ISSN: 0973-7510
E-ISSN: 2581-690X
Aflatoxin-B1 (AFB1) is a common contaminant for staple foods during the storage process. Chronic exposure to AFB1 is widely known to induce the development of hepatocellular carcinoma (HCC). However, there is a lack of understanding of AFBi role in HCC mechanism. This research aims to identify protein(s) in HCC that might interact with AFB1 and to predict the pathway effected by AFB1. Analyses were performed using bioinformatics tools. SMILES notation of AFB1 was submitted into Swiss Target Prediction. Interaction among predicted proteins were analyzed by using STRING. The 3D structure of target protein was constructed by homology modeling. Reverse docking was performed, and the result was ranked based on binding affinity score. Furthermore, protein interaction network was constructed and analyzed by using Cytoscape. Results showed that three protein groups were predicted as target of AFB1, such as kinases, phosphatases, and G protein-coupled receptor with probability of 46.7%, 20%, and 6.7%, respectively. Seven proteins of kinases were strongly related to HCC, including RAF1, MAPK1, MAPK3, AKT1, EGFR, GSK3B, and mTOR. Reverse docking considered the AKT1-AFB1 as the most potential complex with the lowest affinity score -10.2 kcal.mol-1. It has hydrophobic bonds in Trp80, Val270, Tyr272, Asp292, Thr211, Leu210, Leu264, and Lys268 residues, whereas hydrogen bond in Ser205 residues. Moreover, further analysis demonstrated that interaction of AKT1-AFB1 is related to the metastasis pathway in HCC mechanism.
Cancer, Kinase Protein, Protein Interaction, Protein Pathway, Toxin
Aflatoxin is one of secondary metabolites produced by Aspergillus flavus and Aspergillus parasiticus. In general, there are several types of aflatoxins, such as aflatoxin-B1 (AFB1), aflatoxin-B2 (AFB2), aflatoxin G1 (AFG1), and aflatoxin G2 (AFG2). Aspergillus flavus typically produces AFB1 and AFB2, while Aspergillus parasiticus produces all type of aflatoxins. Among those aflatoxins, AFB1 is considered as the most common type which contaminates the staple foods and agricultural product. This mycotoxin is highly produced within 25°-30°C of temperature and 85% of relative humidity during distribution and storage processes.1 The prolonged consumption of food contaminated by AFB1 is considered as one of the major factors for hepatocellular carcinoma (HCC) development.2
HCC is dedicated as a cancer with the highest mortality across the globe. It is also classified as the most common cancer in liver which is indicated by epithelial neoplasm in hepatocyte.3,4 The highest occurrence of the disease is spread across Asia, Africa, and America. Recent studies reported that hepatic tumor is observed in mice exposed by AFB1.5 However, the role of AFB1 within this HCC mechanism remains unclear. Therefore, in this research, we aim to identify and explore the protein(s) which might interact with AFB1 and to predict the pathway effected by AFB1. This is considered as an essential study which could potentially be developed as a foundation for further understanding of relation between AFB1 and HCC. In addition, the analysis of mechanism pathway is part of system biology which integrated to understand the system function in organism through protein-protein interaction network (PPIN).6
Collection of AFB1
The Aflatoxin B1 (AFB1) molecule was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) with ID number CID186907. The notation of canonical simplified molecular-input line-entry system (SMILES) was copied, and its 3D structure was collected in SDF format. The minimization process was conducted by using the Open Babel plug-in in PyRx software (The Scripps Research Institute).7 This process was essential for further analysis related to molecular docking.
Target protein prediction
The target protein of AFB1 was predicted by submitting the canonical SMILES to the Swiss Target Prediction (http://www.swisstargetprediction.ch/). Mus musculus was selected as the organism target, which is defined as the representative research object. Furthermore, top three target class of protein were selected as potential target protein. All predicted target proteins which belong to each class of proteins were listed.
Analysis of AFB1 interaction to the predicted target protein
The obtained target proteins candidates of AFB1 were used as input to the webserver Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org). Collection was performed using multiple protein category with Mus musculus as an organism target. The result was analysed based on the KEGG pathway enrichment analysis by selecting hepatocellular carcinoma. Furthermore, data was linked and further analysed by using Cytoscape software (Institute for System Biology, USA). This process aimed to determine the protein interaction network (PIN) that involved in hepatocellular carcinoma (HCC) disease.
Protein modeling of predicted target protein
The proteins involved in HCC disease interactions were accessed through the Uniprot database (www.uniprot.org). Those amino acid sequences of each protein were retrieved in FASTA format for homology modelling process. Furthermore, it was submitted into SWISS MODEL (http://swissmodel.expasy.org) to get the 3D structure of each protein. The water molecules and other ligands were eliminated by using PyMOL so that the pure protein structure was obtained.
Molecular docking of AFB1 with predicted target protein
The screening process by molecular docking was carried out between each predicted target protein with AFB1 on the specific site. The process was performed by using Autodock Vina on the PyRx software (The Scripps Research Institute).7 Furthermore, the binding affinity score and the position of each screening result were collected and compared. The results of molecular docking were then analyzed by using LigPlot software to identify the chemical bonds of amino acid residues.8
Visualization of AFB1 and selected target protein
The representative results of AFB1 with each of predicted target protein were visualized by using PyMol software.7 It aimed to determine the detailed structure of proteins. Moreover, it depicted the position interaction between AFB1 with the protein.
Protein interaction network (PIN) analysis of predicted target protein of AFB1 in HCC mechanism
Protein interaction network (PIN) which is constructed from predicted target protein of AFB1 was analysed by using topological analysis. It was performed by Network Analyzer application which is integrated into Cytoscape software (Institute for System Biology, USA). There were various parameters which could be selected for analysis. However, number of edges and betweenness parameters were prominent selected to identify the most essential protein in HCC mechanism. Moreover, predicted target protein of AFB1 was also subjected for analysis of protein interaction pathway in HCC mechanism.
Aflatoxin B1 (AFB1) was retrieved from PubChem database with ID number ID186907. It described as a small molecule with molecular formula C17H12O6 and molecular weight 312.27 g/mol. The notation of canonical SMILES COC1=C2C3=C(C(=O)CC3)C(=O)OC2=C4C5C=COC5OC4=C1 was collected from database and utilized as a sample for analysis.
Protein target of AFB1 was predicted by submitting the notation of canonical SMILES into web server Swiss Target Prediction. The process was performed based on the ligand-based target prediction, whereas AFB1 as the center of query molecule. Gfeller et al.,9 stated that predicted protein target is identified based on the similarity of another protein with its known ligand that resemblance to the query molecule.9 As it is shown in Table 1, there were three groups of protein which is predicted as a target of AFB1, such as kinase, phosphatase, and G protein-coupled receptor with probability score 46.7%, 20%, and 6.7%, respectively. In detail, there are 22 kinase proteins predicted as the compatible target protein of AFB1. According to protein target prediction, it showed that AFB1 had several target proteins such as AKT/mTOR and MAPK.
Table (1):
The top 3 predicted target proteins group of AFB1 in Mus musculus.
Group of protein |
Target protein |
Probability score |
|---|---|---|
Kinase |
MAPK1, GSK3B, DYRK1A, ABL1, RAF1, FLT1, KIT, JAK3, RET, MAPK14, LCK, MAPK3, AURKB, LYN, MAPK9, AKT1, AURKA, PRKDC, EGFR, ALK, CDK1, mTOR |
46.7% |
Phosphatase |
PTPN1, CDC25B |
20.0% |
G protein-coupled receptor |
TAAR1, TRH-R2, TRHR, ADORA2A, HTR2C, ADORA, ADORA2B |
6.7% |
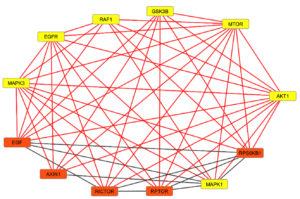
The STRING analysis was conducted to identify the specific candidate of kinase protein for AFB1. The result exhibited that 29 predicted AFB1 target proteins interacted to each other through 97 edges. However, there are only seven proteins which are closely involved in HCC mechanism, such as RAF1, MAPK1, MAPK3, AKT1, EGFR, GSK3B, and mTOR. Furthermore, protein interaction was constructed by using Cytoscape software. Seven proteins related to HCC mechanism were indicated by yellow-colored nodes and the interactions among proteins were exhibited by the grey-colored edges (Figure 1). However, the association among proteins do not necessarily mean that those proteins are directly binding each other. It merely showed that those candidate proteins are strongly related each other of the HCC mechanism.
Figure 1. Interaction of seven proteins related to hepatocellular carcinoma (HCC) based on Cytoscape software. Predicted target protein (yellow).
In brief, to understand the role of seven proteins related to HCC mechanism, the function of each target protein is needed to be identified. Epidermal growth factor receptor (EGFR) belongs to receptor tyrosine kinase (RTK) which is found in plasma membrane, and it has a function to regulate the cell growth.10 Generally, the downstream signaling cascade of EGFR includes two main pathways, such as RAS-RAF-MAP kinase pathway and PI3K-PTEN-Akt pathway. In cancer case, the level of EGFR is frequently elevated or mutated.
Furthermore, AFB1 is estimated to have an ability to interact with one of seven predicted proteins within HCC mechanism. Therefore, molecular docking would be conducted between AFB1 and those selected proteins. Whilst the 3D structures of each protein were constructed by homology modelling. The result of homology modelling showed that all of protein is considered valid according to the similarity score and validation score. The highest similarity score was exhibited by MAPK1, while the lowest score was showed by EGFR (Table 2). However, both similarity scores are still in the requirement range about 80-90% similar with the protein template.
Table (2):
Homology modelling and 3D structure of protein related to hepatocellular carcinoma based on the SWISS MODEL.
Target Protein |
Uniprot ID |
Template ID |
Similarity score (%) |
Validation score (%) |
|---|---|---|---|---|
mTOR |
Q9JLN9 |
6bcu.1.A |
96,63 |
91,94 |
EGFR |
Q01279 |
3gop.1.A |
88,73 |
93,55 |
GSK3B |
Q9WV60 |
1j1b.1.B |
99,05 |
95,58 |
AKT1 |
P31750 |
6hhg.1.A |
97,53 |
94,77 |
MAPK3 |
Q63844 |
4qtb.1.A |
98,56 |
97,36 |
MAPK1 |
P63085 |
3zuv |
100,0 |
94,49 |
RAF1 |
Q99N57 |
3omv.1.A |
98,31 |
86,13 |
Seven proteins which are selected as predicted target protein of AFB1 and related to HCC pathway were selected to be identified about their interaction with AFB1. Each protein, such as RAF1, MAPK1, MAPK3, AKT1, EGFR, GSK3B, and mTOR was examined by reverse docking with AFB1. The molecular docking result showed that among seven proteins, complex of AKT1-AFB1, MAPK3-AFB1, and MTOR-AFB1 are demonstrated to have the lowest binding affinity about -10.2 kcal.mol-1, -9.3 kcal.mol-1, and -9.1 kcal.mol-1, respectively. However, the lowest binding affinity energy is exhibited by the interaction between AFB1 and AKT1 (Table 3).
Table (3):
List of binding affinity based on reverse docking between AFB1 and seven predicted target proteins in hepatocellular carcinoma.
| Ligand | Target Protein | Position of grid docking | Binding energy (kcal. mol-1) | |||||
|---|---|---|---|---|---|---|---|---|
| Center (Å) | Dimension (Å) | |||||||
| X | Y | Z | X | Y | Z | |||
| AFB1 | AKT1 | 269.5 | 214.5 | 239.8 | 150.0 | 99.1 | 170.4 | -10.2 |
| MAPK3 | 33.2 | 46.5 | 56.8 | 61.2 | 48.6 | 69.0 | -9.3 | |
| mTOR | 269.5 | 214.5 | 239.8 | 150.0 | 99.1 | 170.4 | -9.1 | |
| GSK3B | 27.7 | -6.5 | -38.0 | 76.3 | 60.2 | 60.2 | -8.9 | |
| MAPK1 | 21.4 | -25.5 | -9.0 | 63.1 | 67.6 | 80.0 | -7.8 | |
| EGFR | -9.0 | -21.0 | -37.8 | 59.4 | 72.0 | 74.3 | -7.4 | |
| RAF1 | 6.2 | 18.1 | 43.9 | 58.8 | 55.8 | 54.7 | -6.7 | |
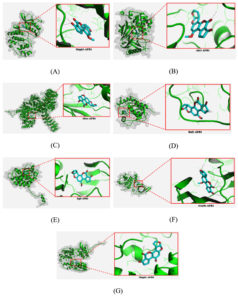
The binding site position of AFB1 on each target protein was also listed in detail. The result showed that AFB1 has different site position on each protein (Table 3), which is vividly depicted in Figure 2. This different position of binding site is exhibited since AFB1 as a ligand was exposed to the whole surface of target protein without any prior specific site. This process is performed to explore the favourable site and pose of AFB1 on target protein. Complex of AKT1-AFB1 was depicted by the lowest binding energy during the molecular docking analysis. Therefore, it is predicted to have a strong bond and can trigger the biological responses.
Figure 2. Interaction of AFB1 with several predicted protein: MAPK3 (A), AKT1 (B), mTOR (C), RAF1 (D), EGFR (E), GSK3B (F), and MAPK1 (G).
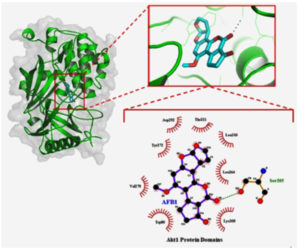
The interaction of protein-ligand in the AKT1-AFB1 complex is pursued for further analysis to find the detailed interaction by using LigPlus. The results showed that AFB1 interacted in the protein domain AKT1 by using hydrophobic and hydrogen bonds. In detail, the interaction of hydrophobic bonds is formed through Trp80, Val270, Tyr272, Asp 292, Thr211, Leu 210, Leu 264, and Lys 268 residues, whereas hydrogen bond is only found in Ser205 residues. On the results visualization, the AFB1 structure is showed in the purple colour, the green dotted line showed the hydrogen bonds with amino acid residue, and half circle pattern in red demonstrated the interaction of hydrophobic bonds (Figure 3).
Further analysis was conducted to identify protein pathway of AKT1-AFB1 in HCC mechanism. Constructed protein interaction network (PIN) in HCC mechanism (Figure 1) was analysed by topological analysis based on number of edges and betweenness parameters score.
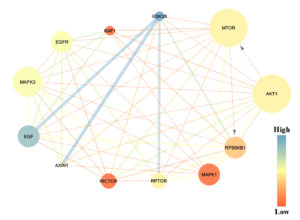
The number of edges is indicated by the size of protein node, whereas the betweenness score is indicated by colour and thickness of the edges (Figure 4). The result showed that AKT1 protein had a lower betweenness score compared to GSK3B protein. AKT1 protein had thin edges which interacted to several proteins, while GSK3B had the thickest blue edges which directly interacted to EGF, AXIN1, and RPTOR with score 4.33, 4.0, 3.5, and 2.5, respectively (Table 5). According to another parameter, the analysis demonstrated the different result. Compared to other protein, AKT1 and mTOR protein exhibited the biggest size of protein node within a network (Figure 4) with 11 edges interacted to various proteins (Table 4). Further analysis showed that AKT1 had an interaction to another protein especially mTOR and RPS6KB1 protein which is indicated by the arrow (Figure 4).
Table (4):
Number of edges of protein within protein interaction network in HCC mechanism.
Protein name |
Number of edges |
Number of In-degree |
Number of Out-degree |
|---|---|---|---|
AKT1 |
11 |
10 |
1 |
mTOR |
11 |
9 |
2 |
MAPK3 |
10 |
1 |
9 |
EGFR |
9 |
2 |
7 |
EGF |
9 |
3 |
6 |
MAPKAP1 |
9 |
7 |
2 |
RPS6KB1 |
9 |
5 |
4 |
RPTOR |
8 |
3 |
5 |
RICTOR |
8 |
8 |
0 |
RAF1 |
7 |
0 |
7 |
GSK3B |
7 |
2 |
5 |
AXIN1 |
6 |
2 |
4 |
Table (5):
Top three betweenness score of protein interaction within protein interaction network in HCC mechanism.
Protein name |
Edge Betweenness score |
|---|---|
GSK3B – AXIN1 |
4.33 |
GSK3B – EGF |
4.00 |
GSK3B – RPTOR |
3.50 |
GSK3B – RPS6KB1 |
2.50 |
Protein target of AFB1 was identified as shown in Table 1. There were three groups of protein predicted as a target of AFB1, such as kinase, phosphatase, and G protein-coupled receptor. In detail, there are 22 kinase proteins predicted as compatible target protein of AFB1.
Kinases are a family of enzyme which has a main function to catalyze the phosphorylation process. In general, kinases transfer a phosphoryl group to the target protein and play essential role for maintaining various cellular functions, such as proliferation, cell growth, motility, and apoptosis.11,12 According to protein target prediction, it showed that AFB1 had several target proteins such as AKT/mTOR and MAPK, identified as signaling pathway which regulate various cellular pathway related to malignancies. It is known that hundreds of kinases are commonly activated in any tumor development.
Based on the Bhullar et al.13 kinase protein is a common targeted protein of cancer drug after G protein-coupled receptor.13 Since 1980, there were more than 30 kinase inhibitors received by FDA for treatment. Moreover, about 150 drugs for kinase-targeted are developed following clinical trials. The target protein prediction of AFB1 provides the future potential for kinase-targeted therapeutics agent. Therefore, a specific target protein is needed to be explored.
Based on STRING analysis, there were 29 predicted AFB1 target proteins interacted each other through 97 edges. However, only seven proteins were predicted to be closely involved in HCC mechanism, such as RAF1, MAPK1, MAPK3, AKT1, EGFR, GSK3B, and mTOR. Although there are association among proteins, it does not mean that those proteins are directly binding each other. It merely showed that those candidates’ proteins is strongly related each other to the HCC mechanism.
To understand the role of seven proteins related to HCC mechanism, the function of each protein needs to be spotted. Epidermal growth factor receptor (EGFR) is belonged to receptor tyrosine kinase (RTK), which is found in plasma membrane, and it has a function to regulate the cell growth.10 Generally, the downstream signaling cascade of EGFR includes two main pathways, such as RAS-RAF-MAP kinase pathway and PI3K-PTEN-Akt pathway. In cancer case, the level of EGFR is frequently elevated or mutated. The alteration of EGFR promotes the constitutive activation of downstream signaling and leads to the unlimited proliferation of cells during G1/S phase.14 In brief, once the EGFR is activated, it will phosphorylate its substrate, which led to the downstream pathway related to AKT activation. Then, AKT will phosphorylate the mammalian target of rapamycin (mTOR). It is one of tumour survival kinase which is commonly overexpressed in tumour for apoptosis escape and metastasis.15,16 Moreover, after activation, AKT will also be translocated into nucleus and regulate the gene responsible for cellular proliferation.17
Furthermore, AFB1 is estimated to have the ability to interact with one of seven predicted proteins within HCC mechanism. Therefore, molecular docking would be conducted between AFB1 and those selected proteins. Result of homology modelling showed that all of protein is considered valid according to the similarity score and validation score. The highest similarity score was exhibited by MAPK1 while the lowest score was showed by EGFR (Table 2). However, both similarity scores are still in the requirement range about 80-90% similar with the protein template.
According to Park and Cho,18 the reverse docking had the opposite process as compared to virtual screening docking.18 Commonly, docking is performed to screen the protein against several compatible ligands. Nevertheless, in reverse docking, target proteins are listed and ranked against selected ligand. It also commonly known as ligand-based docking. The molecular docking result showed that among seven proteins, complex of AKT1-AFB1, MAPK3-AFB1, and MTOR-AFB1 are demonstrated to have the lowest binding affinity about -10.2 kcal.mol-1, -9.3 kcal.mol-1, and -9.1 kcal.mol-1, respectively. However, the lowest binding affinity energy is exhibited by interaction between AFB1 and AKT1 (Table 3).
Result showed that AFB1 has different site position of AFB1 on each protein (Table 3) which vividly depicted in Figure 2. This different position of binding site is exhibited since AFB1 as a ligand was exposed to the whole surface of target protein without any prior specific site. This process is performed for exploring the favourable site and pose of AFB1 on target protein.
Complex of AKT1-AFB1 was depicted by the lowest binding energy during the molecular docking analysis. Therefore, it is predicted to have a strong bond and able to trigger the biological responses. Results showed that AFB1 interacted in the protein domain AKT1 by using hydrophobic and hydrogen bonds. In detail, the interaction of hydrophobic bonds is formed through Trp80, Val270, Tyr272, Asp 292, Thr211, Leu 210, Leu 264, and Lys 268 residues, whereas hydrogen bond is only found in Ser205 residues. On the results visualization, the AFB1 structure is showed in the purple colour, the green dotted line showed the hydrogen bonds with amino acid residue, and half circle pattern in red demonstrated the interaction of hydrophobic bonds (Figure 3).
Further analysis was conducted to identify protein pathway of AKT1-AFB1 in HCC mechanism. Constructed protein interaction network (PIN) in HCC mechanism (Figure 1) was analysed by topological analysis based on number of edges and betweenness parameters score. The number of edges indicated by the size of protein node, whereas the betweenness score indicated by colour and thickness of the edges (Figure 4). Result showed that AKT1 protein had lower betweenness score as compared to GSK3B protein. AKT1 protein had thin edges interacted to several proteins, while GSK3B had the thickest blue edges which directly interacted to EGF, AXIN1, and RPTOR with score 4.33, 4.0, 3.5, and 2.5, respectively (Table 5). According to Simos et al.,19 protein with high score of betweenness score is considered to have the ability to control the other protein interaction within a network.19 However, the controlled interaction of protein still needs further studies.
According to another parameter, the analysis demonstrated the different result. As compared to other protein, AKT1 and mTOR protein exhibited the biggest size of protein node within a network (Figure 4) with 11 edges interacted to various proteins (Table 4). It is widely known that protein with high number of edges or direct interaction is determined as protein which have the most critical role within network due to their abroad interaction.19 Based on that analysis, it showed that AKT1 and mTOR are considered as the essential protein within HCC mechanism. Therefore, interaction of AFB1 to AKT1 protein is suggested as potential strategy in promoting the HCC progression.
Further analysis showed that AKT1 had an interaction to another protein especially mTOR and RPS6KB1 protein which indicated by the arrow (Figure 4). The mTOR protein is phosphorylated by AKT1 protein, then it activated the protein downstream termed RPS6KB1, which is strongly involved in the cancer survival mechanism.20 RPS6KB1 is known as the major substrate of mTOR, which plays an essential role in cell growth, survival, and metastasis by cell cycle regulation. Recent research reported that RPS6KB1 is commonly high expressed in cancer related to the tumour size and metastasis progression. Moreover, overexpression of RPS6KB1 is also considered for exhibiting the worse condition of HCC case in patients.21 Another in vitro result exhibited that AKT1 signalling had oncogenic effect for promoting the hepatocarcinogenesis. Akt is a central of PI3K/Akt signaling pathway which regulates the cells proliferation and cells survival ability.
Based on all analysis, it is suggested that AFB1 have interaction to the most potential protein target in HCC mechanism that is AKT1 protein. The interaction of AKT1-AFB1 involved in the HCC progression by activated protein cascade of AKT1-mTOR-RPS6KB1, which generated the metastasis mechanism. This indicated that the interaction between AFB1 and AKT had essential implications for HCC development.
AFB1 is predicted to interact with several proteins of hepatocellular carcinoma (HCC) major factors. Kinase’s proteins, such as RAF1, MAPK1, MAPK3, AKT1, EGFR, GSK3B, and mTOR exhibited to have close related to HCC development. However, among those target proteins, Akt1 had strong interaction AFB1 with binding energy -10.2 kcal.mol-1 and supported by hydrophobic bonds through Trp80, Val270, Tyr272, Asp292, Thr211, Leu210, Leu264, and Lys268 residues, also hydrogen bond with Ser205 residue. Further analysis demonstrated that the interaction AKT1-AFB1 is related to metastasis pathway in HCC mechanism. This finding would provide the understanding about HCC mechanism, which can be used as a basic development of cancer therapeutic agent.
ACKNOWLEDGMENTS
The authors would like to thank University of Surabaya, Indonesia and Universitas Airlangga, Indonesia for supporting this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Kew MC. Aflatoxins as a cause of hepatocellular carcinoma. J Gastrointestin Liver Dis. 2013;22(3):305-310. PMID: 24078988
- Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893-2917.
Crossref - Pimenta JR, and Massabki PS. Carcinoma hepatocellular: a clinical outlook. Rev Bras Clin Med. 2010;8:59-67.
- Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol. 2018;48:43(1):13-25.
Crossref - Farombi EO. Aflatoxin contamination of foods in developing countries: Implication for hepatocellular carcinoma and chemopreventive strategies. Afr J Biotechnol. 2006;5:1-14.
- Tavassoly I, Goldfarb J, Iyengar R. System biology primer: The basic methods and approaches. Essay Biochem. 2018;62(4):487-500.
Crossref - Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243-250.
Crossref - Rajan MSD, Balajee R. Molecular docking and simulation studies of farnesyl transferase with the potential inhibitor theflavin. J Appl Pharm Sci. 2011;1(8):141-148.
- Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecule. Nucleic Acids Res. 2014;42(Web Server Issue):1-7.
Crossref - Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12(1):3-20.
Crossref - Wang Z, Cole PA. Catalytic mechanism and regulation of protein kinases. Methods Enzymol. 2014;548:1-21.
Crossref - Cicenas J, Zalyte E, Bairoch A, Gaudet P. Kinases and cancer. Cancers (Basel). 2018;10(3):63.
Crossref - Bhullar KS, Lagaron NO, McGowan EM, et al. Kinase-targeted cancer therapies: progress, challenges, and future directions. Mol Cancer. 2018;17(1):48.
Crossref - Saletti P, Molinari F, De Dosso S, Frattini M. EGFR signaling in colorectal cancer: a clinical perspective. Gatrointest Cancer. 2014;5:21-38.
Crossref - Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5(8):671-688.
Crossref - Kumar S, Purohit P, Dagar S. A review: status of genetic modulated nonsmall cell lung cancer targets and treatment (current updates in drugs for non-small cell lung cancer treatment). Asian J Pharm Clin Res. 2018;11(8):40-55.
Crossref - McCubey JA, Steelman LS, Chappell WH, et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3(9):954-987.
Crossref - Park K, Cho AE. Using reverse docking to identify potential targets for ginsenosides. J Ginseng Res. 2017;41(4):534-539.
Crossref - Simos T, Georgopoulou U, Thyphronitis G, Koskinas J, Papaloukas C. Analysis of protein interaction networks for the detection of candidate hepatitis B and C biomarker. IEEE J Biomed Health Inform. 2015;19(1):181-189.
Crossref - Zhang Y, Ni H, Cheng D. Prognostic value of phosphorylated mTOR/RPS6KB1 in non-small cell lung cancer. Asian Pacific J Cancer Prev. 2013;14(6):3725-3728.
Crossref - Li PD, Zhang WJ, Zhang MY, et al. Overexpression of RPS6KB1 predicts worse prognosis in primary HCC patients. Med Oncol. 2012;29(5):3070-3076.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.