) | Sensitive | 72 | 55 |
| Resistant | 58 | 45 | |
| Rifampicin (RD) | Sensitive | 117 | 90 |
| Resistant | 13 | 10 | |
| Vancomycin (VA) | Sensitive | 129 | 99.2 |
| Resistant | 1 | 0.8 | |
As strains of SA with reduced susceptibility to many antibiotics continue to emerge, with increasing the rates of morbidity and death in the hospitals and community, there is a clinical need to fully characterize them and conduct a well-designed research and epidemiological studies. The current MRSA strains in hospitals and community are alarming condition to the clinicians19.
Development of resistance to antibiotics in developing countries, like ours, seems to be mainly related to the unreasonable usage of antibiotics due to their easy availability without prescription at the drug stores, non-optimal use in hospitals, animal husbandry, fisheries and agriculture20.
The accurate and rapid diagnosis of MRSA infections is of major importance. Susceptibility testing of MRSA may be problematic owing to the phenotypic heterogeneous resistance (heteroresistance) displayed by many isolates to anti-Staphylococcal b-lactams. Consequently, many laboratory methods have been developed to increase the resistance expression, including prolonging the incubation period to 24 hours and the supplementation of media with NaCl21.
This study aimed to estimate the role of MRSA as a causative agent of nosocomial infections in MUHs. During the study period, a total of 642 SA isolates were isolated. One hundred and thirty MRSA isolates were isolated representing 20% of all SA nosocomial infections. This result is nearly like the result of Viswanathan et al.22 who have reported that MRSA accounts for 20-40 % of all SA infections, and of Saunders and Holmes23 who reported that this percentage reached up to 30%.
Also, it was stated by Dhanalakshmi, et al.24 that methicillin resistance represented 31.3% of SA isolates. Moreover, Abd El-Baky et al.20 found that methicillin resistance represented 25.4% of SA isolates in the study conducted for documentation of VRSA in the Minia University, Egypt.
Higher rates were reported in U.S.A. hospitals by Diekema et al.25 who mentioned that MRSA accounts for 30-50% of all nosocomial SA isolates and Wisplinghoff et al.26, who found that the proportion of MRSA increased from 22% in 1995 to 57% in 2001.
Higher rates were reported in the Kingdom of Saudi Arabia by Baddour et al.27 who reported that MRSA accounted for 77.5% of all SA nosocomial infections in a study conducted in several hospitals in Riyadh, and by Alzolibani et al.28 who reported that 90% of SA strains were resistant to methicillin in a study conducted in the Qassim region for documentation of VRSA among children with atopic dermatitis. These higher rates could be attributed to the vulnerable study group.
In this study, MRSA constituted 6.48% of the total nosocomial infections. This result is not coming with the higher rates that were reported by Hsueh et al.29 who stated that a rapid emergence of nosocomial MRSA infection (from 26.3% in 1986 to 77% in 2001) was found in a university hospital in Taiwan.
The prevalence of MRSA infection shows marked variation. The difference in the MRSA rates is likely related to differences in the populations of the studies and variations in the infection control measures applied. Some investigators used active surveillance cultures, and others used only culture of clinical specimens. In addition, infection control measures for patients with MRSA colonization or infection in some hospitals are stricter than in other hospitals.
Out of the 130 MRSA isolates, MRSA was isolated alone from 77 cultures (59.2%) while MRSA was isolated with another organism from 53 cultures (40.8%). Gram negative bacilli were common co-pathogens that are resistant to VA required for the MRSA treatment, so they may need to be treated by b-lactam antibiotics which may induce VA resistance among MRSA isolates30,31.
SSIs were by far the commonest sites for MRSA infections in this study, followed by blood stream infections. Similar results have been obtained by Carla et al.32 who found that the most common sites infected by MRSA were surgical wounds (21%), intra venous sites (18%), and bacteremia (13%).
These results do not agree with the results coming from United Kingdom, Ireland and Greece that reported one of the highest rates of MRSA from bloodstream isolates (44%) in 200433.
In the present study, MRSA was noticed to be more prevalent in surgical patients (58.46%) than in medical patients (41.54%). In agreement with our results, Gordon and Lowy34 reported that MRSA was isolated from surgical patients, ICU patients and from medical patients represented (40%, 27%, 33% respectively).
Regarding the risk factors, it was revealed that recent antibiotic therapy (especially b-lactam antibiotics) and prolonged hospital stay for > 7 days are important risk factors for the expansion of MRSA infections (90.0% & 84.6% of cases, respectively). This was supported by many reports35-37. This may be owing, in part, to the more likelihood over time of becoming colonized with MRSA from either horizontal nosocomial transmission or endogenous emergence of resistance.
Similarly, Raygada and Levine38 have demonstrated a close association between recent antibiotic usage and the development of subsequent antibiotic resistance in both Gram positive and negative bacteria.
In the current study, b-lactams alone were the antibiotics prescribed for 73.1% of patients. Other drugs prescribed ranged among fluoroquinolones, monobactams, macrolides and aminoglycosides. These findings are consistent with Borg et al.39 who stated that mecA expression is either constitutive or inducible by some b-lactam antibiotics. Also, extensive antibiotics usage within a hospital may partly explain differences among hospitals in transmission rates of resistant organisms40.
During the study, it was noticed that invasive indwelling devices (such as IV lines, urinary catheters and wound drains), ICU admission and presence of surgical sutures were also important risk factors for the emergence of MRSA infections. This agreed with data mentioned by Ricarda et al.41
Moreover, the greatest incidence of acquisition of MRSA infections occurred when³ 2 different devices were used for the same patient. Likewise, Sadoyama and Gontijo-Filho42 concluded that most nosocomial MRSA infections occur in persons with multiple risk factors for infection.
Concerning the antibiotics sensitivity patterns of the MRSA isolates, it was noticed that, all isolates were resistant to penicillin, ampicillin and cephradine antibiotics. These findings agreed with Noto et al.43 who stated that more than 95% of patients with SA infections worldwide do not respond to first-line antibiotics such as penicillin or ampicillin.
During the study, MRSA was found to be resistance to ciprofloxacin (65%), trimethoprim-sulfamethoxazole (71%), clindamycin (89%) and erythromycin (88%). Similarly, Adwan et al.44 reported that up to 82.1% of nosocomial MRSA isolates were resistant to erythromycin and therefore, the macrolides cannot be considered first line therapy for serious Staphylococcal infections.
However, in a study done by Al-Tawfiq45, nosocomial MRSA isolates showed lower rates of resistance to trimethoprim-sulfamethoxazole (68%), clindamycin (76.6%) and erythromycin (68%) but showed higher rates of ciprofloxacin resistance (76.6%).
In studying sensitivity to gentamicin, MRSA isolates were resistant in 80% of cases. This does not correlate with results obtained in Cyprus by Gourni et al.46, who found that MRSA were gentamicin resistant in 18.75 only.
In this study, thirteen MRSA isolates were found to be resistant to rifampicin (10%). This agreed with the Turkish study that reported the emergence of rifampicin-resistant MRSA in 3 wards of a university hospital in Turkey47.
All MRSA isolates detected by Colakoglu et al.48 were found to be sensitive to fusidic acid, rifampicin and tetracycline, but, some of the isolates that were detected in our study were resistant to these antibiotics.; for fusidic acid (64%), rifampicin (10%) and for tetracycline (92%) were resistant.
The variations in the antibiotics resistance patterns among different studies can be explained by selection pressure of certain drugs used according to the local hospital policy.
Glycopeptides antibiotics are the treatment of choice of MRSA infections. Appearance of different degrees of VA resistance occurred due to its widespread use to treat MRSA and other Gram-positive infections. The first strain of SA with reduced susceptibility to VA and teicoplanin (glycopeptide-intermediate SA [GISA]) was reported in the 1997 from Japan, whereas VRSA isolates were first reported in the 2002 from the USA, Jordan and Brazil10.
The reasons for decreased VA susceptibility may be owing to non-optimal utilization of VA and other antibiotics or due to use of antimicrobial agents in food-producing animals as documented in Saudi Arabia in Qassim area28. These strains represent a crucial challenge for antimicrobial therapy, testing antimicrobial susceptibility and infection control in hospitals20.
Fortunately, one hundred and twenty-nine of MRSA (99.2%) in the current study, were VA sensitive, which agreed with Sievert et al.49 and Antonanzas et al.50 who included VA in antibiotic susceptibility testing of MRSA.
The present study encountered one VRSA strain (0.8%) with VA MIC 32µg/ml. Many studies encountered VRSA strains at higher frequencies than that reported in our study; Saderi et al.51 reported that 3.5% of SA isolates were VRSA and Hakim et al.52 reported VISA (13%). On the other hand, Dhanalakshmi, et al.24 reported that no VISA or VRSA were found among MRSA strains.
The VRSA case reported in our study31 showed multi-drug resistance to penicillin, amoxicillin/clavulanic, ampicillin/sulbactam, cefazolin, cefuroxime, gentamicin and ciprofloxacin but showed susceptibility to linezolid. Multi-drug resistance of VRSA was also reported by many studies3,28,53.
SA infections account for substantial morbidity and mortality in MUHs. The large scale of spread of methicillin resistance has been considered as a fearsome threat to the already challenging treatment of staphylococci54.
Minimizing the risk factors exposure with rapid MRSA diagnosis are essential for early initiation of appropriate therapy and limitation of the non-optimal usage of glycopeptides and the number of deaths. Antibiotic prescription practices should be based on the in vitro antibiotic susceptibility testing and should be reviewed by the hospital administers with implementation of policies aiming at reduction of their non-optimal use. Attention to MRSA prevention and control must remain a constant team effort, concerning all health care professionals, because the new therapeutic agents alone will not provide the long-term solution55. Adherence to the CDC recommendations56 for preventing device- and procedure-associated infections is needed to further prevent SA infections
ACKNOWLEDGEMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
FUNDING
None
AUTHORS’ CONTRIBUTION
AET designed and performed the research work. MFB, FEE and EH supervised and performed the laboratory techniques. AET wrote the manuscript. MFB, FEE and EH revised the manuscript.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections — United States. MMWR Morb Mortal Wkly Rep., 2019; 68: 214–219.
Crossref - Lamers RP, Stinnett JW, Muthukrishnan G, Parkinson CL and Cole AM. Evolutionary Analyses of Staphylococcus aureus Identify Genetic Relationships between Nasal Carriage and Clinical Isolates. PLoS ONE, 2011; 6(1): e16426.
Crossref - Peacock SJ and Paterson GK. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu Rev Biochem., 2015; 84: 577-601.
Crossref - Hirao Y, Ikeda-Dantsuji Y, Matsui H, Yoshida M, Hori S, Sunakawa K, et al. Low level β-lactamase production in methicillin resistant staphylococcus aureus strains with β-lactam antibiotics-induced vancomycin resistance. BMC Microbiology, 2012; 12: 69.
Crossref - Johnson AP. Methicillin resistant Staphylococcus aureus: the European landscape. J. Antimicrob. Chemother., 2011; 66(Suppl 4): iv43-48.
Crossref - Boucher H, Miller LG and Razonable RR. Serious infection caused by methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis., 2010; 51(Suppl 2): S183–S179.
Crossref - Sakai F, Hanaki H, Barada K, Hirao Y, Inamatsu T, Nakae T, et al. A 25-year trace of methicillin-resistant Staphylococcus aureus dissemination in a geriatric hospital in Japan. Internl. J. Gen. Med., 2010; 3: 399–405.
Crossref - Cunha BA. Vancomycin revisisted: a reappraisal of clinical use. Crit. Care. Clin., 2008; 24: 393–420.
Crossref - Fridkin SK, Hageman JC and Morrison M. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med., 2005; 352: 1436-1444.
Crossref - Al-Obeid S, Haddad Q, Cherkaoui A, Schrenzel J and Francois P. First detection of an invasive Staphylococcus aureus strain (D958) with reduced susceptibility to glycopeptides in Saudi Arabia. J. Clin. Microbio., 2010; 6: 2199-2204.
Crossref - Clinical and Laboratory Standards Institute; CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; CLSI document M100-S24. Wayne, PA; 2014.
- Cheesbrough M. Microbiological tests. In Cheesbrough M. (Ed.), District Laboratory Practice in Tropical Countries. New York: Cambridge University Press; 2006; 62-70.
Crossref - Forbes BA, Sahm DF and Weissfeld AS. Staphylococcus, micrococcus and similar organisms. Baily and Scott’s Diagnostic Microbiology (12th ed.): MOSBY Elsevier; 2007.
- Aushbel F, Brent R, Kingstone R, Moore D, Seidman J, Smith J, et al. Preparation of genomic DNA from bacteria. Cited by: Cuurent Protocols in Molecular Biology, Aushbel F, Brent R, Kingstone R, Moore D, Seidman J, Smith J and Struhl K. (eds.). Green and Wiley; New York, 1990; 241.
- Bignardi GE, Woodford N, Chapman A, Johnson AP and Speller DC. Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J. Antimicrob. Chemother., 1996; 37: 53-63.
Crossref - Davis LG, Dibner MD and Battey JF. Agarose gel electrophoresis. Cited by: Davis LG, Dibner MD and Battey JF. (eds.), Basic Methods in Molecular Biology. Elsevier science publishing Co., New York. 1986; 58-61.
Crossref - Sarker SD, Nahar L and Kumarasamy Y. Microtitre plate based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods, 2007; 42(4): 321-324.
Crossref - Biswas PP, Dey S, Adhikari L and Sen A. Virulence markers of vancomycin resistant Enterococci isolated from infected and colonized patients. Jour. of Global. Infec. Dis., 2014; 6(4): 157.
Crossref - Holloway K. Antimicrobial resistance: the facts. Essential Drug Monitor WHO. 2000; 28&29: 7-8.
- Abd El-Baky RM, Ahmed HR and Gad GFM. Prevalence and Conjugal Transfer of Vancomycin Resistance among Clinical Isolates of Staphylococcus aureus. Adv. in Research (AIR), 2014; 2(1): 12-23.
Crossref - Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM and Qian Q. Methicillin-resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J. Clin. Microbiol., 2001; 39(1): 3946-3951.
Crossref - Viswanathan K, Frey KM, Scocchera EW, Martin BD, Swain III PW and Alverson JB. Toward New Therapeutics for Skin and Soft Tissue Infections: Propargyl-Linked Antifolates Are Potent Inhibitors of MRSA and Streptococcus pyogenes. PLoS ONE, 2012; 7(2): e29434.
Crossref - Saunders NA and Holmes A. Multi-locus Sequence Typing (MLST) of Staphylococcus aureus. In, Methicillin Resistant Staphylococcus aureus (MRSA) protocols. Ji Y. (eds). Humana press; 2007; 71-85.
Crossref - Dhanalakshmi TA, Umapathy BL, Mohan DR. Prevalence of methicillin, vancomycin and multi drug resistance among Staphylococcus aureus. J Clin Diag Res., 2012; 6: 974-977.
- Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis., 2001; 32(Suppl 2): S114-S132.
Crossref - Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP and Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004; 39(3): 309-317.
Crossref - Baddour MM, Abuelkheir MM and Fatani AJ. Trends in antibiotic susceptibility patterns and epidemiology of MRSA isolates from several hospitals in Riyadh, Saudi Arabia. Ann. Clin. Microb. Antimicrob., 2006; 5:1–11.
- Alzolibani AA, Robaee AAA, Shobaili HAA, Bilal JA, Ahmad MI and Bin Saif G. Documentation of vancomycin-resistant Staphylococcus aureus (VRSA) among children with atopic dermatitis in the Qassim region, Saudi Arabia. Acta Dermatovenerol APA, 2012; 21: 51-53.
- Hsueh PR, Teng LJ, Chen WH, Pan HJ, Chen ML, Chang SC, et al. Increasing prevalence of methicillin resistant Staphylococcus aureus causing nosocomial infections at a university hospital in Taiwan from 1986 to 2001. Antimicrob. Agents Chemother., 2004; 48(4): 1361-1364.
Crossref - Taha AE, Badr MF, El-Morsy FE and Hammad E. Vancomycin-Resistant MRSA Induced by b-Lactam Antibiotics in Mansoura University Hospitals. Int. J. Curr. Microbiol. App. Sci., 2017; 6(12): 3606-3619.
Crossref - Taha AE, Badr MF, El-Morsy FE and Hammad E. Report of b-lactam antibiotic–induced vancomycin-resistant Staphylococcus aureus from a university hospital in Egypt. New Microbe and New Infect., 2019; 29: 100507.
Crossref - Carla W, Erin L, Jeannie R, Troy M and Donald P. Diagnostic and therapeutic evaluation of community acquired methicillin resistant Staphylococcus aureus (MRSA) skin and soft tissue infections in the emergency department. Jemermed., 2011; 03: 009
- EARSS. European Antimicrobial Resistance Surveillance System (EARSS) annual report 2004. EARSS, Bilthoven, Netherlands, 2005.
- Gordon RJ and Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis., 2008; 46(Suppl 5): S350–S359.
Crossref - Mahamat A, Kenzie F, Brooker K, Monnet D and Daures J. Gould I. and Impact of infection control interventions and antibiotic use on hospital MRSA: a multivariate interrupted time-series analysis. Intern. Jour. of Antimicrob. Ag., 2007; 30: 169–176.
Crossref - Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J. Infect. Dis., 2008; 197: 1226–1234.
Crossref - Tobias W and Mathias W. Antimicrobial treatment of nosocomial methicillin-resistant Staphylococcus aureus (MRSA) pneumonia: current and future options. Intern. Jour. of Antimicrob. Ag., 2010; 36: 391–400.
Crossref - Raygada JL and Levine DP. Managing CA-MRSA Infections: Current and Emerging Options. Infect. Med., 2009; 26(2): 29-58.
- Borg M, Kraker M and Scicluna E. Prevalence of methicillin resistant Staphylococcus aureus (MRSA) in invasive isolates from southern and eastern Mediterranean countries. J. Antimicrob. Chemother., 2007; 60: 1310-1315.
Crossref - Shopsin B, Mathema B, Martinez J, Ha E, Campo ML and Fierman A. Prevalence of methicillin-resistant Staphylococcus aureus in the community. J. Infect. Dis., 2000; 182: 359-362.
Crossref - Ricarda D, Anne K, Alexander M, Knut G, Norbert S, Frank K, et al. Evidence for cost reduction based on pre-admission MRSA screening in general surgery. Int. J. Hyg. Environ. Health., 2008; 211; 205–212.
Crossref - Sadoyama G and Gontijo-Filho PP. Risk factors for methicillin resistant and sensitive Staphylococcus aureus infection in a Brazilian university hospital. Braz. J. Infect. Dis., 2000; 4(3): 135-143.
- Noto MJ, Fox PM and Archer GL. Spontaneous Deletion of the Methicillin Resistance Determinant, mecA, Partially Compensates for the Fitness Cost Associated with High-Level Vancomycin Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother., 2008; 52(4): 1221–1229.
Crossref - Adwan K, Abu-Hasan N, Adwan G, Jarrar N, Abu-Shanab B and Abu-Zant A. Nosocomial infection caused by methicillin resistant Staphylococcus aureus in Palestine. Microb. Drug Resist., 2005; 11(1): 75-77.
Crossref - Al-Tawfiq JA. Incidence and epidemiology of methicillin-resistant Staphylococcus aureus infection in a Saudi Arabian hospital. Infect Control Hosp Epidemiol., 2006; 27: 1137–1139.
Crossref - Gourni M, Kontou M, Hadjipanayiotou C and Protopapa P. Multicenter surveillance of antibiotic resistance in nosocomial Staphylococcus aureus in Cyprus. East Mediterr. Health J., 2001; 7(4-5): 744-749.
- Tosun I, Udo EE, Noronha B, Caylan R, Aydin F, Yetiskul S, et al. Emergence of rifampicin resistance in methicillin resistant Staphylococcus aureus isolated at a Turkish university hospital. Microb. Drug Resist., 2005; 11(1): 48-52.
Crossref - Colakoglu S, Hikmet A, Suheyla S, Tuba T, Yusuf Z and Hande A. Performance of MRSA ID chromogenic medium for detection of methicillin-resistant Staphylococcus aureus directly from blood cultures and clinical specimens. Jour. of Clini Micro., 2007; 53: 345-55.
- Sievert DM, Wilson ML, Boulton ML, Gillespie BW and Wilkins MJ. Resistant Staphylococcus aureus Infections in the United States: A New Classification, a New Resistance and the Implications for Surveillance, Prevention, and Control. 2008.
- Antonanzas F, Lozano C, Torres C. Economic features of antibiotic resistance: the case of methicillin-resistant Staphylococcus aureus. Pharmacoeconomics, 2015; 33: 285-325.
Crossref - Saderi H, Owlia P, Shahrbanooie R. Vancomycin resistance among clinical isolates of Staphylococcus aureus. Arch. Iranian. Med., 2005; 8: 100–103.
- Hakim ST, Arshed S, Iqbal M, Javaid SG. Vancomycin sensitivity of Staphylococcus aureus isolates from hospital patients in Karachi, Pakistan. Libyan J. Med., 2008; 176-179.
Crossref - Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Medical Microbiol., 2008; 57: 72–79.
Crossref - El-Said El-Banna T, Sonbol FI, Abd El-Aziz AA, El-Ekhnawy E. Prevalence and Antimicrobial Susceptibility of Vancomycin Resistant Staphylococci in an Egyptian University Hospital. J. Med. Microb. Diagn., 2018; 7: 272.
Crossref - Huang SS, Singh R, McKinnell JA, Park S, Gombosev A, Eells SJ, et al. Decolonization to reduce postdischarge infection risk among MRSA carriers. N. Engl. J. Med., 2019; 380: 638–650.
Crossref - CDC. Healthcare Infection Control Practices Advisory Committee (HICPAC). Guidance documents. Atlanta, GA: US Department of Health and Human Services, CDC; 2018.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.